Multiple-decked Gd(III) complexes induced by hydrogen bonds depending on anions†‡
Kyoung-Tae
Youm
a,
Hyun Kyung
Woo
a,
Jaejung
Ko
*b and
Moo-Jin
Jun
*a
aDepartment of Chemistry, Yonsei University, Seoul, 120-749, Korea. E-mail: mjjun@yonsei.ac.kr; Fax: +82 2 364 7050; Tel: +82 2 2123 2639
bDepartments of Chemistry, Korea University, Chochiwon, Chugnam, 339-700, Korea
First published on 20th November 2006
Abstract
Hydrogen bonds assembled aquo Gd(III) moieties and btp into a discrete triple-decked complex and a 1-D propagating multiple-decked complex, depending on the anions.
Hydrogen bonds have been considered to be of importance in terms of crystal engineering to control and understand how building motifs make themselves interact or orient in the solid state.1 Hydrogen-bonded networks of coordination compounds have been extensively studied over the past years.2 When anions are incorporated in the self-assembly process they may affect the topology of the networks depending on the size and coordinating power.3 Supramolecular complexes containing lanthanide have been investigated for specific architectures by the topological control of the coordination sphere around lanthanide ions on the grounds that the lanthanide ions have a high coordination number with low stereochemical preference as compared with transition metals.4,5 The design of multifunctional ligands with a specific positional orientation is especially crucial to achieve this goal. As examples, well-designed N donor ligands have been employed to prepare the transition metal–lanthanide heterometallic complexes with specific molecular topologies.4
Here we represent how btp (=2,6-bis(N-1-1,2,4-triazolyl)pyridine)6 ligands are assembled with aquo Gd(III) moieties to be the extended architectures through noncovalent interaction (strong hydrogen bonds and π–π interaction) as well as how the subtle difference of polyatomic anions has an effect on the assembly of aquo Gd(III) moieties with btp. We have recently presented that the btp ligand could act as a good structural motif for noncovalent interaction on the grounds that it possesses not only hydrogen bond acceptors (aromatic N atoms) but also hydrogen bond donors (acidic C–H).7 Therefore, the architectures with high connectivity can occur when the btp ligands relate with aquo lanthanide motifs known as good hydrogen bond donors.5
The reaction§ of btp with Gd(NO3)3·6H2O afforded a dimeric structure induced by hydrogen bonds. The btp ligands interact with Gd(III) ions in the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mole ratio. Even if 1
1 mole ratio. Even if 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ligand-to-metal ions ratios were employed only 3
1 ligand-to-metal ions ratios were employed only 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio complex of {[Gd(btp)(H2O)4(NO3)2]·(NO3)·2btp·H2O·MeOH} (1) was obtained.
1 molar ratio complex of {[Gd(btp)(H2O)4(NO3)2]·(NO3)·2btp·H2O·MeOH} (1) was obtained.
Single crystal X-ray structure analysis¶ represents that the asymmetric unit of 1 is composed of one Gd(III), five H2O molecules, one MeOH, three nitrate ions and three btp molecules. Gd(III) ion adopts a tricapped trigonal prismatic geometry comprising of four H2O ligands and one N atom of btp.(Fig. 1) The rest are two nitrate ions chelating Gd(III) ion in a η2-mode. Only one btp ligand coordinates Gd(III) ion in a monodentate mode. The rest of btp ligands are incorporated with aquo Gd(III) motifs through hydrogen bonds. The symmetry operation on the inversion centre of 1 leads to a dimer architecture which is induced by strong hydrogen bonding (O–H⋯N and O–H⋯O).
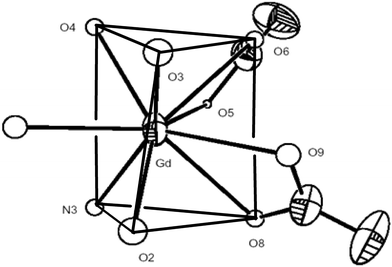 | ||
| Fig. 1 Ennea-coordinated environment around Gd ion in 1 (tricapped trigonal prismatic structure). | ||
Most interestingly, four H2O (O1, O2, O3, and O4) bound with Gd(III) ion play an important role in keeping the dimeric structure through strong hydrogen bonds. 1 has a triply decked architectures comprising six btp ligands in which two btp ligands make each deck. (Fig. 2) Two Gd(III) ions are located in the middle layer. One N atom and aquo ligand held the deck through a coordination bond and hydrogen bond (O3–H⋯N9). In addition, the remaining binding site of O3 is capped by MeOH through the hydrogen bond (O3–H⋯O14). The upper and bottom decks are held by the hydrogen bond of aquo ligand (O2, O4) with triazolyl N atoms of btp, respectively. The free nitrate anions are in the cavity, originating from the structural geometry of btp, through the hydrogen bonds with H2O ligand (O1) (Fig. 3a). Consequently, all btp ligands are stacked layer by layer within the π–π interaction distance (Fig. 3b). Detailed structural parameters of the hydrogen bonds in 1 are presented in Table 1.
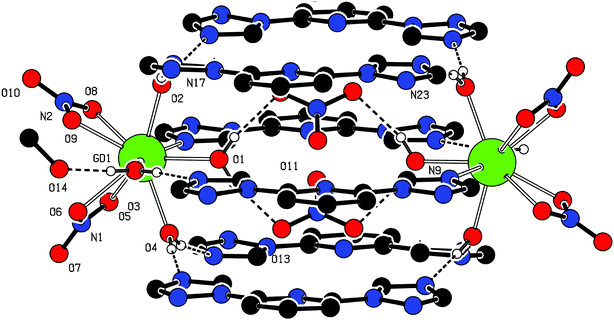 | ||
| Fig. 2 Triply decked dimeric structure of 1 induced by a hydrogen bond. | ||
![Top (a) and side (b) views of a space filling diagram of 1 presenting encapsulation of [NO3]− and a π–π stacking pattern of btp.](/image/article/2007/CE/b614511f/b614511f-f3.gif) | ||
| Fig. 3 Top (a) and side (b) views of a space filling diagram of 1 presenting encapsulation of [NO3]− and a π–π stacking pattern of btp. | ||
| Donor(D)–H⋯Acceptor(A) | H⋯A/Å | D⋯A/Å | D–H⋯A/° |
|---|---|---|---|
| a Symmetry operation codes: #1 −x + 1,−y + 1, −z + 1; #2 −x + 2, −y + 1, −z + 1; #3 x − 1, y, z. | |||
| O(1)–H(1OA)⋯O(12)#1 | 1.96(2) | 2.79(1) | 169(4) |
| O(1)–H(1OB)⋯O(13) | 2.00(5) | 2.81(6) | 162(5) |
| O(2)–H(2OA)⋯N(16)#2 | 1.94(2) | 2.76(4) | 166(4) |
| O(2)–H(2OB)⋯N(17) | 1.99(4) | 2.82(2) | 169(5) |
| O(3)–H(3OA)⋯N(9)#1 | 1.91(5) | 2.74(6) | 169(4) |
| O(3)–H(3OB)⋯O(14) | 1.77(4) | 2.61(4) | 177(5) |
| O(4)–H(4OA)⋯N(23)#1 | 1.94(2) | 2.75(8) | 163(5) |
| O(4)–H(4OB)⋯N(10)#3 | 1.95(2) | 2.78(8) | 173(5) |
TGA indicates that 1 begins losing hydrates and is completely dehydrated below 100 °C, which relates to one MeOH and four H2O ligands. Each second and third abrupt weight loss corresponds to the decomposition of one and two of three btp ligands. The anhydrous Gd(NO3)3 decomposes between 242 °C and over 500 °C. (Fig. 4) The variable-temperature magnetic susceptibility of 1 in the range 5–300 K reveals that there is no magnetic interaction between Gd(III) centres. The XMT of 7.83∼7.89 emu K mol−1 per Gd(III) of 1 is, within error, identical to the spin-only value of 7.88 emu K mol−1. These data are in agreement with the large separation distances between the neighboring magnetic centres (10.66 Å).
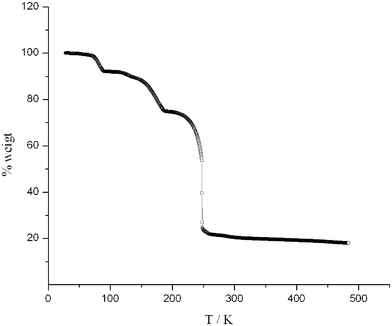 | ||
| Fig. 4 TGA analysis of 1. | ||
When Gd(NO3)3·6H2O reacted with btp with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 molar ratio in the presence of excess NaClO4 the anion-exchanged complex of {[Gd(btp)(H2O)3(MeOH)(NO3)2]·(ClO4)·2btp·MeOH} (2) was isolated.§ Single crystal X-ray structure analysis¶ reveals that the asymmetric unit of 2 is composed of one Gd(III), three H2O molecules, two MeOH, two nitrate ions, one perchlorate ion and three btp molecules. In 2, Gd(III) ion also adopts the tricapped trigonal prismatic geometry like 1, which is bounded with three H2O ligands and one MeOH ligand instead of only four H2O ligands (Fig. 5). The rest of the nine coordination bonds are completed by two nitrate anions in a η2-mode and one N atom of btp. The coordinated aquo and MeOH ligands play an important role in forming dimeric architectures (Fig. 6). Aquo ligand (O8) makes the hydrogen bond with two oxygen atoms (O11, O12) of perchlorate anion in a bifurcated mode and with another perchlorate anion (O14) related by an inversion symmetry, to participate in associating a dimeric structure. Therefore, perchlorate anions appear in the middle of the cavity of the dimer. The hydrogen bonds related with the rest of H2O and MeOH also induce the monomeric unit to create a dimer with decked structures. MeOH (O10) also makes a hydrogen bond with N atom (O10–H⋯N16). More interestingly, O1S (from MeOH) bridges the O7 and N9 atoms by consecutive hydrogen bonds (Gd1–O7–H⋯O1S–H⋯N9). And then, the last aquo ligand (O9) plays an important role in connecting these dimeric units through the O9–H⋯N17 and O9–H⋯N23. Eventually, all btp ligands are well piled up with one another in a layer by layer stacked mode with the π–π interaction, to result in a 1-D framework propagating along the z-axis (Fig. 7). In comparison with 1, Gd ions lie in different btp layers, which could be due to such different features of anions as their intrinsic size and geometries.8 Detailed structural parameters of hydrogen bonds in 2 are tabulated in Table 2.
3 molar ratio in the presence of excess NaClO4 the anion-exchanged complex of {[Gd(btp)(H2O)3(MeOH)(NO3)2]·(ClO4)·2btp·MeOH} (2) was isolated.§ Single crystal X-ray structure analysis¶ reveals that the asymmetric unit of 2 is composed of one Gd(III), three H2O molecules, two MeOH, two nitrate ions, one perchlorate ion and three btp molecules. In 2, Gd(III) ion also adopts the tricapped trigonal prismatic geometry like 1, which is bounded with three H2O ligands and one MeOH ligand instead of only four H2O ligands (Fig. 5). The rest of the nine coordination bonds are completed by two nitrate anions in a η2-mode and one N atom of btp. The coordinated aquo and MeOH ligands play an important role in forming dimeric architectures (Fig. 6). Aquo ligand (O8) makes the hydrogen bond with two oxygen atoms (O11, O12) of perchlorate anion in a bifurcated mode and with another perchlorate anion (O14) related by an inversion symmetry, to participate in associating a dimeric structure. Therefore, perchlorate anions appear in the middle of the cavity of the dimer. The hydrogen bonds related with the rest of H2O and MeOH also induce the monomeric unit to create a dimer with decked structures. MeOH (O10) also makes a hydrogen bond with N atom (O10–H⋯N16). More interestingly, O1S (from MeOH) bridges the O7 and N9 atoms by consecutive hydrogen bonds (Gd1–O7–H⋯O1S–H⋯N9). And then, the last aquo ligand (O9) plays an important role in connecting these dimeric units through the O9–H⋯N17 and O9–H⋯N23. Eventually, all btp ligands are well piled up with one another in a layer by layer stacked mode with the π–π interaction, to result in a 1-D framework propagating along the z-axis (Fig. 7). In comparison with 1, Gd ions lie in different btp layers, which could be due to such different features of anions as their intrinsic size and geometries.8 Detailed structural parameters of hydrogen bonds in 2 are tabulated in Table 2.
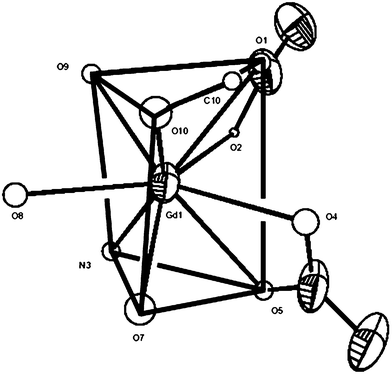 | ||
| Fig. 5 Ennea-coordinated environment around Gd ion in 2 (tricapped trigonal prismatic structure). | ||
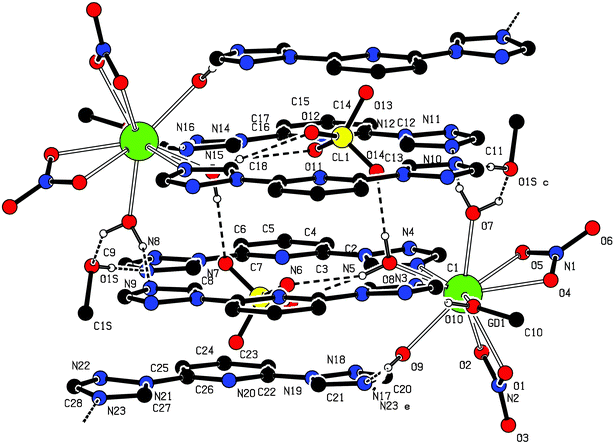 | ||
| Fig. 6 Hydrogen bonds patterns in 2. | ||
![(a) Top view presenting the encapsulation of [ClO4]− and (b) one-dimensional π–π stacking in 2.](/image/article/2007/CE/b614511f/b614511f-f7.gif) | ||
| Fig. 7 (a) Top view presenting the encapsulation of [ClO4]− and (b) one-dimensional π–π stacking in 2. | ||
| Donor(D)–H⋯Acceptor(A) | H⋯A/Å | D⋯A/Å | D–H⋯A/° |
|---|---|---|---|
| a Symmetry operation codes: #1 −x + 1,−y + 1,−z + 1; #2 −x,−y + 1,−z + 1. | |||
| O(7)–H(7OA)…N(10) | 2.02(5) | 2.74(6) | 145(7) |
| O(7)–H(7OB)…O(1S)#1 | 1.98(6) | 2.58(9) | 129(7) |
| O(8)–H(8OA)…O(14) | 1.99(5) | 2.83(5) | 178(6) |
| O(8)–H(8OB)…O(11)#1 | 2.34(3) | 3.11(3) | 154(6) |
| O(8)–H(8OB)…O(12)#1 | 2.45(4) | 3.16(7) | 144(6) |
| O(9)–H(9OA)…N(17) | 1.92(4) | 2.75(1) | 168(6) |
| O(9)–H(9OB)…N(23)#2 | 1.96(2) | 2.77(3) | 162(6) |
| O(10)–H(10)…N(16)#1 | 1.93 | 2.70(7) | 152(7) |
| O(1S)–H(1S)…N(9) | 1.94(4) | 2.77(5) | 170(9) |
We are investigating the subtle effects and roles of polyatomic anions on the self-assembly of Ln complexes with btp ligand depending on the size and negative charges.
Acknowledgement
This work was financially supported by the Korean Science and Engineering Foundation (Grant No. RO1-2001-00053).Notes and references
- For recent review see: M. D. Ward, Chem. Commun., 2005, 5838–5842 Search PubMed
; K. Biradha, CrystEngComm, 2003, 5, 374–384 RSC
; G. R. Desiraju, Chem. Commun., 2005, 2995–3001 RSC
; M. Nishio, CrystEngComm, 2004, 6, 130–158 RSC
; D. Braga, L. Maini, M. Polito and F. Grepioni, Struct. Bonding, 2004, 111, 1–32 RSC
; A. D. Burrows, Struct. Bonding, 2004, 108, 55–96 CAS
.
- L. Brammer, Chem. Soc. Rev., 2004, 33, 476–489 RSC
; A. M. Beatty, Coord. Chem. Rev., 2003, 246, 131–143 CrossRef CAS
; C. B. Aakeroy and A. M. Beatty, Aust. J. Chem., 2001, 54, 409–421 CrossRef CAS
; D. Braga and F. Grepioni, Acc. Chem. Res., 2000, 33, 601–608 CrossRef CAS
.
- B. Hasenknopf, J.-M. Lehn, G. Baum, B. Kneisel and D. Fenske, Angew. Chem., Int. Ed. Engl., 1996, 35, 1838–1840 CrossRef CAS
; B. Hasenknopf, J.-M. Lehn, N. Boumediene, A. Dupont-Gervais, A. V. Dorsselaer, B. Kneisel and D. Fenske, J. Am. Chem. Soc., 1997, 119, 10956–10962 CrossRef CAS
; R. L. Paul, Z. R. Bell, J. C. Jeffery, J. A. McCleverty and M. D. Ward, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4883–4888 CrossRef CAS
; D. A. McMorran and P. J. Steel, Angew. Chem., Int. Ed., 1998, 37, 3295–3297 CrossRef CAS
; C.-Y. Su, Y.-P. Cai, C.-L. Chen, H.-X. Zhang and B.-S. Kang, J. Chem. Soc., Dalton Trans., 2001, 359–361 RSC
; P. J. Steel and C. J. Sumby, Chem. Commun., 2002, 322–323 RSC
.
- For examples see: C. Piguet, J.-C. G. Bunzli, G. Bernardinelli, G. Hopfgartner, S. Petoud and O. Schaad, J. Am. Chem. Soc., 1996, 118, 6681–6697 Search PubMed
; C. Piguet, E. R. Minten, G. Bernardinelli, J.-C. G. Bunzli and G. Hopfgartner, J. Chem. Soc., Dalton Trans., 1997, 421–433 CrossRef CAS
; F. Renaud, C. Piguet, G. Bernardinelli, J.-C. G. Bunzli and G. Hopfgartner, J. Am. Chem. Soc., 1999, 121, 9326–9342 RSC
.
- D.-L. Long, A. J. Blake, N. R. Champness, C. Wilson and M. Schröder, Angew. Chem., Int. Ed., 2001, 40, 2444–2447 CAS
; R. J. Hill, D.-L. Long, N. R. Champness, P. Hubberstey and M. Schröder, Acc. Chem. Res., 2005, 38, 335–348 CrossRef CAS
; L. Chunha-Silva, A. Westcott, N. Whitford and M. J. Hardie, Cryst. Growth Des., 2006, 6, 726–735 CrossRef
.
- Y. Kim, S.-J. Kim and A. J. Lough, Polyhedron, 2001, 20, 3073–3078 CrossRef CAS
.
- K.-T. Youm, J. Ko and M.-J. Jun, Polyhedron, 2006, 25, 2717–2720 CrossRef CAS
.
- M. A. Withersby, A. J. Blake, N. R. Champness, P. Hubberstey, W.-S. Li and M. Schröder, Angew. Chem., Int. Ed. Engl., 1997, 36, 2327–2329 CrossRef CAS
; H. P. Wu, C. Janiak, G. Rheinwald and H. Lang, J. Chem. Soc., Dalton Trans., 1999, 183–190 RSC
; O. S. Jung, Y. J. Kim and J. K. Hong, Inorg. Chem., 2003, 42, 844–850 CrossRef CAS
.
Footnotes |
| † Caution: Perchlorate salts of metal complexes with organic ligands are potentially explosive! Only small amount of materials should be prepared and these should be handled with great care. |
| ‡ Electronic supplementary information (ESI) available: Additional figures for structural description and SQUID diagram (Fig. S1–S5). See DOI: 10.1039/b614511f |
| § Synthesis of 1: Gd(NO3)3·6H2O (0.046 g, 0.100 mmol) and btp (0.021 g, 0.100 mmol) in CH2Cl2/MeOH(20 ml/10 ml ) were stirred under nitrogen for 1 h. The solution was filtered and subsequently allowed to be evaporated slowly without a disturbance at room temperature. In three days or so colorless needle crystals appeared (0.096 g, 90%). Anal. Calc. For C29H35GdN24O14: C, 31.63; H, 3.20%; N, 30.53% Found: C, 31.05%; H, 2.84%; N, 30.81%. 2: Gd(NO3)3·6H2O (0.046 g, 0.100 mmol) and btp (0.021 g, 0.100 mmol) in CH2Cl2/MeOH(20 ml/10 ml ) were stirred under nitrogen for 1 h. The solution was filtered and subsequently added to methanol (5 mL) solution of excess NaClO4, which was allowed to evaporate slowly without disturbance at room temperature. In seven days or so colorless needle crystals appeared. (0.058 g, 51%). Anal. Calc. For C29H35ClGdN23O15: C, 30.56%; H, 3.10%; N, 28.30% Found: C, 30.19%; H, 2.84%; N, 28.77%. |
¶ Crystal data for 1: C29H35GdN24O14, M = 1101.04, triclinic, a = 10.3789(2), b = 1 5.428(3), c = 16.034(3) Å, α = 61.427(2), β = 80.926(3), γ = 72.591(3)°, V = 2151.1(6) Å3, T = 233(1) K, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (No. 2), Z = 2, μ(Mo Kα) = 1.634, 19231 reflections measured, 10017 unique (Rint = 0.0396) which were used in all calculation, R1 = 0.0461, wR2 = 0.00938 [I > 2σ(I)] and R1 = 0.0596, wR2 = 0.00993 for all data. All the hydrogen atoms were located in the calculated positions on the riding model. Nitrate anions are disordered in two sites. 2: C29H35ClGdN23O15, M = 1138.50, triclinic, a = 10.8325(6), b = 15.4222(6), c = 15.6315(7) Å, α = 109.225(3), β = 105.122(3), γ = 105.674(3)°, V = 2191.28(2) Å3, T = 150(2) K, space group P (No. 2), Z = 2, μ(Mo Kα) = 1.634, 19231 reflections measured, 10017 unique (Rint = 0.0396) which were used in all calculation, R1 = 0.0461, wR2 = 0.00938 [I > 2σ(I)] and R1 = 0.0596, wR2 = 0.00993 for all data. All the hydrogen atoms were located in the calculated positions on the riding model. Nitrate anions are disordered in two sites. 2: C29H35ClGdN23O15, M = 1138.50, triclinic, a = 10.8325(6), b = 15.4222(6), c = 15.6315(7) Å, α = 109.225(3), β = 105.122(3), γ = 105.674(3)°, V = 2191.28(2) Å3, T = 150(2) K, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (No. 2), Z = 2, μ(Mo Kα) = 1.668 mm−1, 22190 reflections measured, 9876 unique (Rint = 0.1059) which were used in all calculation, R1 = 0.0610, wR2 = 0.1323 [I > 2σ(I)] and R1 = 0.1073, wR2 = 0.1560 for all data. CCDC reference numbers 612131 for 1, 612130 for 2. For crystallographic data in CIF or other electronic format see DOI: 10.1039/b614511f (No. 2), Z = 2, μ(Mo Kα) = 1.668 mm−1, 22190 reflections measured, 9876 unique (Rint = 0.1059) which were used in all calculation, R1 = 0.0610, wR2 = 0.1323 [I > 2σ(I)] and R1 = 0.1073, wR2 = 0.1560 for all data. CCDC reference numbers 612131 for 1, 612130 for 2. For crystallographic data in CIF or other electronic format see DOI: 10.1039/b614511f |
| This journal is © The Royal Society of Chemistry 2007 |

