Hot off the press
Hot off the Press highlights recently published work for the benefit of our readers. Our contributors this month have focused on protein labelling of DNA and RNAi affecting a behavioral change in fruit flies. New contributors are always welcome. If you are interested please contact molbiosyst@rsc.org for more information, we'd like to hear from you.
Enzymatically aided labeling of proteins with DNA
Reviewed by: Ljiljana Fruk, University of Dortmund, Germany.
DNA–protein conjugates are important building blocks in the design of a range of nanostructures which can, among other applications, be used to arrange biocatalysts in structured motifs. Although some successful methods for the preparation of such conjugates are known, the lack of control over regioselectivity and stoichiometry of the reactions used often poses a problem. Now Duckworth et al. have demonstrated that a fusion-based strategy could be used for covalent labeling of proteins at the C-terminus with single stranded DNA. The advantage of such a strategy lies in the precise control over the DNA position on the protein which can ease the design of nanostructures. This principle is based on the use of enzyme protein farnesyltransferase (PFTase) which catalyses the reaction of a protein containing a C-terminal tetrapeptide tag (CVIA tag) with azide-modified isoprenoid diphosphate. In a first set of experiments, green fluorescent protein (GFP) was used as a model protein and expressed to contain a C-terminal CVIA tag which was subsequently incubated with azide-modified diphosphate and PFTase to yield azide-functionalised GFP. The azide group can then be reacted with alkyne DNA using standard click chemistry resulting in GFP–DNA conjugates which can be further used to hybridise to complementary DNA strands. In experiments that followed four different GFP–DNA conjugates were prepared and successfully assembled to form GFP tetrahedrons with varying numbers of DNA and GFP–DNA strands. The GFP content in such nanostructures was investigated using fluorescence correlation spectroscopy and the final results provided convincing evidence that the nano-tetrahedrons can be assembled to contain different number of GFPs by using the hybridisation ability of DNA–protein conjugates. Finally, it was demonstrated that other proteins can also be labeled with DNA using same methodology indicating that in the future a range of novel protein–DNA hybrid structures could be designed, leading to a further advance in the field of nanostructuring.
B. P. Duckworth, Y. Chen, J. W. Wollack, Y. Sham, J. D. Mueller, T. A. Taton, and M. D. Distefano, A Universal Method for the Preparation of Covalent Protein–DNA Conjugates for Use in Creating Protein Nanostructures, Angew. Chem., Int. Ed. 2007, 46, 8819–8822.
Screening study solves sex peptide mechanism
Reviewed by Thomas Kodadek, UT Southwestern Medical Center, Dallas, Texas, USA.
A fascinating area of modern biological research is the chemical underpinning of animal behavior. Increasingly, powerful molecular techniques are being combined with surprisingly powerful whole-animal screening methods to identify molecules involved in regulating behavior.
This paper by Yapici et al. is an excellent example of this trend. The goal of their study was to further define the molecular basis for a striking behavioral change in the female fruit fly Drosophila melanogaster. Virgin females are generally quite receptive to courting males. Moreover, they retain their eggs. In contrast, female flies that have already mated respond to new suitors quite differently. They are unreceptive to advances by the males and generally lay their eggs. Clearly, this behavioral switch is to the advantage of the male fly that has already mated with the female since it reduces the risk of dilution of his genetic material in the subsequent population. Therefore, it perhaps not surprising, in an evolutionary sense, that previous research had shown a 36 residue peptide present in male seminal fluid, called sex peptide (SP), is responsible for this change in the behaviour of the female after mating. However, the mechanism of action of SP was unknown. This study demonstrated that there is, in fact, a specific SP receptor (SPR) and that the presence of this receptor in specific neurons of the Drosophila brain is essential for this behavioral switch.
To identify the gene or genes involved in transmitting the SP signal in the female, the authors carried out a genome-wide RNA interference (RNAi) screen. This technology allows the reduction or elimination of essentially all of the genes in a cell or organism in a step-wise fashion. In this particular case, the authors monitored the egg laying behavior of thousands of transgenic females carrying transgenes that expressed a particular small interfering RNA (siRNA) in a pan-neuronal fashion. They found that the siRNA targeting a gene that they named SP receptor (SPR) resulted in a marked reduction in egg laying. They then compared the behavior of virgin wild-type females and those treated with the SPR-targeted siRNA in an assay where they were first exposed to a male. Those females that mated were then allowed to lay eggs for two days before being presented with a second male (different from the first). In the initial mating assays, the wild-type and SPR siRNA females were about equally receptive to the male. But whereas the control wild-type females laid many eggs and rejected the second suitor, the SPR siRNA flies laid few eggs and were as receptive to the second male as to the first. This is a remarkable example of a single gene knock-out having a clear-cut effect on a striking behavioral switch.
The authors went on to show that the SPR gene indeed encodes a G protein coupled receptor that recognizes the SP and that this receptor is expressed in specific neurons that were known previously to be involved in mating behavior. It is interesting that the SPR gene is highly conserved throughout the insect world and thus might be an interesting target in pest control strategies.
N. Yapici, Y.-J. Kim, C. Riberio and B. J. Dickson, A receptor that mediates the post-mating switch in Drosophila reproductive behaviour, Nature, 2008, 451, 33–38.
Hot off the RSC press
Microfluidics prove in-gene-ious
Reviewed by: Laura Howes, Royal Society of Chemistry, Cambridge.
Analysing genes in individual cells is now simpler, cheaper and more effective thanks to US researchers.
Many diseases are linked to abnormal cellular events, which are governed by the cell's genes. But while the human genome has now been sequenced, the functions of these genes and how they interact is still largely unknown. Current techniques to analyse cells' genes use either bulk measurements, for an average from many cells, or expensive and labour intensive single cell measurements. Now, a new microfluidic processor can be used to profile gene–gene interactions at the single cell level simply and more efficiently.
Genes are essentially a code, read by RNA polymerase enzymes, to make messenger RNA (mRNA), small fragments of RNA which are then used to assemble proteins. The level of an individual mRNA, therefore, depends upon the activity of its coding gene. The new microfluidic processor, developed by Jiang Zhong at the University of Southern California in Los Angeles and colleagues, was designed to study these mRNAs in large numbers of single cells.
 | ||
| Fig. 1 Using a microfluidic processor to monitor mRNA levels in single cells helps scientists to analyse gene activity. | ||
The cellular mRNAs are studied indirectly: cells are broken down to capture the mRNA which is then used to make complementary DNA (cDNA). The cDNA can then be amplified and it is this that is detected. Zhong explained that existing methods to study single cells' mRNA ‘suffer from low efficiency in converting mRNA to cDNA,’ as well as being expensive and difficult. By extracting the mRNA and producing the cDNA on the same device in nanolitre volumes, the processor is much simpler and more efficient to use.
‘Microfluidic and nanolitre scale biochemical reactions will greatly facilitate biological studies, particularly single cell analysis,’ said Zhong. The team suggests that the processor could be used to investigate many different areas, from organ formation to aging and disease development, and is already making the device easier for biologists to use in the lab by introducing computer automation.
J. F. Zhong, Y. Chen, J. S. Marcus, A. Scherer, S. R. Quake, C. R. Taylor and L. P. Weiner, A microfluidic processor for gene expression profiling of single human embryonic stem cells, Lab Chip, 2008
DOI: 10.1039/b712116d
Enzyme-powered delivery vehicles
Reviewed by: Freya Mearns, Royal Society of Chemistry, Cambridge.
Dutch scientists have made nanotubes move using enzyme-powered motors.
Ben Feringa and co-workers from the University of Groningen, The Netherlands, have designed engines for nanomachines that could potentially be used in the body.
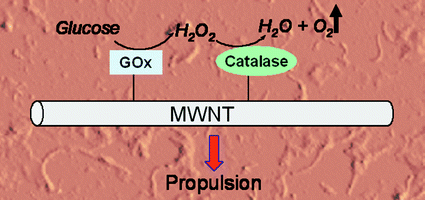
Hydrogen peroxide has proven useful as a chemical fuel for powering microscopic motors but its practicality is somewhat limited by its inherent reactivity, said Feringa. To get around this problem the team have used two enzymes in tandem as the engine for their nanomachine. They explained that by coupling glucose oxidase with catalase, relatively stable glucose can be used as the primary fuel instead. ‘This fuel is already present in the body,’ said team-member Wesley Browne, ‘and it is completely inert.’
 | ||
| Fig. 2 Oxygen gas produced by enzyme reactions causes the nanotube to move. | ||
The glucose oxidase converts glucose and oxygen to gluconolactone and hydrogen peroxide. The hydrogen peroxide is then consumed by the catalase to produce water and oxygen. The results are surprising, said the team, because in principle nothing should happen—more oxygen is consumed than is produced. But the glucose oxidase produces high local concentrations of hydrogen peroxide, which in turn results in bursts of oxygen being released as bubbles of dioxygen gas. This is what causes the enzymes (and the nanotubes they are attached to) to move.
‘This work is the first step towards the design of functional nano-machinery working with renewable resources as fuel,’ said Heinz-Bernhard Kraatz, an expert on nano-biomaterials from the University of Western Ontario, London, Canada.
While the nanotubes' movement currently appears erratic, the Dutch team have noticed it is dependent on the shape of the nanotube/enzyme aggregate and the enzymes' position on the nanotube. Although currently only non-directional movement is demonstrated, said Kraatz, ‘it is clear that the system can be upgraded as soon as rational and site directed modifications of nanosized objects becomes available.’
D. Pantarotto, W. R. Browne and B. L. Feringa, Autonomous propulsion of carbon nanotubes powered by a multienzyme ensemble, Chem. Commun., 2008
DOI: 10.1039/b715310d
| This journal is © The Royal Society of Chemistry 2008 |
