Inotilone and related phenylpropanoid polyketides from Inonotus sp. and their identification as potent COX and XO inhibitors
Hilaire V. Kemami Wanguna, Albert Härtla, Trinh Tam Kietb and Christian Hertweck*ac
aDept. Biomolecular Chemistry, Leibniz-Institute for Natural Products Research and Infection Biology, Beutenbergstr. 11a, 07745, Jena, Germany. E-mail: christian.hertweck@hki-jena.de; Fax: INT+3641-656705; Tel: INT+3641-656700
bCentre of Biotechnology, Vietnam National University, 144 Xuan Thuy Street, Hanoi, Vietnam
cFriedrich-Schiller-University, Jena
First published on 24th May 2006
Abstract
By bioassay-guided isolation, phenylpropanoid-derived polyketides, including an unusual 5-methyl-3(2H)-furanone derivative (inotilone) with potent cyclooxygenase (COX) and xanthone oxidase (XO) inhibitory activities were obtained from the fruiting body of the mushroom Inonotus sp.
Introduction
Arthritis is a general term for severe inflammatory processes in joints or joint tissue. Nonsteroidal anti-inflammatory drugs (NSAIDs), such as diclofenac and indomethacin, have emerged as the most commonly used anti-inflammatory agents for the therapy of rheumatoid arthritis.1 Many of these drugs target cyclooxygenases (COX), which catalyze the first two steps in the biosynthesis of the prostaglandins from the substrate arachidonic acid.2,3 In this context, the selective inhibition of enzyme subtypes, COX-1 and COX-2, has become an important goal.4 In contrast to rheumatoid arthritis, gouty arthritis is mediated by the crystallisation of uric acid (UA) in the joints.5,6 Gout can be treated with drugs that either increase the urinary excretion of UA, or with xanthine oxidase (XO) inhibitors that block the terminal step of UA biosynthesis.7,8 The purine analogue allopurinol is currently the only XO inhibitor in clinical use. Unfortunately, it seems to be associated with an infrequent but severe hypersensitivity.9 Thus, the search for new potent inhibitors of these enzymes, which could be useful as lead structures for new anti-inflammatory and anti-arthritic therapeutics, plays a pivotal role. Here we report on the isolation, structural elucidation and biological evaluation of natural anti-inflammatory COX and XO inhibitors from the mushroom Inonotus sp.Results and discussion
Extracts from the fruiting body Inonotus sp. exhibited significant inhibitory activities against key enzymes involved in inflammatory processes: 3α-HSD, COX and xanthine oxidase. Bioassay-guided separation of the combined crude ethanolic and CHCl3/MeOH extracts of the fruiting body using open column and preparative HPLC yielded several phenolic compounds 11 (4 mg), 9 (20 mg), 5 (4 mg) together with the known compounds 4 (500 mg) and 7 (6 mg) (Scheme 1).The main product from Inonotus sp. was identified as the known metabolite hispidin (4) by comparison of MS, IR and NMR data.10 In addition to 4, another compound 5 with the same molecular formula (C13H10O5) was isolated. Also the 1H NMR spectrum of 5 showed signals similar to those of 4.10 However, the 13C NMR spectrum, which showed a signal for a conjugated carbonyl at δ 179.1, clearly established 5 as the tautomeric γ-pyrone (iso-hispidin).
The molecular formula of the second main product (9) was determined as C14H14O6 based on HR-EIMS and its 13C NMR spectrum. Similar to 4 and 5, the 1H-NMR spectrum showed signals attributable to the ABX spin coupling system of a trisubstituted phenyl moiety at δ 6.77 (1H, d, J = 8.1 Hz, H-12), δ 7.02 (1H, dd, J = 8.2, 1.8 Hz, H-13), δ 7.07 (1H, d, J = 1.8 Hz H-9), a trans disubstituted double bond at δ 7.45 (1H, d, J = 15.8 Hz, H-7) and δ 6.50 (1H, d, J = 15.8 Hz, H-6), and two exchangeable phenolic hydroxyl protons at δ 9.15 and 9.65. In addition, a chelated proton at δ 15.20 was detected. Analyses of 13C, DEPT 135 and HMQC NMR spectra of 9 showed 14 carbon signals including six sp2 methines, four quaternary sp2 carbons (three of which are oxygenated), one methylene carbon at δ 45.6, a methoxy carbon at δ 51.8, a carbonyl carbon at δ 191.8, and a carboxyl carbon at δ 167.9. HMBC NMR spectra proved to be very helpful in defining their connectivities. The correlation of the H-9 (δ 7.07) with C-7 (δ 141.0), C-8 (δ 126.2), C-10 (δ 145.6), and C-11 (δ 148.4), the correlation of H-12 (δ 6.77) with H-8, H-10, H-11, and H-13 and the correlation of H-13 (δ 7.02) with C-7, C-8, C-9, C-11 and C-12, revealed an ortho substitution of the phenolic hydroxyl protons. Other important information was obtained from the observed correlation of the methylene protons (H-2) with C-1 (δ 167.9), C-3 (δ 191.8) and C-4 (δ 100.3). Structural deductions from NMR data were supported by the IR spectrum of 9, which showed absorption bands for hydroxyl groups at 3183 cm−1, a conjugated carbonyl (1632 cm−1) a carboxyl group at 1733 cm−1, and aromatic rings (1567, 1513 and 1435 cm−1). Consequently, 9 represents the methyl ester of the open chain derivative of 4 or 5, and was named inonotic acid methyl ester.
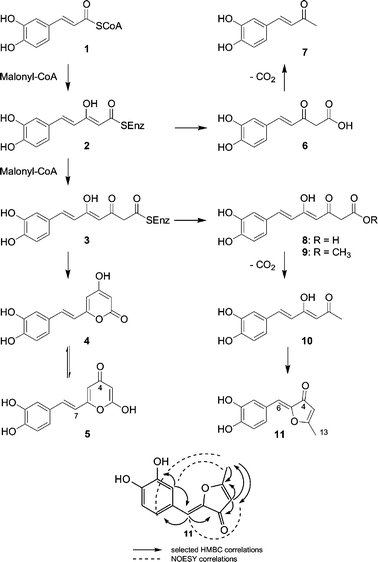 | ||
| Scheme 1 Structures of Inonotus sp. metabolites and model for their biosynthesis. Key HMBC and NOESY correlations of 11. | ||
The molecular formula of compound 11 was determined as C12H10O4 based on HR-EIMS and 13C NMR data. Similar to 4, 5 and 9, the 1H NMR spectrum of 11 showed signals attributable to the ABX spin coupling system of a trisubstituted phenyl moiety. Two olefinic protons at δ 6.49 (1H, s, H-6), δ 5.82 (1H, d, J = 0.6 Hz, H-4) and a methyl group at δ 2.39 (3H, s, H-13) were also observed. Two proton signals were attributable to the phenolic exchangeable hydroxyl protons. The 13C NMR and DEPT 135 spectra of 11 showed 11 sp2 carbon signals including five methines and five quaternary oxygenated carbons including one carbonyl. The occurrence of the carbonyl moiety was confirmed by the 13C spectrum, which showed one signal at δ 186.6. The protonated carbons and their corresponding protons and the full connection of compound 11 were established using HMQC and HMBC experiments, respectively. The correlation of the methyl proton δ 2.39 (3H, s, H-13) with C-2 (δ 180.4), and C-3 (δ 105.4), and the correlation of the olefinic proton H-3 (δ 5.82) with C-4 (carbonyl moiety) and C-5 (δ 144.3) unambiguously revealed a disubstituted dihydrofuranone moiety. The correlation of the olefinic proton H-6 (δ 6.49) with C-4 (δ 186.6), C-5 (δ 144.3), C-7 (δ 122.9), C-8 (δ 117.9) and C-12 (δ 124.7) enabled us to connect the dihydrofuranone moiety with the rest of the molecule. The configuration of the C-5 double bond was established based on molecular modeling and NOESY, which showed a correlation between H-6 (δ 6.49) and H-3 (δ 5.82) and the correlation between the protons H-8 (δ 7.35) and H-12 (δ 7.17) with the methyl protons H-13 (δ 2.39). Thus the structure was established as 2-(3,4-dihydroxybenzylidene)-5-methyfuran-3-one, named inotilone (11). Only recently, related 5-methyl-3(2H)-furanone metabolites have been reported from Phellinus igniarius.11
The structures of compounds 5, 9 and 11, as well as the isolation of the known 4 and 7 suggest that all metabolites share the same biosynthetic origin. All compounds represent linear or cyclized polyketides derived from caffeyl-CoA (1). While 7 appears to be a shunt product resulting from a premature release from the polyketide synthase, 4, 5, 9 and 11 are the result of two rounds of elongation. The structurally unusual 11 could be the product of a decarboxylation-radical ring closure sequence via the known metabolite hispolon 10.12 A related sequence could be involved in the formation of the tri- and tetrahydroxyaurone aglycones of sulfurein and cernuosides.13,14
All compounds were evaluated for their inhibitory activities in hydroxysteroid dehydrogenase (3α-HSD), COX-1, COX-2 and XO enzyme assays according to previously documented procedures. Their inhibitory potencies, expressed as IC50 values, are shown in Table 1 and are compared with those of the references, indomethacin and allopurinol. The results in the present study demonstrated that the phenolic compounds exhibit strong COX inhibitory effects with a prevalence for COX-2 in the case of the compounds 4, 7, 9 and 11. It should be highlighted that hispidin (4) and the novel inotilone (11) selectively inhibit COX-2 at concentrations as low as those of the marketed selective inhibitors meloxicam and nimesulide.3 In all cases, except for compound 11, strong 3α-HSD inhibitory effects were noted, as well as moderate inhibitory effects toward XO, except hispidin (4), which exhibited an inhibitory activity at a level comparable with that of the standard allopurinol. As far as the tautomeric compounds 4 and 5 are concerned, it seems that the α-pyrone is more active than the γ-pyrone.
| Compound | IC50/µM | ||||
|---|---|---|---|---|---|
| 3α-HSD | COX-1 | COX-2 | COX-2/COX-1 | XO | |
| 4 | 8.1 | 0.01 | 8 × 10−4 | 0.08 | 4.4 |
| 5 | 12.1 | 0.05 | 0.13 | 2.6 | 13.8 |
| 7 | 8.9 | 0.03 | 0.01 | 0.3 | 10.1 |
| 9 | 16.1 | 0.46 | 0.21 | 0.4 | 7.1 |
| 11 | 50.4 | 0.36 | 0.03 | 0.08 | 9.1 |
| Indomethacin | 15.4 | 0.10 | 6.00 | 60 | n.a. |
| Allopurinol | n.a. | n.a. | n.a. | n.a. | 4.4 |
In summary, we have isolated and characterized three new phenylpropanoid polyketides with potent COX and XO inhibitory activities from the mushroom Inonotus sp. Apart from their potent anti-arthritic activities, these metabolites represent new members of caffeyl derived polyketides, out of which the structure of inotilone is most notable.
Experimental
General experimental procedures
IR spectra (film) were recorded on a JASCO FT/IR-4100 spectrometer equipped with an ATR device. UV spectra were measured with a Spericord 200 Carl Zeiss spectrometer. High-resolution electron impact mass spectra (HR-EIMS) were recorded on an AMD 402 double-focussing mass spectrometer with BE geometry. NMR spectra were recorded on a Bruker Avance 500 DRX spectrometer at 300.133 MHz for 1H and 75.475 MHz for 13C in DMSO-d6. Chemical shifts are given in ppm relative to TMS as internal standard. HSQC and NOESY (mixing time 0.7 s) data were obtained in the phase-sensitive mode TPPI. Column chromatography was performed using silica gel (60, Merck; 0.063–0.2 µm) and Sephadex LH-20. HPLC was performed using a Gilson binary gradient HPLC system equipped with a UV detector (UV/VIS-151)(370 nm) using a preparative reverse phase C18 (7 µm) column. TLC was carried out with silica gel 60 F254 plates. Spots were visualized by spraying with vanilline/H2SO4 followed by heating. All solvents used were spectral grade or distilled prior to use.Strains
The fruiting body of Inonotus sp. was collected in Vietnam. Its identity was verified by Prof. Trinh Tam Kiet from the Mycological Research Center, Hanoi State University, Vietnam, where a specimen was deposited.Extraction and isolation
The fruiting body of Inonotus sp. (25 g dry weight) was cut into small species, dried and crushed. The resulting powder was extracted three times with ethanol (2 L) and chloroform–methanol (1 : 1) (3 × 2 L, 3 days each). The extracts were subjected to silica gel chromatography (silica gel 60, Merck, 0.063∼0.1 mm, column 4 × 60 cm), using stepwise CHCl3–MeOH (9 : 1, 8 : 2, 1 : 1 v/v) as eluent. Final purification was achieved by preparative HPLC (Spherisorb ODS-2 RP18, 5 µm (Promochem), 250 × 25 mm, acetonitrile–H2O (83 : 17 v/v), at a flow rate of 10 ml min−1 and UV detection at 372 nm). Yields: 500 mg of 4, 4 mg of 5, 6 mg of 7, 20 mg of 9, and 4 mg of 11.| N° | 5 | 9 | 11 | |||
|---|---|---|---|---|---|---|
| δ1H (J/Hz) | δ13C | δ1H (J/Hz) | δ13C | δ1H (J/Hz) | δ13C | |
| a Recorded in DMSO-d6. | ||||||
| 1 | 167.9 | |||||
| 2 | 165.4 | 3.55 s | 45.6 | 180.4 | ||
| 3 | 4.42 d (1.2) | 86.5 | 191.8 | 5.82 q | 105.5 | |
| 4 | 179.1 | 5.91 s | 100.3 | 186.6 | ||
| 5 | 5.59 d (1.2) | 109.0 | 178.3 | 144.3 | ||
| 6 | 156.1 | 6.50 d (15.8) | 118.6 | 6.49 s | 111.9 | |
| 7 | 6.12 d (15.8) | 118.5 | 7.45 d (15.8) | 141.0 | 122.9 | |
| 8 | 6.87 d (15.8) | 130.8 | 126.2 | 7.35 d (2.0) | 117.9 | |
| 9 | 127.4 | 7.07 d (1.8) | 114.7 | 145.4 | ||
| 10 | 6.94 d (1.5) | 113.5 | 145.6 | 148.1 | ||
| 11 | 145.6 | 148.4 | 6.80 d (8.2) | 115.9 | ||
| 12 | 146.5 | 6.77 d (8.1) | 115.7 | 7.17 dd (8.2, 2.0) | 124.7 | |
| 13 | 6.70 d (8.1) | 115.7 | 7.02 dd (8.1,1.8) | 121.5 | 2.39 s | 15.67 |
| 14 | 6.82 dd (8.1,1.5) | 119.2 | ||||
| 1′ | 3.65 s | 51.8 | ||||
Biological assays
The 3α-hydroxy steroid dehydrogenase assay (3-αHSD) was measured spectrophotometrically, and conducted according to the method described by Penning.15 The inhibitory activities of the test compounds are indicated in terms of IC50. Indomethacin was used as reference.The peroxidative activity of cyclooxygenases I and II was measured using luminol as a specific chemiluminescent substrate according to the method described by Forghani et al.16 The inhibitory activities of the test compounds are given in terms of IC50. Indomethacin was used as reference.
The xanthine oxidase activity was measured using lucigenin as the chemiluminescence substrate, and conducted according to the method described by Pierce et al.17 The inhibitory activities of the test compounds are indicated in terms of IC50. Allopurinol was used as the reference.
Acknowledgements
This project has been financially supported by the European Community in the FP5 EUKETIDES programme. We thank Mrs Schwinger and Mrs Röhrig for their excellent assistance in isolation and assays.References
- S. I. Rennard, Proc. Am. Thorac. Soc., 2004, 1, 282 Search PubMed.
- J. R. Vane and R. M. Botting, Inflammation Res., 1998, 47(Suppl 2), 78 Search PubMed.
- J. R. Vane, Y. S. Bakhle and R. M. Botting, Annu. Rev. Pharmacol. Toxicol., 1998, 38, 97 CrossRef CAS.
- D. L. Simmons, R. M. Botting and T. Hla, Pharmacol. Rev., 2004, 56, 387 Search PubMed.
- N. Dalbeth and D. O. Haskard, Rheumatology (Oxford, U. K.), 2005, 44, 1090 Search PubMed.
- H. K. Choi, D. B. Mount and A. M. Reginato, Ann. Intern. Med., 2005, 143, 499 Search PubMed.
- G. Rastelli, L. Costantino and A. Albasini, J. Am. Chem. Soc., 1997, 119, 3007 CrossRef CAS.
- S. Ishibuchi, H. Morimoto, T. Oe, T. Ikebe, H. Inoue, A. Fukunari, M. Kamezawa, I. Yamada and Y. Naka, Bioorg. Med. Chem. Lett., 2001, 11, 879 CrossRef CAS.
- K. R. Hande, R. M. Noone and W. J. Stone, Am. J. Med., 1984, 76, 47 CrossRef CAS.
- L. R. Brady and R. G. Benedict, J. Pharm. Sci., 1972, 61, 318 CrossRef CAS.
- S. Mo, S. Wang, G. Zhou, Y. Yang, Y. Li, X. Chen and J. Shi, J. Nat. Prod., 2004, 67, 823 CrossRef CAS.
- A. A. N. Ali, R. Jansen, H. Pilgrim, K. Liberra and U. Lindequist, Phytochemistry, 1996, 41, 927 CrossRef CAS.
- M. Shimokoriyama and S. Hattori, J. Am. Chem. Soc., 1953, 75, 1900 CrossRef CAS.
- M. K. Seikel and T. A. Geissman, J. Am. Chem. Soc., 1950, 72, 5725 CrossRef CAS.
- T. M. Penning, J. Pharm. Sci., 1985, 74, 651 CrossRef CAS.
- F. Forghani, M. Ouellet, S. Keen, M. D. Percival and P. Tagari, Anal. Biochem., 1998, 264, 216 CrossRef CAS.
- L. A. Pierce, W. O. Tarnow-Mordi and I. A. Cree, Int. J. Clin. Lab. Res., 1995, 25, 93 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2006 |
