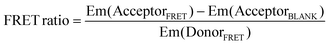Open Access Article
Open Access ArticleFRET probe for detecting two mutations in one EGFR mRNA†
Myat
Thu
a,
Kouta
Yanai
a,
Hajime
Shigeto
 b,
Shohei
Yamamura
b,
Kazunori
Watanabe
a and
Takashi
Ohtsuki
b,
Shohei
Yamamura
b,
Kazunori
Watanabe
a and
Takashi
Ohtsuki
 *a
*a
aDepartment of Interdisciplinary Science and Engineering in Health Systems, Okayama University, Okayama, Japan. E-mail: ohtsuk@okayama-u.ac.jp; Tel: +81-86-251-8218
bHealth and Medical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), 2217-14 Hayashi-cho, Takamatsu, Kagawa 761-0395, Japan
First published on 11th May 2023
Abstract
Technologies for visualizing and tracking RNA are essential in molecular biology, including in disease-related fields. In this study, we propose a novel probe set (DAt-probe and T-probe) that simultaneously detects two mutations in the same RNA using fluorescence resonance energy transfer (FRET). The DAt-probe carrying the fluorophore Atto488 and the quencher Dabcyl were used to detect a cancer mutation (exon19del), and the T-probe carrying the fluorophore Tamra was used to detect drug resistance mutations (T790M) in epidermal growth factor receptor (EGFR) mRNA. These probes were designed to induce FRET when both mutations were present in the mRNA. Gel electrophoresis confirmed that the two probes could efficiently bind to the mutant mRNA. We measured the FRET ratios using wild-type and double-mutant RNAs and found a significant difference between them. Even in living cells, the FRET probe could visualize mutant RNA. As a result, we conclude that this probe set provides a method for detecting two mutations in the single EGFR mRNA via FRET.
Introduction
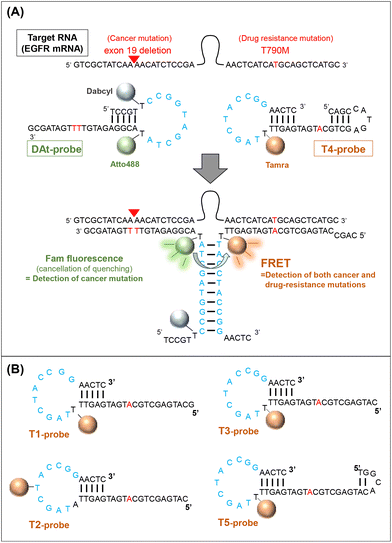
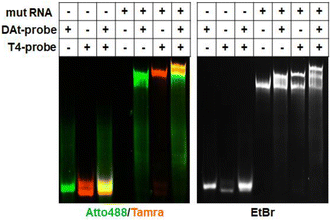
RNAs play various roles associated with their intracellular transport and localization.1–3 Additionally, they are critically involved in regulatory processes in disease occurrence and progression; they are used as diagnostic markers and treatment targets, particularly in cancers.4,5 As a result, RNA detection and imaging have become indispensable technologies in molecular biology, including in disease-related fields. In recent years, many methods for RNA detection and quantification have emerged,6–8 including polymerase chain reaction (PCR)-based methods9,10 that use cell extracts; however, these methods cannot be applied to living cells. Methods applicable to living cells include those using fluorescent probes, such as fluorescent proteins,11–13 RNA aptamers,14,15 and molecular beacons,16–18 which bind to target RNAs. These detection methods are widely used in biological research and medical diagnosis,19,20 but further methodological considerations are required to cope with different cases of medical diagnosis.Many studies have focused on detecting single mutations associated with specific diseases.21,22 However, double mutations have also been reported to be related to the severity and drug resistance of some diseases. For example, a double mutation in the PIK3CA gene is associated with breast cancer.23 Furthermore, many studies have mentioned double mutations in the epidermal growth factor receptor (EGFR) in lung cancer patients.24–26 Globally, the leading cause of cancer death is lung cancer.27,28 It has been reported that about 45% of lung cancer patients have an exon 19 deletion (exon19del) on EGFR mRNA.29 The exon19del mutation in EGFR is a known biomarker for non-small cell lung cancer (NSCLC)30 and this mutation is observed in 54% of patients with EGFR-mutant lung cancer.31 As a first-generation EGFR tyrosine kinase inhibitor (TKI), Gefitinib is effective in this cancer,32 in which the most common EGFR mutations sensitive to TKIs are exon19del and L858R.33 However, the T790M EGFR mutation has been found to be the most common cause of acquired resistance to first-generation TKIs,34 and it renders Gefitinib ineffective.35 In patients with advanced EGFR-mutant NSCLC who acquire resistance to EGFR tyrosine kinase inhibitors, the prevalence of T790M mutation with exon19 is approximately 50%.31,36 As a result, the simultaneous detection of the two mutations would be helpful in considering treatment strategies.
In this study, we designed a probe set (two probes) to detect double mutations in an RNA molecule using fluorescence resonance energy transfer (FRET). EGFR mRNA with a double mutation (exon19del and T790M) was selected as the target of the probe, and a 142-nt partial sequence of the mRNA containing the two mutations was used in this study. The two probes were designed to recognize the respective mutation sites on the mRNA target and generate a FRET signal only when both probes hybridize to the same target (Fig. 1). We attempted to use several FRET pair dyes (donor dyes [Fam and Atto488] and acceptor dyes [Atto590 and Tamra]) among those reported to be preferred,37 and optimized the conditions for double-mutation detection. A probe set for detecting RNA by FRET has been previously proposed,38 but our probe set is novel because it can detect two mutations located far apart in the target RNA sequence. Furthermore, such FRET probes can be used both with cell extracts and in living cells.
Experimental
Preparation of RNAs
For mutant EGFR mRNA (mut RNA) and wild-type EGFR mRNA (wt RNA), DNA templates were prepared through extension of two primers (5′-CCG GGT AAT ACG ACT CAC TAT AGT CGC TAT CAA AA CAT CTC CGA AAG CCA ACA AGG AAA TCC TCG ATG AAG CCT ACG TGA TGG CCA GCG TGG AC-3′ and 5′-GCA TGA GCT GCA TGA TGA GTT GCA CGG TGG AGG TGA GGC AGA TGC CCA GCA GGC GGC ACA CGT GGG GGT TGT CCA CGC TGG CCA TCA C-3′ for mut RNA, and 5′-CCG GGT AAT ACG ACT CAC TAT AGT CGC TAT CAA GGA ATT AAG AGA AGC AAC ATC TCC GAA AGC CAA CAA GGA AAT CCT CGA TGA AGC CTA CGT GAT GGC-3′ and 5′-GCA TGA GCT GCG TGA TGA GTT GCA CGG TGG AGG TGA GGC AGA TGC CCA GCA GGC GGC ACA CGT GGG GGT TGT CCA CGC TGG CCA TCA CGT AGG CTT CAT-3′ for wt RNA) purchased from Eurofins genomics (Japan). The DNA template was transcribed using the AmpliScribe T7-Flash Transcription Kit (Lucigen, USA), according to the manufacturer's instructions. The transcribed RNA was subjected to 7 M urea-denatured 8% polyacrylamide gel electrophoresis (PAGE) to cut out the target RNA. Keeping the gel in 2 mM ethylenediaminetetraacetic acid (EDTA) at 37 °C for 1 h, 2 h, and overnight, RNA was eluted from it. After phenol–chloroform extraction and 2-propanol precipitation, the RNA was dissolved in ultrapure water. Absorbance was measured using a spectrophotometer to determine the purity and concentration of RNA. mutRNA-100 (5′-GTC GCT ATC AAA ACA TCT CCG AAC TCA TCA TGC AGC TCA TGC-3′) lacking the 100 mer between the target sequences was purchased from JBios (Japan).Fluorescence measurements and estimation of FRET efficiencies
Two probes (200 nM each) and the RNA (400 nM, or the concentration indicated in each figure) were mixed in a buffer containing 10 mM Tris-HCl (pH 7.6), 1 mM MgCl2, 150 mM NaCl, and 20% PEG200 (Wako, Japan). The mixture was incubated at 37 °C for 60 min (or the time indicated in each figure). The fluorescence spectra of the mixtures were measured at an excitation wavelength of 475 nm using an FP-6600 spectrofluorometer (JASCO, Japan) equipped with a microcell. Fluorescence intensities were measured at emission wavelengths of 500–700 nm. The FRET ratio was defined as:39where, Em(DonorFRET) and Em(AcceptorFRET) are the fluorescence intensities of the two probe pairs and RNA at 525 nm (donor) and 580 nm (acceptor: Tamra) or 622 nm (acceptor: Atto590), respectively. Em(AcceptorBLANK) is the fluorescence intensity of the acceptor probe and RNA without a donor probe at 580 nm (Tamra) or 622 nm (Atto590).
PAGE analysis
To confirm the specificity of two probes binding to target RNAs by gel electrophoresis (Native 8% PAGE), a reaction solution (5 μL), including 400 nM of the RNA and 200 nM of the probes, was incubated for 60 min at 37 °C. Following that, 2× LS buffer (2 M Tri-HCl (pH 7.6), 500 mM EDTA, and 50% glycerol) was added to the reaction solution at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) to perform native 8% PAGE. The binding ratio of the probe to RNA was calculated using iBright 1500 (Thermo Fisher Scientific, USA), and specificity was confirmed.
1 (v/v) to perform native 8% PAGE. The binding ratio of the probe to RNA was calculated using iBright 1500 (Thermo Fisher Scientific, USA), and specificity was confirmed.
Cell culture, microinjection, and live cell imaging
HeLa cells were used for microinjection and FRET imaging experiments. HeLa cells were obtained from the RIKEN BioResource Research Center, Japan. The cells were maintained in RPMI 1640 medium (Nacalai Tesque, Japan) with 10% fetal bovine serum (Sigma, USA) and 1% antibiotic-antimycotic solution (Gibco, MD, USA) at 37 °C under an atmosphere of 5% CO2. The cells were seeded onto 35 mm glass-based dishes (glass 27ϕ; Iwaki, Japan) coated with poly-L-lysine (Merck, Germany). After 2 d, the medium was replaced with fresh medium without phenol red, and the cells were then microinjected with a mixture containing 10 μM of each of the two probes, 10 μM mut or wt RNA, and 100 μM HEPES-KOH (pH 7.2), which was prepared at 37 °C for 60 min. Microinjection experiments were performed using Femtojet 4i microinjection equipment with femtotips (Eppendorf, Germany) at an injection pressure of 50 hPa and an injection time of 0.3 s. In the microinjected cells, the fluorescence and FRET were imaged through a 60× objective lens using an FV1000 confocal microscope (Olympus, Japan). Excitation and emission wavelengths were 488 nm and 500–684 nm, respectively, for Atto488; 543 nm and 580–680 nm, respectively, for Tamra; and 488 nm and 584–684 nm, respectively, for FRET images.Results and discussion
Designing the probes
To achieve efficient probe–target binding, we designed several oligonucleotide probes with regions complementary to the mutant EGFR mRNA. We designed probe sets composed of (1) a FRET donor probe modified with the fluorophores Atto488 or Fam and (2) an acceptor probe modified with the fluorophores Tamra or Atto590. All probes were purchased from JBios (Japan) (Table 1).| Abbreviation | Sequence (from 5′ to 3′) |
|---|---|
| F-probe | TCC GTT CCG GTA GCT A (T-Fam) A CGG AGA TGT TTT GAT AGC G |
| DF-probe | TCC GT (T-Dabcyl) CCG GTA GCT A (T-Fam) A CGG AGA TGT TTT GAT AGC G |
| DAt-probe | TCC GT (T-Dabcyl) CCG GTA GCT A (T-Atto488) A CGG AGA TGT TTT GAT AGC G |
| T1-probe | GCA TGA GCT GCA TGA TGA GTT (T-Tamra) TA GCT ACC GGA ACT C |
| T2-probe | CAT GAG CTG CAT GAT GAG TTA TAG C (T-Tamra) A CCG GAA CTC |
| T3-probe | CAT GAG CTG CAT GAT GAG TT (T-Tamra) TAG CTA CCG GAA CTC |
| T4-probe | CAG CCA TGA GCT GCA TGA TGA GTT (T-Tamra) TA GCT ACC GGA ACT C |
| T5-probe | TGG CAC ATG AGC TGC ATG ATG AGT T (T-Tamra) T AGC TAC CGG AAC TC |
| At-probe | GCA TGA GCT GCA TGA TGA GTT (T-Atto590) TA GCT ACC GGT AAC TC |
Use of a quencher in the donor F-probe is necessary to detect target RNA by FRET
First, we considered the use of a quencher (dabcyl) in a probe with Fam. When the target RNA was detected by FRET from Fam (Emmax = 520 nm) to Tamra (Emmax = 580 nm) using the F-probe and T4-probe, extremely strong Fam fluorescence hindered the observation of the FRET peak at the Tamra emission wavelength (Fig. 2A and B). To reduce the fluorescence of the F-probe not bound to the target RNA, a dabcyl group was added to the F-probe to design the DF-probe, as shown in Table 1. Using the DF-probe and T1-probe, the FRET peak was clearly observed depending on the target mut RNA (Fig. 2C and D). The DF/T1-probe pair had a specificity problem in that FRET was observed to a small extent, even for wt RNA; however, the specificity was greatly increased by the improvement of the T1-probe, as described below.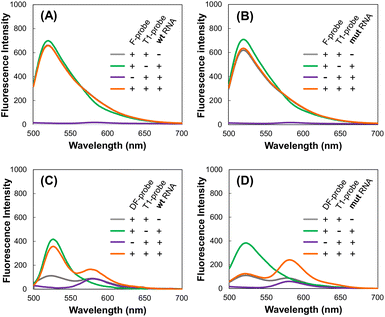 | ||
| Fig. 2 Fluorescence spectra. F- and T1-probes with wt RNA (A) and mut RNA (B). DF- and T4-probes with wt RNA (C) and mut RNA (D). | ||
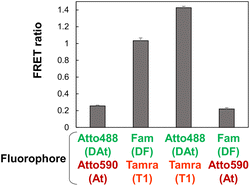
Atto488-Tamra was optimal fluorescent dye pairs
According to literature,37 Atto488 and Atto590 are considered efficient FRET pairs. In this study, the DAt-probe was designed based on the DF probe by replacing Fam with Atto488 (Table 1). The At-probe was designed based on the T1-probe by replacing the Tamra with the Atto590. The FRET efficiencies of the four combinations of donor–acceptor pairs revealed that Atto488-Tamra was the best FRET pair in this RNA detection system (Fig. 3). Unexpectedly, Atto488 and Atto590, which are considered to be an efficient FRET pair by Hirsch et al.,37 resulted in a very low FRET ratio in this study. This discrepancy in the finding could be attributed to the difference in the steric configuration of these dyes—these dyes attached to the end of the dsRNA strand in the study of Hirsch et al.,37 whereas they are attached to the thymidine bases in the dsDNA strand in the present study. The superiority of Tamra over Atto590 can be attributed to the fact that the Em(AcceptorFRET) of Tamra was significantly higher than that of Atto590, regardless of whether the donor was Fam or Atto488. The superiority of Atto488 over Fam can be attributed to the lower Em(AcceptorBLANK) of Atto488 than that of Fam.Improvement of the binding specificity of the T790M-targeting probe
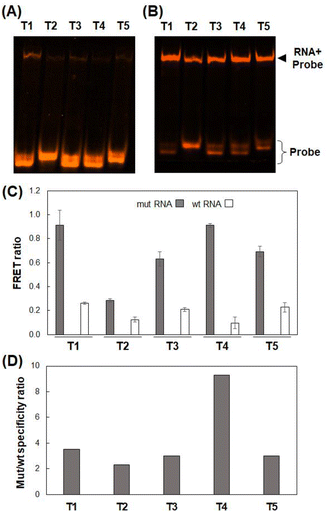
The exon19del-targeting DAt-probe bound efficiently and specifically to the target mut RNA, whereas the T790M-targeting T1-probe bound slightly to the wt RNA (Fig. 4A). As a result, we designed four other Tamra probes (T2–T5) to improve target binding/detection specificity (Fig. 1B). The target RNA binding specificity was improved, particularly with the T4-probe (Fig. 4A and B); the T4-probe bound to mut RNA to the same extent as the T1-probe but hardly bound to wt RNA. The FRET ratio values of the T2- and T4-probes for wt RNA were significantly lower than those of the T1-probe, but that of the T2-probe for mut RNA was also very low (Fig. 4C). In contrast, T4-probe can detect mut RNA by FRET, and its FRET ratio for wt RNA was reduced to 37% compared to T1-probe. As a result, the T4-probe was found to be the best among the five T probes, as indicated by the highest mutant/wild-type specificity ratio of the T4- and DAt-probe pair among five probe pairs (Fig. 4D). The superiority of the T4-probe appears to originate from its ability to weaken the binding to wt RNA (Fig. 4A), rather than from its mut RNA-binding ability, which does not differ significantly among the T1–T5 probes (Fig. 4B). The ability of the T4-probe to avoid binding to wt RNA is probably because the 4 bp stem at the 5′ end masks a part of the RNA-binding sequence, making it impossible for the probe to bind to the RNA without the T790M mutation.Examination of incubation time
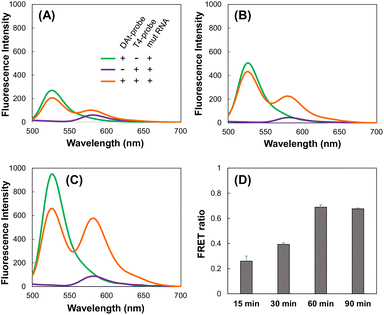
We investigated whether the DAt- and T4-probe pairs could detect the target mut RNA and the amount of time it would take to achieve sufficient detection. Using the DAt-probe alone, the Atto488 fluorescence peak at 525 nm increased depending on the incubation time, indicating the progression of the DAt-probe's binding to mut RNA (Fig. 5A–C, green lines). Using the DAt/T4-probe pair, the FRET peak at 580 nm increased with the incubation time (Fig. 5A–C, orange lines). The FRET ratio increased in a time-dependent manner and reached its maximum at 60 min (Fig. 5D), suggesting that a 60 min incubation is optimal for mut RNA detection.Target RNA detection by the selected DAt- and T4-probe pair
The DAt- and T4-probe pair successfully bound to the target mut RNA (Fig. 6), but not to the wt RNA (Fig. S1†). A significant FRET signal at 580 nm was observed when detecting mut RNA (Fig. 5C, orange line) compared with wt RNA (Fig. S2,† orange line). The FRET ratio of DAt-/T4-probes with mut RNA (0.91, Fig. 7) was much higher than those with wt RNA (0.13) and without RNA (0.13). Moreover, the FRET ratio of this probe set with mut RNA was significantly higher than that of probes with single mutants (Fig. S3†). In addition, the FRET ratio of the DAt- and T4-probes was measured with various RNA concentrations (Fig. S4†). The results demonstrated that 200 nM of this probe set could detect 50 nM of the target RNA, and a lower concentration (10 nM) of this probe set could detect 5 nM of the RNA. Although the intracellular concentration of EGFR mRNA is unknown, it could be detected with this probe set as the mRNA was detected intracellularly by another probe with a detection limit of around 5 nM.40,41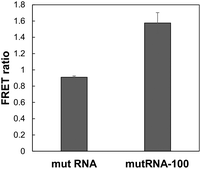 | ||
| Fig. 7 Comparison of mut RNA and mutRNA-100. FRET ratios using DAt- and T4-probes and mut RNA or mutRNA-100 were measured. The data are presented as the mean ± SEM; n = 3. | ||
To determine the effect of the distance between the two mutation sites, we examined fluorescence using DAt4- and T4-probes with mutRNA-100, which is the target RNA lacking a 100-nt sequence between the exon19del and T790M mutations. In the mutRNA-100 sequence, the DAt4- and T4-probes can bind adjacently to each other. Compared to mut RNA, in which the two mutations were far apart, FRET occurred more substantially with mutRNA-100 (Fig. 7).
The FRET ratio of DAt-/T4-probes with mutRNA-100 was 1.7 times higher than that with mut RNA. This result indicated that this probe set was more sensitive in detecting double mutations when the distance between the two mutations was small. However, the probe set can also detect two mutations that are >100 bp apart; the FRET ratio of the probe set with mut RNA was 7 times higher than that with wt RNA.
Detection of exogenously added target RNA in living cells
To investigate whether the selected probes could detect the target RNA in live cells, the two probes and RNA were microinjected into the cytoplasm of HeLa cells. Tamra fluorescence is not affected by RNA binding; therefore, it served as an injection marker, and the marker indicated that the probes and RNA were successfully injected in all experiments (Fig. 8A–C). The DAt probe becomes more fluorescent upon binding to RNA because of dequenching. The stronger fluorescence of Atto488 with mut RNA (Fig. 8A) compared to wt RNA and RNA (−) (Fig. 8B and C) indicated that the DAt probe bound to the target wt RNA in the cell. Notably, a considerably strong FRET signal was observed with mut RNA (Fig. 8A), but not with wt RNA or RNA (−) (Fig. 8B and C). These findings imply that the DAt-/T4 probe pair can detect the target double-mutant RNA in a cell.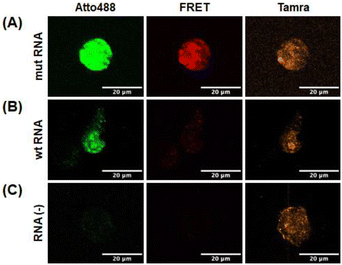 | ||
| Fig. 8 Detection of target RNA in living cells. HeLa cells were microinjected with DAt- and T4-probes with mut RNA (A), wt RNA (B), and no RNA (C). | ||
Conclusions
In this study, we developed a FRET probe (DAt-/T4 probe pair) that can simultaneously detect two mutations in one EGFR mRNA. The first designed probe pair (F and T1 probes) had issues with detection ability and specificity. However, the introduction of a quencher into the FRET donor F-probe, consideration of combinations of donor and acceptor fluorophores, and redesign of the acceptor probe have greatly improved the RNA detection ability and specificity, resulting in the development of the DAt-/T4-probe pair. The two mutations can be detected using this probe pair even if they are located far from each other (e.g., 119 bp in the case of exon19del and T790M). However, the FRET signal was more intense when the two mutations were in close proximity. For more efficient detection of the two distant mutations, a single-molecule FRET probe might be developed based on the present DAt-/T4-probe pair in the future.Herein, we presented an approach that may be adopted for other mutations, but can only be used in specific cases. The present probe can detect not only disease-related mutation (exon19del) but also drug-resistance mutation (T790M) at the same time in EGFR mRNA, and is promising for diagnosis for personalized medicine in lung cancer. In general, the intracellular function of one RNA with two mutations (mutation-1 and mutation-2) would be different from the function caused by the coexistence of the RNA with mutation-1 and that with mutation-2. Thus, the method to detect double mutations is important. This probe design is promising for the detection of double mutations in cellular mRNAs and for diagnosis based on these mutations.
Author contributions
Conceptualization: T. O., H. S., and S. Y.; formal analysis and investigation: M. T. and K. Y.; data curation: M. T. and T. O.; writing—original draft preparation: T. O. and M. T.; writing—review and editing: K. W., H. S., and S. Y.; validation and supervision: T. O. and K. W.; and project administration: T. O.Conflicts of interest
There are no conflicts to declare.References
- K. C. Martin and A. Ephrussi, Cell, 2009, 136, 719–730 CrossRef CAS PubMed.
- C. A. Pratt and K. L. Mowry, Curr. Opin. Cell Biol., 2013, 25, 99–106 CrossRef CAS PubMed.
- L. L. Chen, Trends Biochem. Sci., 2016, 41, 761–772 CrossRef CAS PubMed.
- T. A. Cooper, L. Wan and G. Dreyfuss, Cell, 2009, 136(4), 777–793 CrossRef CAS PubMed.
- H. Wang, Q. Meng, J. Qian, M. Li, C. Gu and Y. Yang, Pharmacol. Ther., 2022, 108123 CrossRef CAS PubMed.
- S. Itzkovitz and A. V. Oudenaarden, Nat. Methods, 2011, 8(Suppl 4), S12–S19 CrossRef CAS PubMed.
- Y. Xia, R. Zhang, Z. Wang, J. Tian and X. Chen, Chem. Soc. Rev., 2017, 46(10), 2824–2843 RSC.
- J. Du, R. Dartawan, W. Rice, F. Gao, J. H. Zhou and J. Sheng, Genes, 2022, 13(8), 1348 CrossRef CAS PubMed.
- C. M. Hindson, J. R. Chevillet, H. A. Briggs, E. N. Gallichotte, I. K. Ruf, B. J. Hindson, R. L. Vessella and M. Tewari, Nat. Methods, 2013, 10(10), 1003–1005 CrossRef CAS PubMed.
- M. Shen, Y. Zhou, J. Ye, A. Maskri, Y. Kang, S. Zeng and S. Cai, J. Pharm. Anal., 2020, 10, 97–101 CrossRef PubMed.
- M. Valencia-Burton, R. M. McCullough, C. R. Cantor and N. E. Broude, Nat. Methods, 2007, 4(5), 421–427 CrossRef CAS PubMed.
- T. Lionnet, K. Czaplinski, X. Darzacq, Y. Shav-Tal, A. L. Wells, J. A. Chao, H. Y. Park, V. D. Turris, M. L. Jones and R. H. Singer, Nat. Methods, 2011, 8, 165–170 CrossRef CAS PubMed.
- H. Y. Park, H. Lim, Y. J. Yoon, A. Follenzi, C. Nwokafor, M. Lopez-Jones, X. Meng and R. H. Singer, Science, 2014, 343, 422–424 CrossRef CAS PubMed.
- Z. M. Ying, H. Y. Xiao, H. Tang, R. Q. Yu and J. H. Jiang, Chem. Commun., 2018, 54, 8877–8880 RSC.
- Y. Furuhata, M. Kobayashi, R. Maruyama, Y. Sato, K. Makino, T. Michiue, H. Yui, S. Nishizawa and K. Yoshimoto, RNA, 2019, 25(5), 590–599 CrossRef CAS PubMed.
- S. Tyagi and F. R. Kramer, Nat. Biotechnol., 1996, 14, 303–308 CrossRef CAS PubMed.
- J. Li, C. Xu, N. Shimada, Y. Miyoshi, K. Watanabe, W. Cong and T. Ohtsuki, Anal. Methods, 2017, 9(20), 2971–2976 RSC.
- Y. Miyoshi, T. Ohtsuki, H. Kashida, H. Asanuma and K. Watanabe, PLoS One, 2019, 14(1), e0211505 CrossRef CAS PubMed.
- S. Zhiwei, W. Yanqiu, G. Fucheng, L. Hui, W. Chuanxin, D. Lutao, D. Lun and J. Yanyan, Acta Biomater., 2023, 155, 80–98 CrossRef PubMed.
- M. Li, F. Yin, L. Song, X. Mao, F. Li, C. Fan, X. Zuo and Q. Xia, Chem. Rev., 2021, 121(17), 10469–10558 CrossRef CAS PubMed.
- X. Liu, T. Jiang, C. Piao, X. Li, J. Guo, S. Zheng, X. Zhang, T. Cai and J. Du, Sci. Rep., 2015, 5(1), 11411 CrossRef CAS PubMed.
- M. Esteva-Socias, et al. , Front. Med., 2020, 7, 594900 CrossRef PubMed.
- N. Vasan, et al. , Science, 2019, 366(6466), 714–723 CrossRef CAS PubMed.
- C. Chakraborty, S. K. Ali and H. Zhu, Sci. Rep., 2014, 4(1), 1–7 Search PubMed.
- P. Ulivi, et al. , Clin. Lung Cancer, 2016, 17(5), 384–390 CrossRef CAS PubMed.
- J. Qin, J. Wang, X. Lin, J. Wang, Z. Xiong, R. Wang, H. Zhao and X. Kong, J. Thorac. Oncol., 2019, 14(4), e65–e68 CrossRef PubMed.
- R. S. Herbst, D. Morgensztern and C. Boshoff, Nature, 2018, 553(7689), 446–454 CrossRef CAS PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jermal and F. Bray, CA Cancer J. Clin., 2021, 71(3), 209–249 CrossRef PubMed.
- Y. Kobayashi and T. Mitsudomi, Cancer Sci., 2016, 107, 1179–1186 CrossRef CAS PubMed.
- D. M. Kim, D. H. Kim, W. Jung, K. Y. Lee and D. E. Kim, Analyst, 2016, 143(8), 1797–1804 RSC.
- Y. H. Huang, et al. , Cancer Res. Treat., 2018, 50(4), 1294–1303 CrossRef CAS PubMed.
- S. Kobayashi, T. J. Boggon, T. Dayaram, P. A. Janne, O. Kocher, M. Meyerson, B. E. Johnson, M. J. Eck, D. G. Tenen and B. Halmos, N. Engl. J. Med., 2005, 352, 786–792 CrossRef CAS PubMed.
- S. V. Sharma, D. W. Bell, J. Settleman and D. A. Haber, Nat. Rev. Cancer, 2007, 7(3), 169–181 CrossRef CAS PubMed.
- L. V. Sequist, et al. , Sci. Transl. Med., 2011, 3(75), 75ra26 Search PubMed.
- E. L. Stewart, S. Z. Tan, G. Liu and M. S. Tsao, Transl. Lung Cancer Res., 2015, 4(1), 67 CAS.
- E. E. Ke, et al. , J. Thorac. Oncol., 2017, 12(9), 1368–1375 CrossRef PubMed.
- M. Hirsch, D. Strand and M. Helm, Biol. Chem., 2012, 393(1–2), 23–35 CAS.
- P. J. Santangelo, B. Nix, A. Tsourkas and G. Bao, Nucleic Acids Res., 2004, 32(6), e57 CrossRef PubMed.
- H. Funabashi, H. Shigeto, K. Nakatsuka and A. Kuroda, Analyst, 2015, 140(4), 999–1003 RSC.
- H. Shigeto, T. Ohtsuki, A. Iizuka, Y. Akiyama and S. Yamamura, Analyst, 2019, 144, 4613 RSC.
- H. Shigeto, E. Yamada, M. Kitamatsu, T. Ohtsuki, A. Iizuka, Y. Akiyama and S. Yamamura, Micromachines, 2020, 11(7), 628 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00554b |
| This journal is © The Royal Society of Chemistry 2023 |