Transcription factor logic using chemical complementation
Jonathan E.
Bronson
,
William W.
Mazur
and
Virginia W.
Cornish
*
Department of Chemistry, Columbia University, New York, NY 10027, USA
First published on 16th November 2007
Abstract
Chemical complementation was used to make a transcription factor circuit capable of performing complex Boolean logic.
Artificial transcription regulation networks are used to quantitatively study biological processes such as quorum sensing, circadian rhythm, cellular memory and biochemical signaling pathways.1–4 In biotechnology, they are used for the biosynthesis of natural products such as resveratrol and the malaria drug precursor artemisinic acid.5,6 The creation of ‘smart cells’, which are engineered to perform sophisticated decision making such as the ability to recognize and invade tumor cells,7 is based on artificial transcription networks as well. Often, these designed networks are treated like electrical circuits with transcription factors functioning as Boolean logic gates.8 Multi-input logic functions, such as AND or OR logic, are currently created using combinations of simpler transcription factors, such as LacI, TetR, cI or LuxR.8,9 This approach to creating logic gates has several drawbacks though. Very few small molecule inducible transcription factors have been well characterized and shown to be robust and orthogonal enough to the cell's genetic machinery to use in artificial networks, so using them in combination quickly limits the size of the networks that can be built. Additionally, regulating promoters with multiple transcription factors can produce unexpected transcription regulation.10 These limitations are most pronounced in eukaryotic systems, which are necessary to study many processes pertinent to human development and disease. We offer a solution to these limitations here, using our previously reported dexamethasone–methotrexate (Dex-Mtx) yeast three-hybrid system,11,12† by showing that chemical complementation can be used to create transcription factor logic gates.
In the yeast three-hybrid system, depicted in Fig. 1a, a DNA-binding domain (DBD) and an activation domain (AD) of a transcriptional activator are genetically separated and fused to two receptor proteins that bind respective ligands with high affinity. A heterodimeric small molecule designed to bind the two receptor proteins effectively dimerizes the DBD and AD, reconstituting the transcriptional activator and activating transcription of a downstream reporter gene. This system builds on previous work on n-hybrid systems and chemical dimerizers.13–16 For this study, a B42–glucocorticoid receptor chimera (B42–GR) was used as the AD, a LexA–dihydrofolate reductase chimera (LexA-DHFR) as the DBD, Dex-Mtx11 and dexamethasone–trimethoprim17 (Dex-Tmp) as the chemical inducers of dimerization (CIDs) and lacZ as the transcription reporter. The chimeras were made from E. coli DHFR and a variant of the hormone-binding domain of the rat GR with two point mutations. Both chimeric proteins were placed under control of the GAL1 promoter. Both small molecules dimerize this three-hybrid system, however, Dex-Tmp has a higher KD for DHFR than does Dex-Mtx.18 Although Dex-Mtx and Dex-Tmp both dimerize this three-hybrid system, Mtx and Tmp have significantly different binding affinities for eukaryotic DHFRs and should be functional distinguishable molecules in other environments.19 Dimerization of the transcription factor can be disrupted by the presence of 10 µM Mtx without an observable decline in cell viability.11
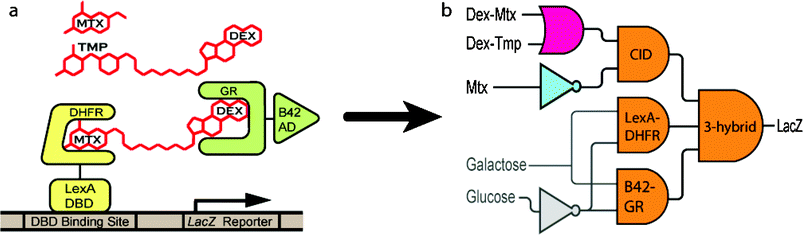 | ||
| Fig. 1 (a) The three-hybrid system. A heterodimeric ligand (Dex-Mtx or Dex-Tmp (red)) bridges a DNA bindingprotein –receptor protein chimera (LexA-DHFR (yellow)) and a transcriptional activation protein –receptor protein chimera (B42-GR (green)) effectively reconstituting a transcriptional activator and stimulating transcription of a lacZ reporter gene. Transcription can be disrupted by the small molecule Mtx (red)). (b) The three-hybrid system viewed as a three-input AND gate. LexA-DHFR and B42-GR are further regulated by the GAL1 promoter, creating a two transcription step circuit. AND, NOT and OR gates directly involved in the three-hybrid logic gate are shown in orange, blue and fuchsia, respectively. Inputs regulating production of three-hybrid components are shown in gray. | ||
This three-hybrid system behaves as a three-input Boolean AND gate with LexA-DHFR, B42-GR and Dex-Mtx and/or Dex-Tmp as the inputs. Regulation of the CID is achieved by its presence or absence from the media. To regulate the DBD and AD, we placed both under control of the GAL1 promoter, creating the two transcription step circuit depicted in Fig. 1b. We evaluate the three-hybrid logic gate in the context of this circuit. Note the GAL1 promoter is only active in the presence of galactose and strongly repressed in the presence of glucose.20 This circuit is capable of processing five bits of information: the presence or absence of glucose, galactose, Dex-Mtx, Dex-Tmp and Mtx in the cellular environment. The 32 entry truth table for this circuit is shown in Fig. 2. Only three combinations of inputs, shown in bold in the table, should result in lacZ transcription. The circuit corresponds to the logical expression ((Dex-Mtx OR Dex-Tmp) AND (NOT Mtx)) AND (Gal AND (NOT Glu)).
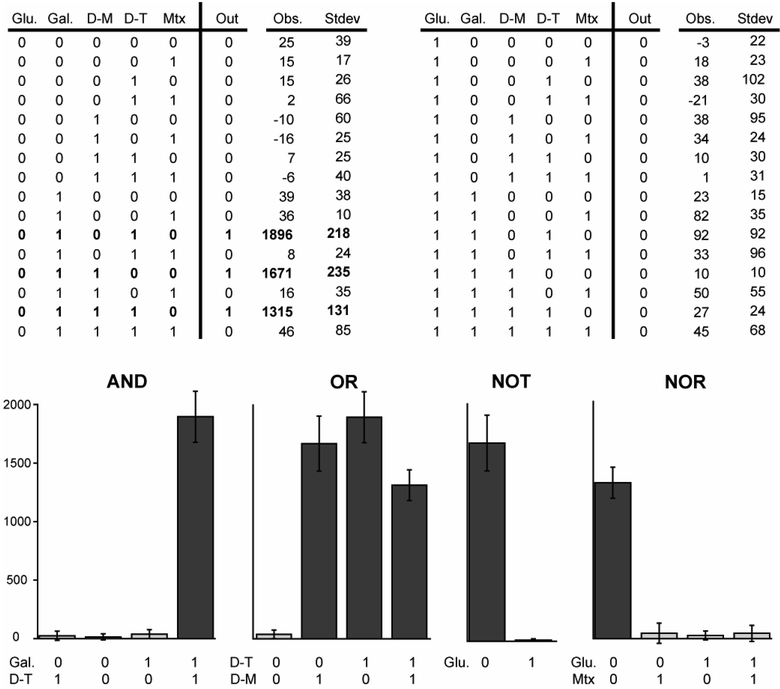 | ||
| Fig. 2 (Top) The 32 entry truth table for the three-hybrid genetic circuit. This circuit obeys the logical expression ((Dex-Mtx OR Dex-Tmp) AND (NOT Mtx)) AND (Gal AND (NOT Glu)). The table was split in two for formatting purposes only. A 0 indicates the absence of the input from the media and a 1 indicates the presence of the input in the media. Combinations of inputs that produce a transcription output are shown in bold. The observed outputs (in Miller units), averaged over four trials, and standard deviations of the measurements are shown to the right of the expected outputs. Inputs (left to right) are: 2% glucose, 2% galactose, 1 µM Dex-Mtx, 10 µM Dex-Tmp and 10 µM Mtx. (Bottom) Graphs demonstrating that when several inputs are held constant, the three-hybrid circuit reduces to simpler one and two bit logic functions. On states are shown in dark gray and off state are shown in light gray. Error bars show standard deviation. Glucose, Dex-Mtx and Mtx were all set to 0 for the AND gate. Glucose and Mtx were set to 0 and galactose was set to 1 for the OR gate. Dex-Tmp and Mtx were set to 0 and galactose and Dex-Mtx were set to 1 for the NOT gate. Galactose, Dex-Mtx and Dex-Tmp were set to 1 for the NOR gate. | ||
The ability of this circuit to perform the expected logical operations was assessed by growing cells containing the circuit in synthetic complete media with 2% raffinose and all 32 combinations of the inputs. Transcription of lacZ was determined using a standard ONPG hydrolysis assay.20 Each condition was tested in quadruplicate. The averaged values and standard deviations are shown in Fig. 2. As expected, all combinations of inputs expected to produce a logical 0 showed activity on the order of 100 or 101 Miller units. All combinations of inputs expected to produce a logical 1 showed activity on the order of 103 Miller units. When both Dex-Mtx and Dex-Tmp are present, the circuit shows a slightly weaker output than it does in the presence of just one or the other, however. This is likely due to the inhibitory effect of high concentrations of chemical dimerizers on the three-hybrid system.11 These results show the circuit behaves as predicted and the on states and off states are easily distinguishable.
If simpler logic gates are desired, this three-hybrid circuit can be converted to an AND, OR, NOT or NOR logic gate by holding several of the inputs constant. AND logic is created when glucose and Mtx are off and galactose and either CID are used as inputs. OR logic is created when glucose and Mtx are off, galactose is on and both CIDs are used as inputs. NOR logic is created when galactose and either CID is on and glucose and Mtx are used as inputs. NOT logic is created when galactose is on, Dex-Mtx or Dex-Tmp is on and either glucose or Mtx is used as the input. The outputs of several of these logic gates are shown in Fig. 2. YES logic (small molecule or protein inducible transcription) may be produced several ways as well.
These results demonstrate that chemical complementation can be used to create multiple input transcription factor logic gates. Both complex circuits and simple one or two bit logic gates can be created. The on states and off states of our genetic circuit behaved robustly and with the expected logics. Although not shown here, increasing levels of logical sophistication can be added by having the cell enzymatically modify the chemical inducer of dimerization or by having multiple three-hybrid systems with different DBD–ligand receptor small molecule pairs or different AD–ligand receptor small molecule pairs.13 Since transcription factors based on chemical complementation are created using known receptor–small molecule pairs and protein chimeras that do not require allosteric interactions, it is possible to rapidly generate new, modular transcription factors. The transcription output of one gate can be an input for another, so chemical complementation logic gates are easily connected to each other. All of these features suggest chemical complementation is a useful platform to build artificial transcription factor networks in yeast.
As it becomes possible to create larger transcription factor networks, more complicated cellular decision making will be possible as well. For example, it might be desirable to create a yeast strain that could monitor the conditions in a fermentor, determine whether they were more favorable for producing ethanol or glycerol and turn on/off the appropriate biosynthetic pathways. It would not be possible to make such a strain without creating a sophisticated genetic circuit inside it. Moving forward, we plan to construct three-hybrid NAND gates and characterize our current system in greater depth to further enhance the utility of three-hybrid transcription factors.
Acknowledgements
We thank Ron Weiss and Milan Stojanovic for their helpful comments while preparing this manuscript. We thank Hening Lin for the synthesis of Dex-Mtx and Sarah Gallagher for the synthesis of Dex-Tmp. This research was supported by the NIH (GM62867-01A1) and NSF (CHE 99-84928).Notes and references
- M. T. Chen and R. Weiss, Nat. Biotechnol., 2005, 23, 1551–1555 CrossRef CAS.
- M. B. Elowitz and S. Leibler, Nature, 2000, 403, 335–338 CrossRef CAS.
- T. S. Gardner, C. R. Cantor and J. J. Collins, Nature, 2000, 403, 339–342 CrossRef CAS.
- L. C. You, R. S. Cox, R. Weiss and F. H. Arnold, Nature, 2004, 428, 868–871 CrossRef CAS.
- J. Beekwilder, R. Wolswinkel, H. Jonker, R. Hall, C. H. R. de Vos and A. Bovy, Appl. Environ. Microbiol., 2006, 72, 5670–5672 CrossRef CAS.
- D. K. Ro, E. M. Paradise, M. Ouellet, K. J. Fisher, K. L. Newman, J. M. Ndungu, K. A. Ho, R. A. Eachus, T. S. Ham, J. Kirby, M. C. Y. Chang, S. T. Withers, Y. Shiba, R. Sarpong and J. D. Keasling, Nature, 2006, 440, 940–943 CrossRef CAS.
- J. C. Anderson, E. J. Clarke, A. P. Arkin and C. A. Voigt, J. Mol. Biol., 2006, 355, 619–627 CrossRef CAS.
- J. Hasty, D. McMillen and J. J. Collins, Nature, 2002, 420, 224–230 CrossRef CAS.
- M. Kaern, W. J. Blake and J. J. Collins, Annu. Rev. Biomed. Eng., 2003, 5, 179–206 CrossRef CAS.
- Y. Setty, A. E. Mayo, M. G. Surette and U. Alon, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 7702–7707 CrossRef CAS.
- H. N. Lin, W. M. Abida, R. T. Sauer and V. W. Cornish, J. Am. Chem. Soc., 2000, 122, 4247–4248 CrossRef CAS.
- K. Baker, C. Bleczinski, H. N. Lin, G. Salazar-Jimenez, D. Sengupta, S. Krane and V. W. Cornish, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16537–16542 CrossRef CAS.
- Directed Molecular Evolution of Proteins: or How to Improve Enzymes for Biocatalysis, ed. S. Brakmann and K. Johnsson, Wiley-VCH, Weinheim, 2002, pp. 127–158 Search PubMed.
- S. Fields and O. K. Song, Nature, 1989, 340, 245–246 CrossRef CAS.
- E. J. Licitra and J. O. Liu, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 12817–12821 CrossRef.
- D. M. Spencer, T. J. Wandless, S. L. Schreiber and G. R. Crabtree, Science, 1993, 262, 1019–1024 CAS.
- S. S. Gallagher, L. W. Miller and V. W. Cornish, Anal. Biochem., 2007, 363, 160–162 CrossRef CAS.
- S. J. Benkovic, C. A. Fierke and A. M. Naylor, Science, 1988, 239, 1105–1110 CrossRef CAS.
- D. P. Baccanari, S. Daluge and R. W. King, Biochemistry, 1982, 21, 5068–5075 CrossRef CAS.
- A. Adams, D. E. Gottschling, C. A. Kaiser and T. Stearns, Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Plainview, 1998, pp. 123–125 Search PubMed.
- K. Baker, D. Sengupta, G. Salazar-Jimenez and V. W. Cornish, Anal. Biochem., 2003, 315, 134–137 CrossRef CAS.
Footnote |
| † Standard protocols for yeast genetics were used.20 Synthetic defined media were purchased from Qbiogene. ONPG, amino acids, D-raffinose and D-galactose were purchased from Sigma-Aldrich. D-Glucose was purchased from Mallinckrodt Chemicals. Yeast was grown in U-bottomed 96-well plates (VWR) while shaking at 200 rpm in a 30 degree incubator for two days before taking measurements. Spectroscopic measurements were taken with a SpectraMaxPlus 384 spectrophotometer (Molecular Devices). The yeast strain used in this study was the V781Y strain previously described by Baker et al.21 It contains Pgal1-LexA-eDHFR and 8lexAop-lacZ integrated into the chromosome at the ade4 and ura3 loci, respectively, as well as a 2µ plasmid containing Pgal1-B42-(GSG)2-rGR2 and a tryptophan auxotrophy marker. Synthesis of Dex-Mtx is described by Lin et al.11 Synthesis of Dex-Tmp is described in Gallagher et al.17 |
| This journal is © The Royal Society of Chemistry 2008 |
