Diversity of hepatotoxic cyanobacteria in the Nile Delta, Egypt
Ranya
Amer†
a,
Beatriz
Díez†
b and
Rehab
El-Shehawy
*b
aDepartment of Environmental Biotechnology, GEBRI, Mubarak City for Scientific Research and Application Technology (MuCSAT), Egypt
bDepartment of Botany, Stockholm University, Sweden. E-mail: rehab@botan.su.se; Fax: +46 (8) 165525; Tel: +46 (8) 163918
First published on 3rd December 2008
Abstract
Optimized denaturing gradient gel electrophoresis (DGGE) fingerprinting and real-time PCR were performed to identify and quantify the potential hepatotoxic microcystin- and nodularin-producing cyanobacteria present in freshwater samples collected during different seasons at three different sites from the Nile River Delta. The combined use of molecular gene markers (targeting the aminotransferase domain of the hepatotoxin synthetase modules and the 16S rRNA gene) and light microscopy demonstrated the dominance of different freshwater Microcystis phylotypes, including the potential hepatotoxin producers M. wesenbergii and M. aeruginosa, several Synechococcus and Cyanobium phylotypes, as well as the presence of Nodularia spumigena and Cylindrospermopsis raciborskii in the samples “collected during summer 2006 and winter 2007”. Certain seasonal changes (summer and winter) in Microcystis microdiversity were detected. Real-time PCR revealed no difference in the quantities of potential hepatotoxin-producing cyanobacterial communities between summer and winter, but did show differences between the three sampling sites investigated. The expression of the aminotransferase domain analyzed by DGGE fingerprinting demonstrated that all Microcystis phylotypes present in the samples might have been active at the time of the sampling. Statistical analysis showed a significant effect of TP, and not TN, on the relative abundance of the potentially hepatotoxic cyanobacterial community.
Introduction
In light of the 3-domain concept of life, cyanobacteria are a diverse group of photosynthetic bacteria that exist almost everywhere on the globe.1 Many cyanobacterial species are known to produce toxins, for example, species of the genera Microcystis, Nodularia, Anabaena, Aphanizomenon, Cylindrospermopsis, and Planktothrix (Oscillatoria).2–4 Cyanotoxins are classified according to their mode of action in vertebrates as hepatotoxins, neurotoxins, cytotoxins, dermatotoxins, and irritants.5 Hepatotoxins are widely distributed and commonly found in eutrophic water bodies. They act by inhibiting protein phosphatases 1 and 2 (PP1 and PP2A).6,7Microcystin is a hepatotoxin that is commonly found in eutrophic lakes, ponds, and reservoirs worldwide. The biosynthesis of microcystin, as described in the genus Microcystis8 was later found to be produced by several other genera, including Anabaena, Nostoc, Nodularia, and Planktothrix. It is a cyclic heptapeptide produced non-ribosomally by a multifunctional enzyme complex consisting of peptide synthetase (PS)-polyketide synthase (PKS) modules and tailoring enzymes. The PS-PKS is encoded by the mcy gene cluster. Another hepatotoxin is nodularin, which is produced only by the cyanobacterium Nodularia and is synthesized by a PS-PKS module encoded by the nodularin synthetase gene cluster, nda.9
Blooms of toxic cyanobacterial species may contain both toxic and non-toxic strains, and morphological discrimination between the two types is not possible.10 Therefore, PCR-based techniques, such as real-time PCR targeting the toxin biosynthesis genes, have become popular for detecting and quantifying toxic species due to their specificity, sensitivity, and speed.10–12 PCR-based methods are valuable and effective as early warning tools for monitoring drinking water to prevent acute and chronic exposure to cyanotoxins, for understanding the geographical and seasonal distribution of toxic cyanobacteria, and for analyzing environmental factors that influence bloom proliferation and toxin production. Characterization of the genes responsible for hepatotoxin biosynthesis has made it possible to design general PCR primers that target the aminotransferase (AMT) domains of the mycE and ndaF genes,13 allowing detection of microcystin- and nodularin-producing cyanobacteria.
The Nile River is the main source of drinking water for Egypt, as well as for nine other countries, serving a population of approximately 160 million people. In Egypt, the Nile River suffers from eutrophication and contamination from industrial and/or domestic discharge, resulting in endemic water-borne diseases. Toxin-producing cyanobacteria have been isolated from the southern part of the Nile in Egypt.14–16 These previous studies have relied only on morphological analysis, isolation, and toxicity assays of the isolates; no molecular analysis of the genetic diversity has been undertaken until now.
In this work, we evaluated the presence of potential hepatotoxin-producing cyanobacteria in the Nile Delta region (northern part of the Nile River, Egypt) and analyzed their genetic diversity by using PCR-based identification methods, targeting potentially hepatotoxin-producing species. We used several methodologies and two genetic markers to allow in-detail fingerprinting of potentially hepatotoxic cyanobacterial communities, phylogenetic identification, toxin activity, and quantification of the potentially hepatotoxic genera that occur at three sites along the Nile River Delta during summer and winter. We hypothesized that hepatotoxin-producing cyanobacteria were stable active components of the phytoplankton community of the Delta that have been under-represented in the literature due to eutrophication of the water, and that water and environmental management plans in the region need to consider the presence of these potentially toxic microorganisms. To our knowledge, this is the first study to combine real-time PCR and denaturing gradient gel electrophoresis (DGGE) fingerprinting (by using the same primer pair in both techniques) to identify and quantify these microorganisms.
Materials and methods
Sampling sites
The Nile Delta region is estimated to contain 35% of the population of Egypt and is characterized by heavy agricultural and industrial activity. The quality of the water in the two Nile Branches that form the Delta, Domietta and Rosetta (Fig. 1), is deteriorating due to agricultural and industrial waste effluent. Samples were collected in Cairo (situated south of the Delta region), in Kafr El-Zayat (situated on the Domietta Branch), and in Zefta (situated on the Rosetta Branch; Fig. 1). The sampling sites were chosen to geographically represent the Nile Delta. Samples were collected during the summer (August) of 2006 and winter (January) of 2007. Water surface temperatures during sampling were 19 °C, 18 °C, and 19 °C during winter and 25 °C, 28 °C, and 26 °C, during summer at Cairo, Kafr El-Zayat, and Zefta, respectively. | ||
| Fig. 1 Map of the Nile Delta region (http://www.thenilerivercruises.com/images/nile_river_map.jpg) modified to show the approximate location of the sampling sites. Thick arrow= Kafr El-Zayat, thin arrow = Zefta. | ||
Sample collection
Surface offshore water samples were collected using a 90 µm mesh phytoplankton net, and 500 ml of planktonic concentrate from each site was filtered through 10 µm pore-size polycarbonate filters (Whatman). Five replicas were collected per site and each was treated individually. Filters were stored in XS buffer for DNA extraction (Tillett and Neilan, 2000), in XS buffer with phenol at −20 °C for RNA extraction,17 and in 2.5% gluteraldehyde for microscopic examination.Strains
Axenic cultures of Microcystis aeruginosa PCC 7005 (non-toxic) and PCC 7806 (toxic) were purchased from Pasteur Institute, Paris, France. Axenic culture of Nodularia spumigena strain AV1 was obtained as a gift from Prof. Kaarina Sivonen, University of Helsinki, Helsinki, Finland.Light microscopy and morphological descriptions
Light microscopy was performed on the glutaraldehyde-fixed samples using a Zeiss Axiovert 200 microscope equipped with interference contrast; images were recorded with an AxioCam HRC digital camera. The morphological identification of cyanobacteria was done according to Komárek and Anagnostidis.18–20RT-PCR and PCR amplification
DNA and RNA were isolated from the samples as described by Tillett and Neilan.17 The isolated RNA was treated with RNase-free Dnase I (Qiagen, Valencia, USA) to remove contaminating DNA.cDNA was synthesized from 200 ng RNA using the iScript cDNA Synthesis Kit (Bio-Rad, California, USA) according to the manufacturer's instructions, and quantified via spectrophotometry (NanoDrop Technologies, Biocompare, South San-Francisco, USA). To verify the absence of contaminating DNA in each RNA sample, negative controls were performed using aliquots from each RNA sample as a template (without performing RT reactions) instead of using cDNA.
All PCR was performed using 0.2 U of HotStar Taq DNA polymerase (Qiagen, Valencia, USA) in a 20 µl reaction. The PCR mixture contained 1 × HotStar Taq polymerase buffer, 0.5 pmol of forward and reverse primers, 0.2 mM dNTPs, and 100 ng template DNA.
The cyanobacterial-specific 16S rDNA oligonucleotide primers, CYA106F (with 40 nucleotide GC clamp at the 5′end) and CYA781R,21 were synthesized by (DNA technology A/S, Risskov, Denmark) and used for DGGE analysis. Amplified products were 675 bp. The PCR program was as follows: initial temperature 94 °C for 2 min, 35 cycles of 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min, with a final extension step at 72 °C for 10 min.
The HEP-F and HEP-R oligonucleotide primers targeting the aminotransferase (AMT) domain of the hepatotoxin synthetase mcyE and ndaF genes13 and containing a GC clamp21 at the 5′ end of HEP-F were synthesized (DNA Technology A/S, Risskov, Denmark) and used for DGGE analysis. Amplified products were 472 bp. PCR reactions were performed with an initial temperature of 94 °C for 2 min, followed by 35 cycles at 92 °C for 50 s, 52 °C for 1 min, and 72 °C for 1 min, with a final extension step at 72 °C for 10 min.
To verify the specificity of the HEP-F and HEP-R primer pair and to optimize the PCR and DGGE conditions to reduce the non-specific DGGE bands, we used DNA isolated from the non-toxic strain Microcystis aeruginosa PCC 7005 as a negative control and from the toxic strains Microcystis aeruginosa PCC 7806 and Nodularia spumigena AV1 as positive controls. PCR products were analyzed on 1% agarose gels with 1 × TAE-buffer and were stained with ethidium bromide for 10 min.
Relative quantification of the potentially hepatotoxin-producing species by real-time PCR
Real-time PCR using HEP primers was performed in duplicate22,23 in iCycler iQ (Bio-Rad, California, USA) using a iQ SYBR Green Supermix (Bio-Rad, California, USA). The reaction mixture contained 1× iQ SYBR Green Supermix, 0.5 µM of each primer, and RNAase-free water to 25 µl. The RT-PCR program was 95 °C for 30 s, 95 °C for 3 min, followed by 40 cycles at 94 °C for 45 s, 52 °C for 45 s, and 72 °C for 45 s. A melt curve was automatically generated after each run. The DNA was quantified using a standard curve generated using 10-fold serial dilutions of Microcystis aeruginosa PCC 7806 DNA.Denaturing gradient gel electrophoresis
DGGE of the PCR products was carried out using a Dcode system (BioRad, California, USA). DGGE was run at 75 volts for 16 h in 0.75 mm, 6% polyacrylamide gels (37 : 5 : 1 acrylamide bisacrylamide) submerged in 1 × TAE buffer (40 mM Tris, 40 mM acetic acid, and 1 mM EDTA, pH 7.4) at 60 °C, as described previously.24 A linear gradient of denaturing agents from 45–65% and 45–75% was used to resolve the 16S rRNA gene and AMT-domain PCR products, respectively. After electrophoresis, the gel was stained in 1 × TAE buffer containing SYBRGold Nucleic Acid Stain (1 : 10000 dilution, Molecular Probes Invitrogen AB, California, USA) and the results were recorded using a molecular imager (ChemiDoc XRS system, BioRad, California, USA). The most dominant bands were excised from the gels and submerged in 20 µl DNAase RNAase-free H2O (ultraPURE, Gibco, Invitrogen AB, California, USA) and stored at 4 °C overnight. It was assumed that DGGE bands with same position on the gel correspond to identical base pair sequence and belong to an individual population present in the natural community. In order to confirm such an assumption, more than one band with the same position on the gel were cut, re-amplified and sequenced. An aliquot of the eluted DNA was subjected to an additional round of PCR using the same primers and sequenced on an ABI 3130XL system (DNA technology A/S, Risskov, Denmark).Phylogenetic reconstructions
Partial 16S rRNA and AMT-domain sequences were aligned in Bioedit, version 7.0.4.1 (http://www.mbio.ncsu.edu/bioedit/bioedit.html), using ClustalW. All sequences were subjected to BLAST searches (http://www.ncbi.nlm.nih.gov/blast)25 and the closest relatives from GenBank were included for phylogenetic analysis. Only sequences from published studies or culture collections were included, and the reference taxa were used for phylogenetic inference from distance approximations by the neighbor-joining method and Kimura two-parameter (K2P) in PAUP (version 4.0b10, Sinauer Associates Inc., Sunderland, MA). One thousand bootstrap replicates were performed for both data sets. The 16S rRNA gene sequence of Gloeobacter violaceus PCC 7421 and the mcyE gene sequence of Nostoc punctiforme PCC 73102 were used as outgroups, as appropriate.The sequences generated in this study have been deposited in the GenBank database under accession numbers (16S rRNA-DGGE bands): EU099011–EU099022, (AMT domain-DGGE bands): EU099023–EU099029.
Chemical and hepatotoxin analyses of the water samples
Water samples from each site were passed through a 0.2 µm membrane filter and stored at −20 °C. Total nitrogen (TN) and total phosphorus (TP) were determined as described in Eaton et al.26 Hepatotoxins were analyzed using an ELISA kit (Abraxis, BioScience, Inc, Los Angeles, USA).Statistical analysis
Multiple Regression ANOVA test was performed in order to analyze the effect of TN and TP on the relative abundance of the potential hepatotoxic cyanobacteria at the sampling sites using the R statistical software program, version 2.3.1 for Windows XP (http://www.r-project.org/). Difference was considered to be statistically significant at P < 0.05.Results
Sample collection and concentration procedures were performed based on a preliminary light microscopy evaluation of the community composition and structure. The presence of mucilaginous Microcystis colonies prompted collection of the biomass by filtration through a 10 µm filter following initial concentration of the sample in a 90 µm net. All of the community retained on the 10 µm filters was used for further morphological and molecular investigations.Tentative morphological analysis of dominant cyanobacterial species
As seen in Fig. 2, the samples harbored morphologically distinct cyanobacteria; the unicellular Microcystis genus was the dominant organism present. The most abundant Microcystis morphotypes were tentatively identified as M. aeruginosa (Fig. 2 A), M. wesenbergii (Fig. 2C and D) and M. flos-aquae (Fig. 2B). Other freshwater planktonic cyanobacteria in narrow association on the colonial surface of Microcystis were tentatively identified as Synechococcus epigloeicus (Fig. 2C). Taxonomic classification was according to Komárek and Anagnostidis.18–20 | ||
| Fig. 2 Optical light microscopy micrographs of the dominant cyanobacterial morphotypes in the Nile Delta region. (A) Microcystis aeruginosa. (B) M. flos-aquae (C) M. wesenbergii colonies and Synechococcus epigloeicus on the colonial surface of M wesenbergii, and (D) M. wesenbergii. Tentative taxonomic classifications were according to Komárek and Anagnostidis (Komárek & Anagnostidis 1998, 1999, 2005). Scale bar = 10 µm. | ||
Identification of potentially hepatotoxic cyanobacteria by real-time PCR
DNA was extracted from field samples17 using a standard phenol-chloroform method, which is considered the most reliable and accurate method for good coverage of the cyanobacteria genotypes present in water samples.27Except for the genus Planktothrix, the presence of the hepatotoxin-synthetase genes in the genome of cyanobacteria is a molecular marker for the potential toxicity of cells harboring these genes.28–30 Therefore, we used general primers targeting the AMT domain of the hepatotoxin producing cyanobacterial genera to relatively quantify the abundance of the potential hepatotoxic cyanobacterial community.13
As shown in Fig. 3, real-time PCR confirmed the presence of potentially hepatotoxin-producing cyanobacteria at all three sampling sites.13 The relative quantity of the amplified AMT PCR products did not vary between the seasons (summer and winter) or the sample sites except for Kafr El- Zayat (Fig. 3), which demonstrated higher quantities during both summer and winter seasons with the highest peak during summer.
 | ||
| Fig. 3 Real-time PCR-based quantification of the potentially hepatotoxic cyanobacterial community in the water samples collected from the three sampling sites during summer (black columns) and winter (light columns). The DNA quantities are expressed relative to a standard curve (see Material and methods section). Error bars indicate SE, n = 5. | ||
Chemical and hepatotoxin analyses
The amount of total phosphorus was found to be significantly higher in the water samples from Kafr El-Zayat as compared to the two other sites, while the amount of total nitrogen was found to be significantly higher at the three sampling sites during winter (Table 1). Multiple Regression ANOVA test showed no colinearity between TN and TP. Only TP showed a significant effect on the relative abundance of the potentially hepatotoxic cyanobacterial community (r2 = 0.8601, p = 0.000143). TN : TP (molar) ratio at Kafer El-Zayat was found to be 13.05 ± 0.065 and 78.81 ± 2.0 during summer and winter, respectively, while the ratio is >99 at the other sites indicating strong phosphorus limitation.| Concentration (mg/L) | Summer | Winter | ||||
|---|---|---|---|---|---|---|
| Cairo | Zefta | Kafr El-Zayat | Cairo | Zefta | Kafr El-Zayat | |
| TN | 2.87 ± 0.89 | 2.27 ± 1.633 | 4.45 ± 1.51 | 9.2 ± 0.424 | 14.46 ± 1.71 | 16.55 ± 1.34 |
| TP | 0.0125 ± 0.00 | 0.051 ± 0.00 | 0.74 ± 0.22 | 0.02 ± 0.00 | 0.019 ± 0.00 | 0.46 ± 0.045 |
The concentration of hepatotoxins in the water samples was below the detection limit of ELISA and therefore below the limit set by WHO for drinking water (1 µg/L).
Identity and distribution of potentially hepatotoxic cyanobacterial community by DGGE fingerprinting
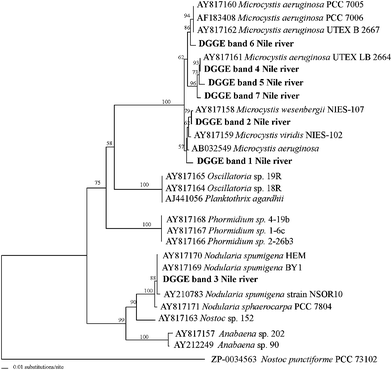
The cyanobacterial sources of the 16S rRNA and AMT-domain DGGE bands (Fig. 4) were identified by sequencing. After preliminary DGGE band identification by Blast comparisons with the GenBank, a more detailed analysis using all 16S rRNA and AMT-domain phylotypes was performed. The resulting phylogenetic reconstructions are shown in Fig. 5 and 6, respectively. | ||
| Fig. 4 Genetic fingerprints of cyanobacteria collected from the Nile Delta at the three sample sites (C = Cairo, K = Kafr El-Zayat, and Z = Zefta) during summer and winter. The partial gene fragments retrieved were visualized using DGGE analyses. (A) 16S rRNA genes bands, and (B) AMT-domain bands. The 16S rRNA and AMT-domain DGGE bands that were excised from the gel for sequencing are numbered and denoted by black arrows. Designations correspond to those shown in the phylogenetic reconstructions in Fig. 5 and 6. *Asterisks denote cDNA winter sample fingerprints obtained by HEP-DGGE analysis. M= Microcystis strain; N= Nodularia strain. | ||
 | ||
| Fig. 5 Phylogenetic affiliations of cyanobacterial 16S rRNA gene DGGE-band sequences from Nile River Delta water samples collected from three different sites during summer and winter. The tree was inferred using neighbor-joining distance algorithm with the Kimura 2P model correction. Sequences from this study are in bold. Bootstrap values >50 are shown. | ||
 | ||
| Fig. 6 Phylogenetic affiliations of AMT gene DGGE-band sequences from hepatotoxic cyanobacteria present in the Nile river water samples collected from three sampling different sites during summer and winter. The tree was inferred using neighbor-joining distance algorithm with the Kimura 2P model correction. Sequences from this study are in bold. Bootstrap values >50 are shown. | ||
AMT-domain DGGE analysis showed a detailed fingerprint of the potentially hepatotoxic cyanobacterial genera in the community. It showed different cyanobacterial dynamics and revealed more genotypes than 16S rRNA gene analysis. A clear succession of different genotypes was apparent between winter and summer, (Fig. 4). In addition, AMT-domain DGGE images confirmed the expression of those genes in several winter samples (see samples designed by an asterisk in Fig. 4). As shown by our phylogenetic clustering positions, all of the AMT-domain DGGE bands retrieved, except for one, were related to Microcystis spp. (DGGE bands 1, 2, 4, 5, 6; Fig. 4 and 6). In particular DGGE band 6 was found to share the same position at the gel with the toxic Microcystis aeruginosa PCC 7806. The only DGGE band not related to Microcystis (DGGE band 3) was related with Nodularia spumigena and confirmed its presence by the DGGE position of the toxic isolate culture of Nodularia spumigena strain AV1 included in the DGGE images (see Fig. 4).
The potentially toxic M. wesenbergii was found to be present in several of the samples according to our DGGE analysis (DGGE band 2 in Fig. 4).
It is notable that AMT-domain DGGE band 4, related to the M. aeruginosa UTEX LB 2664 strain, was present in all of the samples investigated (Fig. 4), indicating that it is probably the most abundant M. aeruginosa strain present at the sites chosen.
The morphotype M. flos-aquae (observed by LM) has never been described as a toxic strain, which explains why it was not detected in our AMT-domain DGGE fingerprinting analysis (Fig. 5 and 6).
In addition, other non-toxic freshwater cyanobacteria, Synechococcus and Cyanobium spp. (DGGE bands 1 to 6, 8 and 9; Fig. 4 and 5) were abundant, as shown by 16S rRNA gene analysis. These picoplanktonic cyanobacteria clearly do not carry any hepatotoxic genes, as evidenced by our AMT-DGGE fingerprinting analysis (Fig. 6).
Furthermore, there are some DGGE bands belonging to summer samples (Fig 4A two diffuse bands in C and Z in a line between bands 8 and 9, and Fig. 4B three bands in a line between bands 4 and 5) that have not been considered. It has been previously reported that DGGE has some methodological biases. Between those, it includes the difficulty of cutting or re-amplifying all the bands observed on the gel images given most of the times to the relative low intensity of them.31 Although relative abundance obtained by DGGE analysis is still under discussion, the relative intensities of the bands obtained and visualized on the gel images included in the present study can give us an estimation of the low contribution that those particular bands (populations) must have had when compared to the total community.
The remainder of the 16S rRNA DGGE bands shown in Fig. 4 were related to potentially toxic and highly adaptable freshwater cyanobacteria, such as the filamentous heterocystous Cylindrospermopsis raciborskii species (band 11; Fig. 4 and 5) and the filamentous non-heterocystous Pseudanabaenaceae family members (DGGE band 10; Fig. 4 and 5). These species were previously identified in Nile water samples.15,32 However, none of those bands were identical in sequence to any band obtained from our AMT-domain DGGE gene analysis (Fig. 6), indicating that these strains are probably non-hepatotoxic.
Although real-time PCR did not detect clear seasonal differences (see section above and Fig. 3), DGGE analysis revealed major seasonal differences in community composition. In summer, the relative abundance of Microcystis phylotypes, detected using 16S rRNA DGGE analysis, disappeared or dramatically decreased. An identical trend was observed when using HEP primers. In that case, several of the slightly different Microcystis phylotypes, including the one retrieved when targeting the 16S rRNA gene, disappeared over the summer (Fig. 4).
Discussion
The presence of potentially hepatotoxic species in water bodies used for drinking purpose is a cosmopolitan problem documented to have caused several incidences of human and cattle poisoning. Nationwide actions are required and need to be unified behind modern tools to sustainably manage the water bodies with the aim of protecting both human health and biodiversity. The Nile River, which is the main source of drinking water for approx. 160 million people living along its shores in nine countries, suffers from eutrophication and the presence of hepatotoxic cyanobacterial species. The potential health threat these species pose on such a massive number of people is worth careful study.Toxic-producing cyanobacterial species were previously isolated and identified in the southern part of the Nile River, Egypt, using morphological identification combined with toxicity assays.14,33 Hamed15 has also morphologically identified cyanobacterial species, including species known to be toxic, at Cairo and the Delta region. Morphological identification is time consuming and it requires high expertise. Accurate identification can be complicated by the deficiency and plasticity of morphological characteristics. In fact, morphological features used for the identification of species such as colonial form, mucilage patterns and cell arrangement in the colony are frequently variable and dependent on the environment.34 Furthermore, the requirement for detailed identification is illustrated by the co-occurrence of toxin producing and non-producing cells that are morphologically indistinguishable.10,35–37
For all these reasons, PCR-based identification and monitoring emerged as an attractive tool that offers sensitivity, specificity and speed. Disagreement between observed morphotypes and presence of toxic genes such as mcyB has been already demonstrated.38–41 However, correspondence between molecular methodologies and for instance immunodetection method, support the applicability of the molecular marker methodology for the monitoring of aquatic ecosystems.42
For comparative purposes with previous studies in the Nile, which focused mainly on the southern region, we have morphologically examined samples collected from the Nile Delta region. Fig. 2 shows potential morphologically distinct cyanobacteria with the unicellular Microcystis genus being the dominant organism present. The most abundant Microcystis morphotypes were tentatively identified as M. aeruginosa (Fig. 2 A), M. wesenbergii (Fig. 2C and D) and M. flos-aquae (Fig. 2B). This is in accord with the study conducted by Hamed,15 which documented the presence of M. aeruginosa and M. flos-aquae in the Nile Delta region. As far as we know, no previous study has documented the presence of M. wesenbergii in the Nile River. It has been argued by several authors that classification of the genus Microcystis is no more valid since features of many morphospecies overlap.43,44 Such overlap might explain the absence of M. wesenbergii from previous studies conducted in the Nile.
Our real-time PCR analysis showed the interseasonal persistence of potentially hepatotoxic cyanobacterial species (Fig. 3). This is not surprising considering that the water in the Nile is eutrophied and relatively warm throughout the year, which can promote the interseasonal persistence of strains. Samples from Kafr El-Zayat showed higher abundance of potentially hepatotoxic species. Concentration of total nitrogen was found to be higher during summer than during winter at the three sampling sites, and total phosphorus concentration was found to be highest at Kafr El-Zayat during both summer and winter, with summer samples showing the highest quantity (Table 1). Our statistical data revealed that phosphorus content of water was the significant variable that affects the abundance of potentially toxic species. In support to our findings Håkanson et al.45 argued that no simple relationship between TN : TP and cyanobacterial abundance could be found, and that generally, variation in TP rather than TN seems to be more important to predict variations in cyanobacterial abundance among systems. Klausmeier et al.46 argued that the Redfield ratio of TN : TP equal to 16 is not a universal biochemical optimum but an average of species-specific ratio that can vary between 8.2 to 45.0. This could explain the high abundance of hepatotoxic cyanobacterial species found at Kafr El-Zayat with TN : TP falling within or close to the range suggested.
The level of hepatotoxins in the water samples was found to be below the limit set by the WHO for drinking water. This is in accordance with the study conducted by Mohamed et al.,33 and indicates that, despite the presence of potentially hepatotoxin-producing genera in the Nile River, there was no obvious direct potential health threat at the time and places of sampling, because cyanobacterial toxins are not released unless the cells lyse. The effect of chronic exposure to such very low toxin levels needs to be further investigated. In addition, these results are in accordance with Mohamed,47 suggesting that the governmental procedures for treating the River water for drinking purposes needs to be adjusted in order to avoid cyanobacterial cell lysis and toxin release.
Fingerprinting techniques such as denaturing gradient gel electrophoresis (DGGE) provide very informative results with less time-consumption. For genotypic analysis of cyanobacteria, the use of DGGE analysis of the intergenic transcribed spacer (rRNA-ITS) has increasingly being applied. Several studies have made use of that methodology to study diversity and to distinguish between toxic and non-toxic strains of the cyanobacterial genera such as Microcystis, that could not be distinguished microscopically, and their relative band intensities in the DGGE profile also enabled semi-quantitative estimates of the abundances of these strains.48–51 The population dynamics deduced from the DGGE analysis were confirmed by independent measurements of changes in the total microcystin concentration.51 Nevertheless, it has been shown that targeting the mcy genes by PCR is more reliable for identifying hepatotoxic genera than targeting other genes, such as the 16S rRNA gene.41 This explains our 16S rRNA gene-DGGE analysis that provided no evidence for the presence of more than one Microcystis phylotype (DGGE band 7; Fig. 4 and 5).
Microcystis spp. are colony-forming, unicellular, fresh water cyanobacteria capable of forming blooms in eutrophied water bodies during warm months.52 The blooms of Microcystis usually consist of different morpho- and genotypes,18 which was confirmed by our DGGE analysis. M. aeruginosa has caused incidences of poisoning of domestic animals and wildlife worldwide. Whether there has been such an incidence in the Nile River Delta or valley is not known.
The dominance of Synechococcus and Cyanobium genera in association with the genus Microcystis has been reported in other freshwater systems.53,54 The genus Synechococcus was also identified morphologically (by LM), in narrow association with Microcystis colonies (Fig. 2B). The abundance and diversity of these picocyanobacteria suggests that they could play an important role as primary producers in the system. Detailed exploration of the picoplanktonic fraction and the diversity of Synechococcus and Cyanobium clusters in freshwater ecosystems continue to reveal new ecotypes and genotypes.55–58
The detection of C. raciborskii by 16S rRNA DGGE analysis and not by AMT-domain DGGE analysis raises concerns about the universal application of the HEP primers set used in this study for the detection of all cyanobacterial hepatotoxic species and deserves further investigation.
Our results suggest a community dynamic in which the relative abundance of the various genotypes varies seasonally while a relatively stable quantity of the overall cyanobacterial phytoplankton community is maintained. Included in the stable community are some potentially toxic strains that persist due to scant variation in abiotic factors, such as temperature and nutrients (Table 1).
Conclusion
This work is the first to use a genetic approach to analyze the cyanobacterial planktonic community composition in depth and, specifically, to identify potentially hepatotoxic species in the Nile River. We showed the presence and persistence of a rich planktonic cyanobacterial community dominated by Microcystis phylotypes, such as the potential hepatotoxin producers M. wesenbergii and M. aeruginosa, together with several Synechococcus and Cyanobium phylotypes. The quantity of the potentially hepatotoxic members was generally stable but their microdiversity varied between summer and winter. We also showed that although some of the hepatotoxic Microcystis spp. might have been active during sampling, the amount of hepatotoxin detected was below the WHO limit for drinking water, indicating that either little or no cell lysis was occurring at the time of sampling or that the amount of the hepatotoxin released was small enough to be diluted in the water body. We recommend that governmental procedures for treating the Nile River water for drinking purposes need to take into consideration the presence of these potentially toxic organisms. Our study also represents the first time that real-time PCR and DGGE fingerprinting analyses have been optimized and co-applied using the same primer pair to study hepatotoxic genes in environmental samples, providing a direct method for qualitative and quantitative analysis of hepatotoxic microbial communities present in a fresh water body.Acknowledgements
The authors wish to thank Dr E. Dittmann (Humboldt University, Germany), Dr A. Sanjurjo (Facultad de Biología, Spain) and Dr R. Tavera (Universidad Nacional Autónoma de México, Mexico) for helping with the morphological identification. Travel grant (MENA-programme) for RA and RE from the Swedish Research council (VR) is also acknowledged.References
- C. R. Woese, O. Kandler and M. L. Wheelis, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 4576–4579.
- O. M. Skulberge, W. W. Carmichael, G. A. Codd and R. Skulberg, in Algal Toxins in Sea food and Drinking water, ed. I. R. Falconer, Academic Press, London, 1993, pp. 145–164 Search PubMed.
- P. V. Lakshmana Rao, R. Bhattacharya, N. Gupta, M. M. Parida, A. S. B. Bhaskar and R. Dubey, Arch. Toxicol., 2002, 76, 227–235 CrossRef CAS.
- S. Haider, V. Naithani, P. N. Viswananthan and P. Kakkar, Chemosphere, 2003, 52, 1–21 CrossRef CAS.
- W. W. Carmichael, Hum. Ecol. Risk Assess., 2001, 7, 1393–407 CrossRef.
- C. MacKintosh, K. A. Beattie, S. Klumpp, P. Cohen and G. A. Codd, FEBS Lett., 1990, 264, 187–192 CrossRef CAS.
- J. Goldberg, H. B. Huang, Y. G. Kwon, P. Greengard, A. C. Nairn and J. Kuriyan, Nature, 1995, 376, 745–753 CrossRef CAS.
- E. Dittmann, B. A. Neilan, M. Erhard, H. von Dohren and T. Borner, Mol. Microbiol., 1997, 26, 779–787 CAS.
- C. Moffitt and B. Neilan, Appl. Environ. Microbiol., 2004, 70, 6353–6362 CrossRef CAS.
- E. Dittmann and C. Wiegand, Mol. Nutr. Food Res., 2006, 50, 7–17 CrossRef CAS.
- A. J. Ouellette and S. W. Wilhelm, Front. Ecol. Environ., 2003, 1, 359–366.
- K. Furukawa, N. Noda, S. Tosunida, T. Saito, T. Itayama and Y. Inamori, J. Biosci. Bioeng., 2006, 102, 90–96 CrossRef CAS.
- A. Jungblut and B. A. Neilan, Arch. Microbiol., 2006, 185, 107–114 CrossRef CAS.
- S. Brittan, Z. A. Mohamed, J. Wang, V. K. B. Lehmann, W. W. Carmichael and K. L. Rinehart, Toxicon., 2000, 38, 1759–1772 CrossRef.
- A. F. Hamed, Acta Bot. Hungar., 2005, 47, 117–136 Search PubMed.
- Z. A. Mohamed, H. M. El Sahrouny and W. S. M. Ali, Toxicon., 2006, 47, 584–590 CrossRef CAS.
- D. Tillett and A. Neilan, J. Phycol., 2000, 36, 251–258 CrossRef CAS.
- J. Komárek and K. Anagnostidis, in Cyanoprokaryota. ed. H. Ettl, G. Gartner, H. Heynig and D. Mollenhauer, S_sswasserflora von Mitteleuropa. Gustav Fischer, Jena, 1998 Search PubMed.
- J. Komárek and K. Anagnostidis, in Cyanoprokaryota. ed. H. Ettl, G. Gartner, H. Heynig and D. Mollenhauer, S_sswasserflora von Mitteleuropa. Gustav Fischer, Jena, 1999 Search PubMed.
- J. Komárek and K. Anagnostidis, in Cyanoprokaryota. ed. H. Ettl, G. Gartner, H. Heynig and D. Mollenhauer, S_sswasserflora von Mitteleuropa. Gustav Fischer, Jena, 2005 Search PubMed.
- U. Nübel, F. Garcia-Pichel and G. Muyze, Appl. Environ. Microbiol., 1997, 63, 3327–3332 CAS.
- J. Huggett, K. Dheda, S. Bustin and A. Zumla, Genes Immun., 2005, 6, 279–284 CrossRef CAS.
- M. L. Wong and J. F. Medrano, Biotechniques, 2005, 39, 75–85 CrossRef CAS.
- S. F. Altschul, T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller and D. Lipman, Nucleic Acids Res., 1997, 25, 3389–3402 CrossRef CAS.
- B. Díez, C. Pedrós-Alió, T. L. Marsh and R. Massana, Appl. Environ. Microbiol., 2001b, 67, 2942–2951 Search PubMed.
- A. D. Eaton, M. A. Franson, American Public Health Association, American Water Works Association and Water Environment Federation, Washington, DC, 2005, pp. 4-14–4-153.
- E. Schober and R. Kurmayer, Lett. Appl. Microbiol., 2006, 42, 412–417 CrossRef CAS.
- R. Kurmayer, G. Christiansen, J. Fastner and T. Börner, Environ. Microbiol., 2004, 6, 831–841 CrossRef CAS.
- G. Christiansen, R. Kurmayer, Q. Liu and T. Börner, Appl. Environ. Microbiol., 2006, 72, 117–123 CrossRef CAS.
- L. N. Sangolkar, S. S. Maske and T. Chakrabarti, Water Res., 2006, 40, 3485–3496 CrossRef CAS.
- B. Díez, C. Pedros-Alio and R. Massana, Appl. Environ. Microbiol., 2001a, 67, 2932–2941 Search PubMed.
- Z. A. Mohamed, FEMS Microbiol. Ecol., 2007, 59, 794–761.
- Z. A. Mohamed, H. M. El-Sahrounyand and W. S. Ali, Arch. Environ. Contam. Toxicol., 2007, 52, 489–495 CrossRef CAS.
- S. Otsuka, S. Suda, R. H. Li, S. Matsumoto and M. M. Watanabe, J. Gen. Appl. Microbiol., 2000, 46, 39–50 CrossRef CAS.
- K. Sivonen and G. Jones, in Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management, ed. I. Chorus and J. Bartram J., E & FN Spon, New York, NY, 1999, pp. 41–111 Search PubMed.
- J. Fastner, M. Erhard and H. von Döhren, Appl. Environ. Microbiol., 2001, 67, 5069–5076 CrossRef CAS.
- I. Janse, W. E. A. Kardinaal, M. Meima, J. Fastner, P. M. Visser and G. Zwart, Appl. Environ. Microbiol., 2005, 70, 3979–3987.
- B. A. Neilan, E. Dittmann, L. Rouhiainen, R. A. Bass, V. Schaub, K. Sivonen and T. Börner, J. Bacteriol., 1999, 181, 4089–4097 CAS.
- T. Nishizawa, M. Asayama, K. Fujii, K.-I. Harada and M. Shirai, J. Biochem., 1999, 126, 520–529 CAS.
- S. Otsuka, S. Suda, R. H. Li, M. M. Watanabe, H. Oyaizu, S. Matsumoto and M. M. Watanabe, FEMS. Microbiol. Lett., 1999, 172, 15–21 CrossRef CAS.
- D. Tillett, D. L. Parker and B. A. Neilan, Appl. Environ. Microbiol., 2001, 67, 2810–2818 CrossRef CAS.
- M. Bittencourt-Oliveira, Harmful Algae, 2003, 2, 51–60 CrossRef.
- S. Otsuka, S. Suda, S. Shibata, H. Oyaizu, S. Matsumoto and M. M. Watanabe, Int. J. Syst. Evol. Microbiol., 2001, 51, 873–879 Search PubMed.
- R. Kondo, T. Yoshida, Y. Yuki and H. Shingo, Int. J. Syst. Evol. Microbiol., 2000, 50, 767–770 Search PubMed.
- L. Håkanson, A. C. Bryhn and J. K. Hytteborn, Science Total Environ., 2007, 379, 89–108 CrossRef CAS.
- C. A. Klausmeier, E. Litchman, T. Daufresne and S. A. Levin, Nature, 2004, 420, 171–174 CrossRef.
- Z. A. Mohamed, Water Res. Manag., 2001, 15, 213–221 Search PubMed.
- I. Janse, M. Meima, W. E. A. Kardinaal and G. Zwart, Appl. Environ. Microbiol., 2003, 69, 6634–6643 CrossRef CAS.
- I. Janse, J. Bok and G. Zwart, J. Microbiol. Methods, 2004a, 57, 279–281 Search PubMed.
- I. Janse, W. E. A. Kardinaal, M. K. Agterveld, M. Meima, P. M. Visser and G. Zwart, Environ. Microbiol., 2005, 7, 1514–1524 CrossRef CAS.
- W. E. A. Kardinaal, I. Janse, M. K. Agterveld, M. Meima, J. Snoek, L. R. Mur, J. Huisman, G. Zwart and P. M. Visser, Aquat. Microb. Ecol., 2007, 48, 1–12 Search PubMed.
- H. O. Utkilen, M. Skulberg, B. Underdal, N. Gjølme, R. Skulberg and J. Kotai, Phycologia., 1996, 35, 189–197.
- A. J. A. Ouellette, S. M. Handy and S. W. Wilhelm, Microb. Ecol., 2006, 51, 154–165 CrossRef CAS.
- E. Kolmonen, K. Sivonen and J. Rapala, Aquat. Microb. Ecol., 2004, 36, 201–211 Search PubMed.
- N. V. Ivanikova, R. M. L. McKay, G. S. Bullerjahn and R. W. Sterner, J. Phycol., 2007, 43, 475–484 CrossRef.
- N. D. Crosbie, M. Pöckl and T. Weisse, Appl. Environ. Microbiol., 2003, 69, 5716–5721 CrossRef CAS.
- A. Ernst, S. Becker, U. I. A. Wollenzien and C. Postius, Microbiol., 2003, 149, 217–228 CrossRef CAS.
- B. R. Robertson, N. Tezuka and M. M. Watanabe, Int. J. Syst. Evol. Microbiol., 2001, 51, 861–871 Search PubMed.
Footnote |
| † Shared first-authorship |
| This journal is © The Royal Society of Chemistry 2009 |
