A new role for cisplatin: probing ribosomal RNA structure
Keshab
Rijal
and
Christine S.
Chow
*
Department of Chemistry, Wayne State University, Detroit, Michigan, 48202, USA. E-mail: csc@chem.wayne.edu; Fax: +01 313-577-8822; Tel: +01 313-577-2594
First published on 10th November 2008
Abstract
In this study, cisplatin, the well-known anticancer drug, was used to probe accessible purine residues in E. coli16S ribosomal RNA (rRNA), free or in the context of 30S subunits or 70S ribosomes, both in vitro and in vivo.
The aquated form of cisplatin, cis-[Pt(NH3)2Cl2], reacts preferentially with purine bases, to generate several types of DNA adducts, such as 1,2-GpG-intrastrand cross-links, which are believed to contribute to the known anticancer activity of the compound.1 The interactions of cisplatin with RNA have received much less attention in the literature.2,3RNA has a tendency to fold into complex 3D structures, and large RNA molecules can form a wide range of tertiary structures and microenvironments with varying solvent and small-molecule accessibilities.4 Here, a new use of cisplatin to proberibosomal RNA structure is reported. The ribosome is the protein synthesis machinery in all organisms and serves as an ideal target for certain classes of antibacterials.5 High-resolution X-ray crystal structures of 70S ribosomes and 30S subunits have greatly expanded our understanding of the overall folding of ribosomal RNA (rRNA) when assembled with proteins.6 The formation of charged pockets, which might occur during various stages of ribosome assembly and function, as well as influence small molecule interactions, is difficult to examine experimentally due to a lack of appropriate methods.
A number of chemical probes have been used to obtain structural information on RNA, but have limited utility because of an inability to penetrate cell walls, high toxicity, or challenges in detecting their cleavage or modification sites.7 The formation of platinum adducts on the ribosome could provide information about small molecule accessibility under a range of cellular conditions, as well as lead to identification of new target sites for potential antimicrobial compounds. This approach has several advantages, such as easy detection of the platinum adducts and the ability to work in vivo.
Inside cells, cisplatin is monoaquated to its active form, cis-[PtCl(NH3)2(OH2)]+ (1) (Fig. 1).1 The positive charge of 1 facilitates favourable interactions with the polyanionic nucleic acids.2,3 Adducts with RNA are formed when the labile ligands (Cl and/or H2O) are replaced with the N7 of purine bases. We investigated reactions of 1 with 16S rRNA, free or in the context of 30S subunits or 70S ribosomes. Ribosomes were isolated from E. coli DH5α strain by the sucrose gradient method8 and 16S rRNA was isolated from 30S subunits by phenol–chloroformextraction. Freshly prepared complex 12,3 was incubated with rRNA or ribosomes in 10 mM phosphate, 5 mM NaCl, 10 mM MgCl2, pH 6.5, in the dark for 5 h. The ratio of 1 to nucleotides was 1 : 15 in all cases, except for in vivo studies. After 5 h, the reactions were quenched with NaCl (200 mM) and immediately frozen. Proteins were removed from 30S subunits and 70S ribosomes by phenol-chloroformextraction followed by ethanolprecipitation. A 32P-5′-end-labeled DNA primer (5′-CCAAGTCGACATCGTTT-3′) was annealed at positions 831–814 of 16S rRNA (E. coli numbering), followed by reverse transcription.9 This region corresponds to helix 24 (h24, or 790 loop) of 16S rRNA, and was chosen because of its location near the ribosome decoding site and predicted solvent accessibility based on X-ray crystal structures.6 To locate the binding sites of 1, dideoxy sequencing reactions were carried out and compared directly to the reverse transcription reactions on 8% denaturing polyacrylamide gels.9 The nucleotide positions modified by 1 were detected by reverse transcriptase stops or pauses.
![A partial secondary structure map of 16S rRNAh24 (residues 769–820) (left) shows the primer-binding site and major (***) or minor (*) reactive sites of cis-[PtCl(NH3)2(OH2)]+ (1) (middle). The autoradiogram (right) shows reverse transcriptase stops (indicated by arrows) on E. coli 16S rRNA, 30S subunits, and 70S ribosomes due to 1 binding (+ lanes were treated with 1; − lanes are controls). Dideoxy sequencing lanes are labeled as C, U, A and G. Identical sites in the in vitro and in vivo experiments with 1 are indicated with symbols (◆, ●).](/image/article/2009/CC/b816633a/b816633a-f1.gif) | ||
| Fig. 1 A partial secondary structure map of 16S rRNAh24 (residues 769–820) (left) shows the primer-binding site and major (***) or minor (*) reactive sites of cis-[PtCl(NH3)2(OH2)]+ (1) (middle). The autoradiogram (right) shows reverse transcriptase stops (indicated by arrows) on E. coli 16S rRNA, 30S subunits, and 70S ribosomes due to 1 binding (+ lanes were treated with 1; − lanes are controls). Dideoxy sequencing lanes are labeled as C, U, A and G. Identical sites in the in vitro and in vivo experiments with 1 are indicated with symbols (◆, ●). | ||
Cisplatin is known to coordinate with N7 of guanines.1–3Reverse transcriptase was expected to pause or stop at the nucleotide on the 3′ side of the platinum reactive site, as is observed with dimethyl sulfate modification of purine residues.8 As shown in Fig. 1, a strong stop occurs at U801, which is adjacent to two consecutive G residues, G799 and G800, in the internal loop region. The sites are denoted on the secondary structure map10 of 16S rRNA (Fig. 1). Another strong stop is observed at U804, which is also adjacent to a G in the closing base pair of the internal loop. Minor stop sites are observed at A787, G799, G800, A802, G803 and C810, which all correspond to G residues or nucleotides on the 3′ side of a G. Minor bands are observed in all lanes, including the control lane, due to the presence of secondary structures in the 16S rRNA template that block reverse transcriptase.
Reverse transcription with 16S rRNA that was reacted with 1 and then isolated from 30S subunits or 70S ribosomes shows a strong and prominent stop at U801 (Fig. 1). This major stop site is the same as that observed in free 16S rRNA, indicating that consecutive G799 and G800 residues of 16S rRNA comprise the preferred target site for 1 within 30S subunits and 70S ribosomes. Thus, this site appears to be accessible to the aquated platinum complex whether 16S rRNA is free or complexed with the ribosomal proteins. In contrast, U804 is no longer a prominent reactive site of 1, revealing a difference in the structure or accessibility of that site in free vs. complexed 16S rRNA.
Cisplatin is believed to enter cells by passive diffusion and undergo hydration to the aquated species;1 therefore, we hypothesized that the parent compound could also be used for in vivo probing of bacterial ribosomes. Formation of cisplatin adducts in the ribosome that are mapped by reverse transcription would indicate accessible rRNA residues in vivo, and could reveal new target sites for novel antibacterials. After E. coli DH5α was grown to 0.2 to 0.3 OD, cisplatin was added to a final concentration of 100 to 200 μg ml−1 of cells and further incubated for 2 h. After cooling on ice for 15 min, the cells were pelleted by centrifugation. The cells were washed with buffer (3 × with 50 mM Tris-HCl, 100 mM NH4Cl and 10 mM MgCl2) and total RNA was isolated using lysozyme.11 The RNA component was extracted with phenol-chloroform followed by ethanolprecipitation to remove proteins. Although several stops are observed in the background (16S rRNA without cisplatin treatment, lane 0, Fig. 2), reverse transcription with the h24 primer shows a prominent stop at U801, the nucleotide on the 3′-side of the consecutive guanines in the internal loop of h24, only on cisplatin-treated 16S rRNA (Fig. 2, lanes 100 and 200 μg ml−1). These results are consistent with the in vitro probing studies. Minor stop sites are also observed at A792 and G791 in the loop region (Fig. 1 and 2).
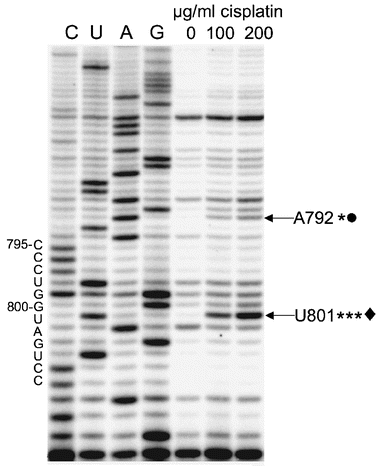 | ||
| Fig. 2 An autoradiogram shows reverse transcription mapping of in vivo reactive sites (indicated by arrows; ***major, *minor) of cisplatin on 16S rRNA/70S ribosomes (the concentrations of cisplatin are given for each lane, 0–200 μg ml−1 of cells). Dideoxy sequencing lanes are labeled as C, U, A and G. Sites that are identical in the in vitro and in vivo probing experiments are indicated with symbols (◆, ●). | ||
These results demonstrate that cisplatin can be used to probe accessible guanine residues of 16S rRNAin vitro and in vivo. Furthermore, the differences in nucleotide accessibility in free 16S rRNAvs. that in ribosomes can be determined. It should be noted, however, that the in vivo results shown here represent an average of all ribosomes at various stages of protein synthesis in the cell. Our results indicate that the platinum complex is sensitive to variations in ribosome structure, and thus could be employed to examine ribosomes at various stages of protein synthesis, or complexed with various factors, mRNA or tRNAs. Most importantly, we have revealed that a positively charged platinum complex is useful for targeting rRNA, and has the ability to provide key information about helix or loop accessibility in ribosomes. As shown in this study, helix 24 is clearly accessible to the metal complex, and thus should also be accessible to small molecules that could serve as novel drug leads. Sites that differ in platinum reactivity between 30S subunits and 70S ribosomes could reveal compelling regions for mechanism-based drug targeting. Furthermore, the fact that the platinum complex generates stable adducts is significant because the kinetics of the reaction can be monitored,3 which will allow the determination of how positively charged compounds such as aminoglycosides5 identify target sites with favourable electrostatic contributions from a number of possible reactive sites on the ribosome.
We are grateful to Prof. Philip Cunningham and Tek N. Lamichhane for helpful discussions, technical assistance, and use of equipment for the ribosome preparations.
Notes and references
- For a review, see: E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467 Search PubMed.
- P. Papsai, J. Aldag, T. Persson and S. K. C. Elmroth, Dalton Trans., 2006, 3515 RSC.
- M. Hägerlöf, P. Papsai, C. S. Chow and S. K. C. Elmroth, JBIC, J. Biol. Inorg. Chem., 2006, 11, 974 CrossRef CAS.
- For a review see: P. B. Moore, Annu. Rev. Biochem., 1999, 68, 287 Search PubMed.
- F. Franceschi and E. M. Duffy, Biochem. Pharmacol., 2006, 71, 1016 CrossRef CAS.
- (a) M. M. Yusupov, G. Zh. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. D. Cate and H. F. Noller, Science, 2001, 292, 883 CrossRef CAS; (b) B. S. Schuwirth, M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila-Sanjurjo, J. M. Holton and J. H. D. Cate, Science, 2005, 310, 827 CrossRef CAS; (c) M. Selmer, C. M. Dunham, F. V. Murphy IV, A. Weixlbaumer, S. Petry, A. C. Kelley, J. R. Weir and V. Ramakrishnan, Science, 2006, 313, 1935 CrossRef CAS.
- (a) C. Brunel and P. Romby, Methods Enzymol., 2000, 318, 3 CAS; (b) M. Balzer and R. Wagner, Anal. Biochem., 1998, 256, 240 CrossRef CAS; (c) M. Lindell, P. Romby and E. G. H. Wagner, RNA, 2002, 8, 534 CrossRef CAS.
- D. Moazed, S. Stern and H. F. Noller, J. Mol. Biol., 1986, 187, 399 CrossRef CAS.
- S. Stern, D. Moazed and H. F. Noller, Methods Enzymol., 1988, 164, 481 Search PubMed.
- R. R. Gutell, J. C. Lee and J. J. Cannone, Curr. Opin. Struct. Biol., 2002, 12, 301 CrossRef CAS.
- M. Dresden and M. B. Hoagland, Science, 1965, 149, 647 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2009 |
