Origins of life and biochemistry under high-pressure conditions
Isabelle
Daniel
a,
Philippe
Oger
b and
Roland
Winter
c
aLaboratoire de Sciences de la Terre, UMR 5570 CNRS – UCB Lyon1 – ENS Lyon, Bât. Géode, 2 rue Raphael Dubois, F-69622 Villeurbanne cedex, France. E-mail: isabelle.Daniel@univ-lyon1.fr
bLaboratoire de Sciences de la Terre, UMR 5570 CNRS – UCB Lyon1 – ENS Lyon, 46 allée d'Italie, F-69364 Lyon cedex 07, France. E-mail: philippe.oger@ens-lyon.fr
cUniversity of Dortmund, Department of Chemistry, Physical Chemistry I – Biophysical Chemistry, Otto-Hahn Str. 6, D-44227 Dortmund, Germany. E-mail: roland.winter@uni-dortmund.de
First published on 29th August 2006
Abstract
Life on Earth can be traced back to as far as 3.8 billion years (Ga) ago. The catastrophic meteoritic bombardment ended between 4.2 and 3.9 Ga ago. Therefore, if life emerged, and we know it did, it must have emerged from nothingness in less than 400 million years. The most recent scenarios of Earth accretion predict some very unstable physico-chemical conditions at the surface of Earth, which, in such a short time period, would impede the emergence of life from a proto-biotic soup. A possible alternative would be that life originated in the depth of the proto-ocean of the Hadean Earth, under high hydrostatic pressure. The large body of water would filter harmful radiation and buffer physico-chemical variations, and therefore would provide a more stable radiation-free environment for pre-biotic chemistry. After a short introduction to Earth history, the current tutorial review presents biological and physico-chemical arguments in support of high-pressure origin for life on Earth.
 Isabelle Daniel | Isabelle Daniel studied Earth Sciences at the Ecole Normale Supérieure of Lyon, where she obtained a PhD in 1995 in the field of mineralogy under extreme conditions of pressure and temperature. She spent one year at Arizona State University in the group of Prof. P. F. M. McMillan. In 1996 she was appointed assistant professor and in 2004 professor of geology at the University of Lyon 1. Her research interests include experimental measurements at high pressure and high temperature of the physical and chemical properties of materials relevant to the Earth’s interior, including minerals, fluids, and rocks. In collaboration with Philippe Oger, she has also developed an experimental program of in situ measurements on live micro-organisms at high pressure. |
 Philippe Oger | Philippe Oger received his PhD in Plant Pathology in 1995 from the University of Paris, where he determined the alteration induced by the culture of genetically modified plants on the soil bacteria and their metabolism. From there he moved to the University of Illinois at Ubrana-Champaign to study the evolution of carbon metabolism in Agrobacterium tumefaciens. He joined the CNRS and the Laboratoire de Sciences de la Terre in Lyon in 2000. His current research focuses on the influence of high hydrostatic pressure on microbial metabolism in relation to geological processes. Several analytical techniques have been developed, or adapted, for the study in situ under controlled pressures and temperatures of the metabolism of model eukaryotes (yeast and Emiliana huxleyi) and several strains of model piezophilic and piezotolerant bacteria and archaea. |
 Roland Winter | Roland Winter was born in Offenbach in 1954. He studied chemistry at the University of Karlsruhe and obtained his PhD degree in Physical Chemistry in 1982. He then joined Professsor Hensel's group at the University of Marburg as postdoctoral fellow where he worked on liquid matter under extreme conditions and where he received his habilitation and venia legendi in Physical Chemistry. After spending one year as a visiting scientist in Prof. Jonas' laboratory at the Department of Chemistry of the University of Illinois at Urbana-Champaign, where he started his work on high pressure molecular biophysics, he was appointed Professor at the University of Bochum. Since 1993 he has held a Chair of Physical Chemistry (Biophysical Chemistry) at the University of Dortmund. His research interests comprise the study of the structure, dynamics and phase behavior of model biomembranes and proteins. He also addresses pressure effects in molecular biophysics, such as pressure-induced phase transformations of lipid membranes and unfolding, denaturation and aggregation of proteins. |
1. The environmental conditions at the surface of the young Earth
A recent detailed review of the birth and of the infancy of the solar system and the Earth can be found elsewhere.1–3 The present section summarizes this story, and places the emphasis on the physical, chemical, and environmental conditions of the potential initial habitat for life.Among the four aeons defined between the formation of the Earth and present day (Fig. 1), the geological timescale involved in the early evolution of the Earth until life emerged includes the hadean and archean periods. The Hadean is ordinarily defined as that time between the formation of the solar system and early accretion of the planets (4.6–4.5 Ga) and the onset of life. Consequently, the upper limit defined as the Archean has become older as the search for geological traces of life progresses, and it depends on what is taken to constitute evidence for life. It is currently located at around 4.0 ± 0.2 Ga, on the basis of the enrichment in 13C of inorganic carbonates. Alternatively, it could be defined as the end of the period of heavy bombardment in the solar system, i.e. 3.9 Ga, which can be deduced from the impact record of other planets like Mercury or Mars, or the Moon. Then, the Archean covers the early development of life until 2.5 Ga, when the Proterozoic begins.
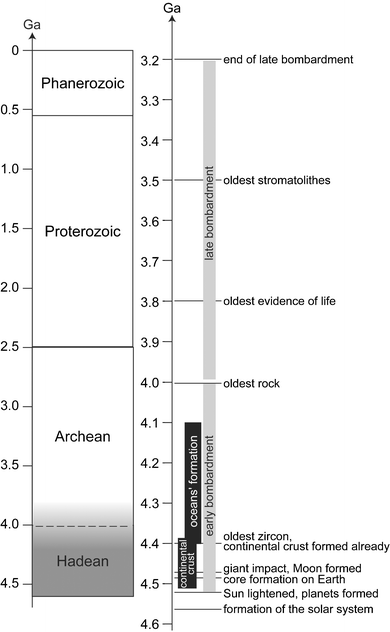 | ||
| Fig. 1 Geological timescales, including the major early events that occurred on the young Earth. 1 Ga is one billion years, 1 Ma is one million years. Geological time is divided into four aeons, as presented in the left timescale. Hadean started 4.6–4.5 Ga ago, with the formation of the Earth, until the origin of life 4.0 ± 0.2 Ga ago. As included in the name, Archean covers the early stages of life to about 2.5 Ga. Proterozoic is from to 2.5 to about 0.56 Ga. Phanerozoic is not yet finished, and started with the appearance of skeletons or shells that could be fossilized in terranes. The onset of Phanerozoic is a time of evolutionary explosion. One should be aware that almost all marine invertebrate phyla appeared at that time. The right time scale details the major geological events that affected the young Earth until mid-Archean. (modified after13) | ||
Formation of the Solar system, including the Earth
The solar system formed 4.5672 ± 0.0006 Ga ago,4 in a dense cluster of the Milky Way, after one or more recent local supernova explosions. The Sun has steadily brightened since it took its place on the Main Sequence 4.52 Ga ago, as the nuclear fire took over. During its first billion years, the Sun brightened by only ca. 3/4 of its current luminosity. The so-called faint young Sun imposes a stringent constraint on the climate of the young Earth. Without the addition of greenhouse gases the young Earth would have been at most times and places frozen over. In contrast to its overall luminosity, the active young Sun was a much stronger source of ultraviolet light, X-rays and solar wind than it is today.The planets formed roughly in the mean time the Sun spent on the pre-main sequence, ca. 40 million years (Ma). Unfortunately, no outcrop has yet been found dating from the first 500 Ma of Earth's history. The oldest ‘real’ rocks are the 4.03 Ga old Acasta gneisses in the north of Canada, while the oldest terrestrial objects are highly refractory zircons, the oldest found to date being 4.40 Ga old.5 Therefore, deducing Earth's infancy relies entirely on isotopic geochemistry of short-lived nuclides, combined with theory and comparison with other solar system objects. Thanks to the improved performance of modern mass spectrometers, the timing of the evolution of early solar system, including the Earth, has been recently revised and it generally appears that it evolved faster than previously thought.3 The accretion of terrestrial planets may be envisaged as a four-stages process, although a variety of theories have been advanced (see Chambers,6 for a recent review, and references therein). First, the circumstellar dusts settle to the middle plane of the disk, within a few thousands of years. Second, planetisimals grow up to ca. 1 to 10 km in size, by collisions occurring at relatively low speed. Third, there is a “runaway” growth of 1,000 km-sized planetary embryos. This is thought to take place within a few hundred thousand years. It is actually possible that Mercury- or Mars-sized objects could originate in this fashion. All recent data for Martian meteorites indicate that accretion and core formation on Mars were extremely fast, maybe less than 1 million years. Consequently, Mars may represent a unique example of a large primitive planetary embryo, with a different accretion history from that of the Earth. In the fourth and late stage, the larger objects like Earth or Venus grew by prolonged collisions of planetary embryos, whose orbits become more elliptic and thereby cross each other as gravitational perturbations occur. Earth and Venus should have gained most of their mass in the first ten million years, but significant accretion continued for much longer. While accretion continued, the Earth's core was formed rapidly. The global differentiation of the silicate Earth was completed 4.53 Ga ago, within ca. 30 million years of Earth's formation.7 The most widely held theory for the formation of the Moon is that there was a giant impact between a Mars-sized planet and the Earth at around 40–50 Ma, when it was approximately 90% of its current mass, but already differentiated. Such a planetary collision would have been catastrophic. The energy released would have been sufficient to increase the temperature of the Earth by thousands of degrees.
Formation of atmosphere
The formation of Earth's atmosphere is usually described as a two-step process, but might actually include several sources of volatile compounds. Compelling arguments are provided by the abundance and the isotopic composition of noble gases. A schematic scenario is currently proposed, built on the comparison between the present atmosphere and objects like meteorites or comets. However the issue is still hotly debated, and further discussion can be found in Kramers.8 A primary atmosphere was captured from the gases of the solar nebula by gravity. It was overwhelmingly hydrogen, and other volatiles were present as hydrides. Minor amounts of He, CO, N2 and H2O were also present. Such a cool reduced primary atmosphere provides a good substrate for prebiotic chemistry. The deficit in rare gas of the atmosphere at the present time, compared to cosmic abundances, suggests that the present-day atmosphere is secondary. It was degassed from within the solid Earth, after a primary atmosphere, if any, was lost. It would contain mostly CO2, H2O and H2S of volcanic origin, and lesser amounts of CH4, CO, N2 and NH3. Amongst these volatile species, water has received the most attention. The most probable source of Earth's water was ice, that could have condensed locally in planetesimals from which the bulk Earth was made, in more distant planetesimals scattered from the actual asteroid belt, or in comets for a small part of the total.Geodynamics of the proto-Earth
Jupiter's volcanically active moon Io provides an interesting analogy for the very early Hadean Earth for a few tens of millions of years after the Moon-forming impact. The heat in Io is generated by tides, and the global mean heat flow is very high. Io cools very efficiently by lava flows. When erupted, the lavas are very hot, >1600 K, suggesting an ultramafic composition, i.e. one rich in Mg. After cooling the old flows sink slowly back into the crust, which overlies a hot magmatic ocean that is not highly differentiated. On Earth, the ultramafic lavas would react with abundant water and carbon dioxide to form hydrous minerals and a large amount of molecular hydrogen and methane. As in present-day recycling, the hydrous ultramafic rocks would dehydrate and become dense enough to sink in the mantle, thereby erasing much of the evidence for this process. If Earth ever entered this state, it did not last long because the necessary high heat flows could not be sustained.Geodynamics of the Hadean Earth, formation and fate of continental crust and of the early ocean
Then, follows a period of time that is fortunately documented by zircons. The chief source of terrestrial data for the Hadean is from ancient detrital zircons found in Archean and Proterozoic quartzites in Australia (Cavosie et al.9 and references therein). The very existence of old zircons implies that there were places near the surface where zircon could be protected from subduction already 4.40 Ga ago for the oldest samples.5 Series of isotopic data on zircons suggest that continents were already a significant presence on Earth's surface before 4.4 Ga and perhaps before 4.5 Ga, only 150 Ma after the formation of the solar system. The age distribution of ancient zircons compared to modern suites indicates that recycling of the Earth's crust was approximately 10 times faster than nowadays. However, the corresponding heat flow would be only ∼3–5 times the present, and consequently the Hadean lithosphere was thick enough to display plate-like tectonics. The plates were certainly smaller than today with a mean lifetime of 20 Ma compared to 60 Ma in modern times.Oxygen isotopes measured in zircons from Jack Hills (Australia) provide convincing evidence that rocks on Earth were being chemically altered at low temperature by liquid water before 4.2 Ga.10 Although controversial, this might suggest that Earth's oceans were already in place by 4.2 Ga, or maybe already by 4.4 Ga, depending on the author. Many Archean volcanic rocks appear to have erupted under water onto a continental substrate, as illustrated at Kambalda (Australia) by pillowed basalts, that contains old zircons and geochemical signature of assimilation of old continental crust. It seems that oceans flooded much of the continental crust during the late Archean, at the time these rocks formed. The actual oceanic level is controlled by several factors, among which, the volume of the oceans, and the buoyancy of the oceanic and continental crusts play a major role. The first explanation is that the volume of the ocean was certainly greater in the Hadean than at present. It may have been up to twice that of today's ocean. Indeed, this ocean probably contained most of the terrestrial water, since the Hadean mantle was hotter and thus dryer than its modern counterpart.
The hot Hadean mantle induced both a tectonic regime and topography for the Hadean Earth that is radically different from the modern one. The convection was vigorous, and Hadean hot spots were enormous. Oceanic crust was produced by a higher degree of partial melting, and it could be about 30 km thick, i.e. 4 times the modern one (7 km), and it could be overloaded by large amounts of magmatic products from hot spots. It reacted with water at the surface (independently of whether it is vapor or liquid) and formed serpentine and H2, which would promptly transform large amounts of CO2 and N2 into CH4 and NH3, respectively. The hydration of oceanic crust triggers that of continental crust. The degassing of CH4 and NH3 is a key point for the Hadean climate and the development of life. The large thickness of the oceanic crust would lead to an ocean floor, which could be shallower than at present time. This provides a second explanation to the conjecture that much of the continental surfaces were probably flooded during the late Archean. The consequence of a thick oceanic crust is also steep subducting slabs.2
Conversely, the continental lithosphere was thin, because both the crust and the lithospheric mantle were thinner than at modern time. The Hadean continental crust was enriched in short-lived radioactive elements, and consequently less viscous. The mantle being hot, the 1200 K isotherm that defines the bottom of the lithosphere was shallow. This also supports the assumption that most of the continental surfaces could be flooded at that time.
The Hadean climate
The Hadean climate was certainly different to the modern one. After large impacts, Earth was certainly infernally hot for limited periods of time. However, the young Sun was much fainter in the Hadean ages than it is now, and the geothermal heat was probably climatologically insignificant after 4.5 Ga. If the evidence for a snowball Earth is supported by the geological record and models for the neoproterozoic period, when the Sun was 96% as bright as it is now, it must be even more plausible when the Sun emitted 25% less than its present-day brightness. The temperature at the Earth's surface was most likely governed by the actual atmospheric content in greenhouse gases, namely H2O, CO2 and CH4. Although H2O is a very efficient greenhouse gas, it is not always necessarily available in large amounts in atmosphere, since it must be mobilized from oceans or ice sheets. In the absence of other greenhouse gases, it takes ∼0.1 MPa CO2 in the atmosphere to provide enough greenhouse warming to stabilize liquid water at the surface. Although this is only about 0.5% of Earth's carbon inventory, it is 3000 times more than today's values. The amount of CO2 in the oceans depends on their pH, which was acidic at that time. Moreover, CO2 may have been massively pumped from the atmosphere by weathering reactions with both the ultramafic volcanic rocks and the impact ejecta. The final amount of atmospheric CO2 depends also on the effective decarbonation reactions within the Hadean subduction zones. If carbonate recycling was efficient, too little CO2 would have been left in the air to avoid a snowball Earth. If CO2 was liberated in subduction zones, and instilled in the atmosphere, the surface temperature might have been as high as 500 K. Methane would be a good greenhouse candidate, provided there are agents and catalysts available to generate it from CO2 and H2O. Although most methane is of biological origin today, it is also produced abiotically by alteration of serpentine minerals by carbonic acid at the bottom of the oceans. Hence, methane could help in keeping the Earth warm once life was born; but it is not yet clear whether it played a significant global role before the onset of life.11From the above discussion, it appears that the Hadean Earth could have been frozen over its surface most of the time, with ice sheets of thickness ranging from 0 up to 100 m, depending on the mean heat flow and its geographical distribution. Two cases can be envisaged: the ice blanket is thick, the sun does not get through, and the ocean is isolated from atmosphere; alternatively, the ice is thin enough that the sun penetrates, and that it can be broken by waves allowing exchanges. In both situations, the ice can be locally broken at hotspots, where intense volcanism might be present, creating warm ponds. Moreover, volcanoes always produce CO2, which will build up in the atmosphere if disconnected from oceans, ultimately leading to massive melting of the ice blanket.
Late-stage bombardment punctuated the Hadean with warm or maybe Inferno episodes. Tens of 10 to 100 km sized asteroids might statistically have hit the Earth ca. 3.9 Ga, after life had already emerged according to isotopic evidence. Impacts were still frequent as late as 3.2 Ga. Geological records, as shown by spherule beds, reveal four impact events between 3.5 and 3.2 Ga. The size of those spherule beds is large compared to the modern ones, and could they be produced by impacts big enough to boil off tens of meters of ocean water. Moreover, at least one impact of an asteroid larger than 500 km is predicted to have occurred between the formation of Earth and ∼3.8 Ga ago, leading to the complete evaporation of the oceans. Such cataclysmic events would have extirpated any life from the ocean. However, they may have favoured life in the deep regions of the Earth, at the ocean floor for instance, in environments protected from impacts. The occasional boiling of the oceans provides a useful explanation for the presence of hyperthermophilic (extreme heat-lovers) and piezophilic (pressure lover) organisms at the root of the tree of life (see below). This does not necessarily require that the first organism was a hyperthermophile or a piezophile, but it rather suggests that hyperthermophiles and piezophiles were privileged during one or several impacts. One should notice that if life emerged in hydrothermal vents on the Earth's ocean floor, life could be widespread in the solar system, wherever such hydrothermal systems are present with appropriate chemistry. Life might also have an extraterrestrial origin; it could have been brought on Earth by comets, meteorites or asteroids. Recent experiments indicate that some bacteria can survive shock pressures as high as 78 GPa.12
2. High-pressure biotopes and microbial diversity
High pressure is ubiquitous in the environment
Pressure increases with depth in both the oceans and underground, at an approximate rate of 3 MPa km−1 in the water column and 10 MPa km−1 beneath rock. The definition of “deep” or “high pressure” requires intelligent choices to be made. To encompass a zone typified for its remoteness from the productive surface waters, the deep sea is conveniently defined as water depths of 1000 m and below.13 Consequently, all environments above 10 MPa qualify for producing “high pressure” biotopes.High pressure waters encompass 88% of the volume of the oceans, that have an average depth of 3800 m, and thus achieve an average hydrostatic pressure ca. 38 MPa. The maximum depth in the trenches can reach 11,000 m (110 MPa), but the volume of seawater below the “abyssal plain” (∼6000 m) is only 0.1% of the total. In the ocean, temperature decreases with depth until an almost constant 3 °C is reached below the thermocline (30–100 m). Thus, the high-pressure ocean is cold. In the continental system, on the contrary, the average geothermal gradient is ca. 25 °C km−1. Considering the actual temperature limit for life, e.g. 121 °C would thus place the “deep” limit for the putative continental biosphere ca. 5 km below ground on average, under maximal pressures of 150 MPa. At the time of emergence of life, the limits in pressure would be even smaller, due to the higher heat flux. Even though the maximal productivity of the high pressure continental or marine biosphere is orders of magnitude lower than that of the surface biotopes, due to their extremely large volume, these high pressure biotopes contribute significantly to the production and recycling of organic carbon on Earth.14
Physical characteristics of high pressure biotopes
The deep, sub-continental biosphere is mostly uncharacterized. It is variable with depth in terms of temperature and pressure, and highly variable in composition as a function of the host rock. It lacks oxygen and light. Potential energy sources include geothermally produced reduced minerals, H2 and CH4. There is so far no evidence that the deep continental biosphere extends outside of the fluid fraction contained in the rock within cracks, fractures, and the intrinsic rock porosity. Based on a 3% average porosity of surface rock and a 5 km average thickness, Gold (1992) estimates its total volume to be ca. 1016 m3.14 Assuming an average organismal content of 1%, consistent with recent reports, the biological productivity of the continental biosphere amounts to up to 1014 tons, which exceeds the production of the surface biosphere. The recognition of the importance of the deep underground biosphere in terms of volume and biological production is fairly recent. Indeed, in 1984 Jannasch estimated the continental biosphere to be only 38 m thick!13 Until today, the microbiological data on the deep continental biosphere is extremely scarce, and has only concerned a few hundred meters in depth. Several fundamental questions regarding the origin of the deep continental biosphere microorganisms, how fast they divide or how they gain energy from the rock matrix remain largely open.The deep ocean is characterized by the lack of sunlight, a stable average temperature of ca. 3 °C, low organic carbon or mineral and a constant oxygen concentration. Theoretical settling rates of phytoplankton, as well as the flocculent aggregates of particulate matter constituting “marine snow”, range from 1.0 to 0.1 m per day, or 5000 m in 1–50 years. It is generally estimated that about 99% of the organic matter produced photosynthetically in the surface waters is recycled in the upper 100–1000 m. Only about 1% of photosynthetically produced organic carbon reaches the deep-sea floor. Thus, the major nutritional characteristic of the deep sea as a habitat is the relatively low input of organic carbon. As a corollary, adaptations to oligotrophy (life with limited amount of nutrients) and psychrophily (optimal life at low-temperature) are common.
Unearthing the biological diversity of the high-pressure biotopes
Until the late 19th century, it was commonly accepted that there was no life in the oceanic waters below 600 m, a position theorized by Edward Forbes in 1840. The capture by the Challenger expeditions (1873–1976) of live deep-sea specimen of fish and invertebrates was the first step towards the discovery of the high pressure ecosystems. However, high-pressure biology did not start until the late 1940s and the pioneering work of ZoBell and Morita, due to the technical difficulties associated with the culture of pressure-dependent organisms.13 Obligate piezophilic and psychrophilic microorganisms that cannot develop at ambient P,T conditions, were isolated and characterized, all of them bacteria. Lastly, the vision that life in the deep is characterized by its dependence upon the remote synthesis of organic carbon by photosynthetic organisms and by a low and constant temperature had to be qualified in 1977 when dense and thriving populations of invertebrates were discovered at hydrothermal vents at about 2600 m depth.15The deep-sea vents were discovered as the result of a systematic search for active volcanism at submarine spreading centers.15 In this zone, the contraction of freshly extruded lava upon cooling allows seawater to penetrate several kilometers downward into the newly formed crust. Reacting with basaltic rock at high pressures and temperatures exceeding 350 °C, the seawater is transformed into an acidic and highly reduced “hydrothermal fluid” enriched in metals, hydrogen sulfide, and molecular hydrogen. Going upwards, the hydrothermal fluid can contact sea water inside the porous basaltic rock and emerge as warm vents (3–50 °C). Alternatively, the fluid can emerge in the open ocean without prior mixing as hot vents (up to 400 °C). Upon mixing with the cold ocean seawater, the minerals present in the hot hydrothermal fluid precipitate to form chimney-like structures and a cloud-like smoke of mineral particles, hence the nickname “smokers”, around which the vent communities are spatially structured. From the time of their discovery, it was obvious that vent organisms were related to surface organisms. Similarly, from the beginning it was clear that vent organisms must obtain their energy from the hydrothermal fluids, since the organic carbon production of the vent organisms exceeded by orders of magnitude the amount of organic material that could possibly sediment from the surface.15
The diversity of bacterial and archaeal species isolated from the hydrothermal environment is large. In contrast to clones isolated from the open ocean, bacteria and archaea isolated from deep-sea hot vents are thermophiles. Most isolates are chimiolithotrophs, e.g. capable of gaining energy from the chemical transformation of dissolved minerals and to realize the fixation of dissolved carbonates into organic molecules. Many isolates are aerobes or facultative anaerobes, i.e. they use oxygen as a final electron acceptor. A growing number of obligate anaerobes have been characterized. The vent systems are also characterized by a great diversity of microscopic eukaryotes (ciliates, flagellates, nematodes, fungi, etc.), and especially predatory species that are supposed to feed on chimiosynthetic prokaryotes. Last, several vents are inhabited by large invertebrates, such as polykaetes, mussels, crabs or shrimps, which make these ecosystems crowded places, hence their nickname of deep-sea oases. Aside from their architectural resemblance to their closest non-vent cousins, the macro fauna of the vent ecosystems have a unique feeding strategy. This was characterized first in the vent clam, Clyptogena magnifica and in the vent worm Riftia pachyptila, a tube worm lacking a digestive tractus.16 Instead of feeding on smaller prey as would normally be the case, mussels and worms are fed by symbiotic, obligate anaerobic chimiolithotrophic bacteria that occupy a large portion of the animal's body. Bacteria use the energy from the hydrothermal vents fluids to fix dissolved carbon, much like their free-living counterpart. What the benefit to the bacterium would be apart from shelter is so far unclear. So, the vent ecosystems are organized very much like surface ecosystems, except that the vent ecosystem relies on prokaryotic chimiolithotrophs rather than on photosynthetic primary producers.
For several years, it was impossible to disconnect the deep chimiolithotrophic vent ecosystem from the photosynthetic surface ecosystem. Indeed, if oxygen was required as a final electron acceptor in absence of an appropriate anaerobic chimiolithotrophic prokaryotic compartment, then photosynthesis was required to produce the oxygen, and the vent ecosystem was indirectly dependent upon it. When the first obligate anaerobic archaea were isolated by Erauso et al. (1993),17 the demonstration was made that the vent ecosystem could function in the absence of oxygen, and therefore disconnected from sun light as an energy source.
Deep-sea hydrothermal vents and the origins of life
The possibility that the vent ecosystems could function independently from the surface ecosystems has several important implications.First, all ecosystems on Earth depend directly or indirectly upon light as the source of energy for the fixation of atmospheric carbon through photosynthesis. Photosynthesis is a very complex energy harvesting process, which could not have appeared at the beginning of life on Earth. Harvesting chemical energy is a simpler mechanism, and it could have appeared earlier in the evolution of life on Earth. Thus, the vent chimiolithotrophy could represent a remnant energy harvesting mechanism, providing a window on the origin of life on Earth.
Second, the similarity between the top and bottom ocean biosphere clearly indicates that organisms from both environments originate from the same lineage. Whether the hot, anoxic deep-sea environments were colonized recently by surface Bacteria, Archaea and Eucarya, or whether the cold, irradiated surface environment was colonized gradually by microbes from the deep-sea remains an open question.
Third, the physico-chemical conditions in the deep biosphere are stable for geologically long periods, and ionizing radiation is low. In contrast, on the surface radiation is high and the physico-chemical environment variable, more so on the young Earth, when life appeared. Thus, the conditions of the deep biosphere could have been more appropriate for the emergence of life, and the deep biosphere might have witnessed its emergence.18
Fourth, the possibility that life can survive and proliferate in the absence of light, and that it might have emerged under high pressure, greatly expands the possibility that life could exist, or have emerged elsewhere in the Universe. For example, it becomes reasonable to assume that life could have emerged on other celestial bodies within our solar system, such as in the deep-oceans of Europa, or that life is still present within the Martian subsurface.19
3. The LUCA lived under high pressure
What's behind the name?
The LUCA is short for Last Universal Common Ancestor. In the same way as the Darwinian theory of evolution tells us that we and chimps originate from a common ancestor some 10 millions of years ago, all modern life forms share a common history back as far as the split that gave rise to the three “domains” of life: Archaea, Bacteria and Eucarya. Indeed, this will remain one of the greatest discovery of modern biology: the three domains of life depend on the same genetic storage molecule, DNA, the same genetic transcription molecule, RNA, the same catalytic molecules, proteins, and on the same alphabet to code the genetic information to be stored in DNA, the universal genetic code (four bases, 20 codons). These facts alone are sufficient to point out, the unique nature of our ancestor, the LUCA. Thus, since the LUCA already contained nucleic acid–based genetic information and replication, it cannot be the earliest life form that roamed the Earth. Genetic information is organized in genes and transmitted vertically to siblings. Therefore, tracing the genes that are common to all life forms on Earth and tracing their evolution within and between the three kingdoms of life allows us to go back in time. By tracing what functions were already present in the LUCA we can gain information about the environment in which the LUCA used to live. By tracing what functions appeared first, and in which order they appeared can give priceless information on how life originated on Earth.The LUCA was a cellular organism
Comparative genomics and phylogenetic reconstruction enable us to compare full genome sequences, extract genes of interest and reconstruct their ancestral sequence by assuming a defined set of evolutionary hypotheses. One can identify two sets of genes: the ubiquitous gene set (genes that are present in all current life forms), and the minimal gene set (genes necessary and sufficient to sustain a functional organism). To date more than 200 complete genome sequences from the three domains of life are available.The ubiquitous gene pool comprises 70 genes,20 among which 58 encode proteins involved in the translation of RNA into proteins, e.g. ribosomal proteins, amino acyl ARNt synthetases and protein modification enzymes. Therefore, the LUCA did possess a “modern” protein synthesis mechanism, capable of elaborating sophisticated proteins. The universal protein set also comprises the SRP54 protein of the SRP (signal recognition pathway) ribonucleoproteic complex, and Srα its cognate membrane receptor. These two proteins are involved in the translocation through the plasma membrane of proteins during protein synthesis. Thus, the LUCA already had a cytoplasmic membrane, a hypothesis also supported by the presence of two membrane associated ATP synthetases in the ubiquitous gene set. The ubiquitous gene set comprises only 3 proteins involved in the processing of DNA itself, a surprising result since DNA is the genetic storage material of all cells. Mushegian and Koonin have proposed that the LUCA had a RNA-based genome, and that DNA replication was invented twice, leading to the major dichotomy between the Archaea/Eucarya and Bacteria domains.20 In contrast, Forterre proposed that the LUCA had a DNA based genome, and that the separation of the Archaea/Eucarya and Bacteria domains occurred when the host DNA replication was displaced by the simpler, more effective DNA replication of a viral particle.21 The ubiquitous gene set does not contain any metabolic gene. This result is not a surprise, since the phylogenetic studies on metabolic genes clearly demonstrate their recurrent loss and acquisition during evolution.
What did the genome of the LUCA look like?
Two separate reports have given similar estimates of the minimal number of genes that should have been present in the LUCA.22,23 This set comprises a minimum of 500 to 700 genes. If one assumes an average 1 kb size for each gene, the LUCA had a 500 to 700 kb genome. The LUCA would not be photosynthetic, but it could use several organic or inorganic compounds as source of energy. The LUCA appears to have been a complete cell, with a well-established membrane system, including sets of transmembrane transporters/channels for minerals or sugars, as well as several ATPases. The presence of a superoxide dismutase, that is an enzyme involved in the protection of cells from the damages induced by molecular oxygen, suggests that the LUCA could have been an aerobe. Several arguments favor the DNA-LUCA hypothesis. The minimal gene set comprises a variety of DNA replication/repair/recombination enzymes that would suffice for the processing and transmission of the genetic information. The size of the LUCA genome, e.g. 500–700 kb, exceeds the calculated maximum size for an RNA-based genome, based on the fidelity of RNA polymerases. Thus, it appears that our LUCA looked much alike to modern cells. Unfortunately, in the absence of an appropriate fossil record, comparative genomics and phylogenetic studies cannot answer the question of the time at which the three domains of life diverged from the LUCA.Where is the root to the tree of life?
This question, although seemingly insignificant, has generated a heated debate over the last two decades. Carl Woese, who discovered the Archaea, has long considered that the nature of the LUCA is one of the most important problems in biology, since determining who went first in evolution can give clues about what life looked like at the origin. For a long time, it seemed a hopeless quest to reconstruct the early evolution of life, considering the very scarce fossil record available, and the technical difficulties in phylogenetic reconstruction to the origins. However, for the last three decades the prevalent view has been that we have a clear-cut vision of the most ancient history. The topology of the universal tree of life was deduced from the universally conserved ribosomal rDNA gene (Fig. 2a), which depicts a division of the living world into three domains, Archaea, Bacteria, and Eucarya, and a tree root placed between the two prokaryotic domains.24 In this scheme, life arose through a stepwise complexification to the LUCA and then to the eukaryotes, and higher animals. Furthermore, since the ability to grow at high temperature was found in all deep-rooted branches of the tree of life it became clear that the LUCA used to live in, or was derived from other organisms living in a high temperature environment.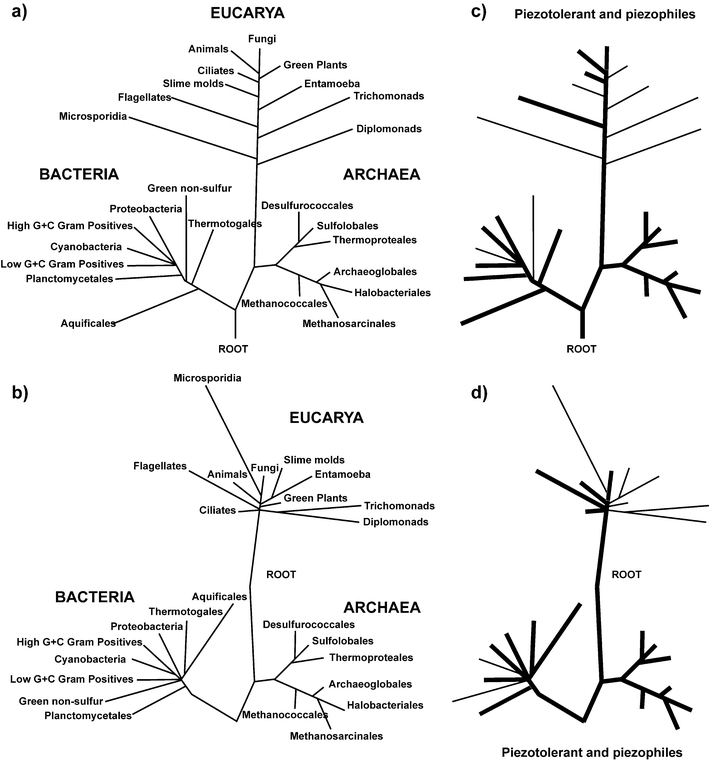 | ||
| Fig. 2 Piezophily in the tree of life. (a) The classical view of the tree of life. The topology of the tree is mainly based on rDNA comparison. (b) A revised topology of the universal tree of life, after correction of phylogenetic pitfalls, such as long branch attraction or the removal of uninformative site from the analysis. (c) and (d) Distribution of piezophilic and piezotolerant organisms in the two trees of life. Thick blacks lines highlight bacterial groups in which piezotolerant and piezophile organisms have been characterized. Thin black lines highlight groups for which only piezotolerant species are known. | ||
However, this consensus view was challenged by complete genome comparison that showed that many protein phylogenies contradict the universal tree of life, in that each domain was a mosaic of the two others in terms of gene contents, and that eukaryotes contained more bacterial genes than archaeal ones, and that archaea contained more bacterial than eukaryotic ones. The novel topologies of the universal tree of life (Fig. 2b) give less support to the high temperature LUCA, and they suggest a possible root between the Archaea and the Eucarya.25
Was the LUCA simple or complex?
As mentioned above, the LUCA already contained a large genome, and a variety of complex metabolic pathways. The LUCA was not simple in itself. The debate remains between those who consider present bacterial mechanisms as streamlined versions of a more complex ancestral one (eukaryotic-like) or as relics of ancestral systems.Both complexification and simplification have occurred during evolution. For example, the evolution from the origin of life to the LUCA obviously must have been from simple (pre-biotic) to complex (DNA/RNA-based cellular organization), whereas the evolution from Gram negative bacteria to mitochondria and chloroplasts is one example of evolution from complex to simple. What happened in the case of the eukaryote/prokaryote transition?
Carl Woese has always argued that the LUCA must be primitive, a progenote, from which all life forms have evolved through stepwise complexification. The counter-intuitive hypothesis of a eukaryotic-like LUCA was proposed by Reanney who considers many RNA molecules typical of eukaryotes to be relics of the RNA world and ought thus to have been present in the LUCA.26 Two additional arguments can be put forward to support the “complex to simple” scenario in which a eukaryotic-like LUCA evolved by simplification to give birth to present-day prokaryotes. First, reductive evolution of central molecular mechanisms still occurs in bacteria. Second, most cellular functions require multiple enzymes physically interacting with one another. It is very difficult to imagine the displacement of a single component by several others simultaneously. In contrast, a replacement of several components by a single one is much easier, if the latter can perform the same task with similar or better efficiency. The occurrence of such events is shown by the well documented displacement of the original proteobacterial RNA polymerase (3 subunits) by a bacteriophage-like RNA polymerase (1 subunit) in the evolution of mitochondria and chloroplasts.
Was the LUCA a piezophile and thermophile?
Regardless of whether the LUCA had a genome organization similar to eukaryotes or prokaryotes, can we infer the growth environment of the LUCA from the different topologies of the universal trees of life and from the minimal gene set ? Thermophily and piezophily are widespread characters in bacterial and archaeal phyla (Fig. 2c and 2d). In addition, in contrast to thermophily, piezophily is well represented in excretal phyla. Several species of animals (various vertebrates and invertebrates), as well as numerous microscopic eukaryotes from the ciliates, flagelates or fungal families inhabit the deep-seas and the hydrothermal vent environments, although no piezophilic members have been reported yet for the photosynthetic phyla. This is not unexpected since high-pressure environments lack light. In addition, the most deeply branching prokaryotes and eukaryotes in the tree of life are thermophilic and/or piezophilic, and the branches leading to extant thermophiles and piezophiles are short. From the thermophile data, Woese concluded that the LUCA was most likely a thermophile.24 Likewise, the LUCA was most likely a piezophile, and grew in a high pressure environment. Although the issue of the thermophilic nature of the LUCA is still debated, its piezophilic nature should not be challenged. Piezophily is evenly distributed within the three domains of life, and therefore insensitive to changes in tree topology or changes in the position of its root. A piezophilic and thermophilic LUCA would be consistent with what is known or conjectured about the early Earth environment and the necessary stability of physico-chemical conditions for life to emerge, which would occur in the depth of the primordial ocean.18If life emerged in the deep sea, then one question remaining would be the colonization of the surface environment. The recent isolation of an obligate photosynthetic green-sulfur bacterium27 from a deep-sea hydrothermal vent might represent the missing link between the ancestral chimiolithotrophic energy harvesting metabolism which emerged in the depth of the ocean, and the photosynthetic light energy harvesting metabolism which eventually moved upward to colonize the ocean and thereafter the land surfaces.
4. Effect of pressure on metabolism
The investigation of the effect of pressure on microbial activities started in 1949, with the pioneering work of ZoBell and Johnson.28 They first defined as barophilic, microorganisms which exhibited enhanced growth at high pressure or required pressure for growth. As already noted in the introduction, the pressure of interest for live microorganisms is several orders of magnitude lower than those usually considered in Earth or material sciences. The deepest microorganisms yet isolated and characterized were sampled at 11,000 m depth or 110 MPa, in the deep-sea sediments of the Marianas trench, where the Pacific oceanic lithosphere subducts into the Earth's mantle. More recently, the prefix “baro” was replaced by “piezo”, and a distinction was made between piezotolerant and piezophile microorganisms (Fig. 3).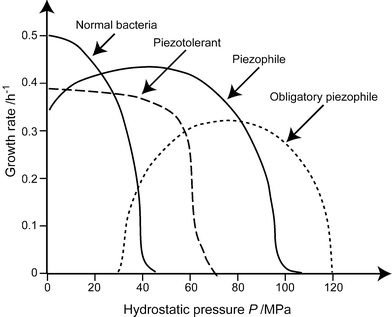 | ||
| Fig. 3 Definitions of the relations between growth rate of microorganisms and pressure. Piezophile microorganisms display a maximum growth at high pressure. They can either grow at atmospheric pressure or not, and are called strictly piezophile in the latter case. Piezotolerant microorganism grow best at atmospheric pressure, but can sustain high pressure, whereas mesophile or piezosensitive microorganisms totally stop growing at 40–50 MPa. (Redrawn after ref. 57.) | ||
A large number of piezophile microorganisms have been isolated and characterized. Pressure-regulated metabolism and gene expression have been explored in piezophiles as well as in mesophiles (lovers of temperate conditions), and in microorganisms as well as in higher eukaryotes.29 The physical and chemical effects of pressure on the major biomolecules found in cells are described in the next section of this review. However, the pressure-induced phenomena that occur in living organisms have not been systematically investigated due to their complexity.
To illustrate the effect of pressure on a microorganism, we can consider the brewing and baking yeast Saccharomyces cerevisiae, because it is the best characterized unicellular eukaryote in terms of genome characterization, and physiology. Yeast has proved to be a good eukaryotic model and evidence exists that mechanisms operating in yeast also occur in complex eukaryotes. Since 1996, the entire genome yeast has been sequenced. Gene features can be found in the Saccharomyces Genome Database (http://www.yeastgenome.org). The response of yeast to pressure has been recently reviewed30,31(Table 1). The prokaryotic counterpart to S. cerevisiae would be the mesophile model Escherichia coli, the behaviour of which at high pressure is reviewed by Welch et al.32 The characteristics of high-pressure adapted deep-sea prokaryotes belonging to Archea and Bacteria were reviewed by Bartlett.29
| Pressure/MPa | Effects |
|---|---|
| 0.1–50 MPa | Arrest of cell growth |
| Metabolic changes | |
| Inhibition of amino acid uptake | |
| Stress-inducible expression | |
| 50 MPa– | Inhibition of ethanol fermentation |
| Internal acidification | |
| Stress-inducible expression | |
| 100 MPa– | Reduction in viability |
| Membrane, and cell wall perturbations | |
| Acquired piezotolerance | |
| Stress-inducible expression | |
| 200 MPa– | Alteration of genome preservation |
| Shrinkage and leakage of cells | |
| Stress-inducible expression |
S. cerevisiae is piezotolerant. Yeast growth and cellular activity in virtually unaffected at pressures lower than 20–30 MPa depending on strain genotype. Higher pressures are however perceived as a stress in this micro-organism. Consequently, S. cerevisiae exhibits adaptation mechanisms through pressure-inducible genes and pressure-induced proteins. The major steps of yeast response to high pressure stress are summarized below.
Yeast viability decreases with increasing pressure and this effect is more pronounced above 100 MPa, until all wild type cells of yeast are killed at 220 MPa. However, a pressure of 50 MPa is neither sufficient to kill nor to alter the yeast cell morphology. Although not directly relevant to S. cerevisiae, this shows that pressures in the range of 200–500 MPa can be employed for sterilization of food. The piezotolerance of yeast cells depends on the duration of high pressure application. Although a short treatment at 50 MPa will not kill the cell, an incubation at 50 MPa for 24 h will result in 100% mortality. Piezotolerance also depends on the position in the cellular cycle. Yeast cells in stationary phase are more resistant to pressure than proliferating cells. A comparison of the yeast cell growth response to pressure stress with that to a classical heat-shock of 40 °C for 30 min, shows that pressurized cells at 50 MPa for 30 min have a slower response and also take longer to recover normal growth. This suggests that they still suffer metabolic changes after pressure is released.
The influence of pressure on gene expression has been recognised. In most cases gene expression is inhibited by pressure, but there are also specific proteins produced upon pressure-shock. Pressure has a specific effect on DNA, and shifts the double DNA helix towards a denser form. Also, protein association with DNA is less stable at high pressure, demonstrating that pressure may interfere with transcription, and alter genome expression. One issue was to determine whether the effect of pressure was mostly due to the relative instability of certain gene promoter binding or of mRNAs with pressure, or to a well defined stress response. Among the 6200 known or predicted genes of S. cerevisiae, 131 were induced more than 2-fold and 143 repressed greater than 2-fold by pressure, although the up-regulated genes are largely unknown. The genomic response of yeast to high pressure is a typical stress response. Genes involved in stress defence and carbohydrate metabolism are highly induced by pressure, while genes involved in cellular transcription, protein synthesis and cell cycle regulation are down-regulated. The stress response of yeast to pressure is similar to, but fundamentally different from, that of heat shock. Heat-shock induces a set of Heat Shock Proteins (HSPs), and activates the metabolism of trehalose (a nonreducing disaccharide), whose function is to prevent unfolding or to promote refolding of proteins, in order to keep the cell machinery working. The HSP genes induced by pressure in yeast are slightly different. For instance, high molecular weight HSPs or trehalose 6-phosphate synthetase genes are strongly induced by heat-shocks but are indifferent to pressure. In contrast, the small HSP26 gene is strongly induced by pressure. This gene codes for a small protein with a molecular chaperone activity, and like other members of the HSP family, it protects proteins from irreversible aggregation. The HSP26p complex probably dissociates under pressure, thus ensuring a chaperone activity of the protein, like at high temperature. After returning to ambient pressure, the complex probably reverts to the associated form and loses its chaperone activity. A number of yeast genes regulated by pressure also include a large set of Cold-Shock specific genes. This tends to show that yeast cells possess a mechanism to sense the stress of pressure, and to activate the appropriate gene expression machinery.
Protein synthesis is one of the most piezosensitive cellular functions, probably due to the disassembly of ribosome as a function of pressure. In contrast, RNA synthesis is maintained at higher pressures. Hence, even if the genes responsible for stress-induced proteins are up-induced, those proteins cannot be produced due to the inactivation of the protein synthesis apparatus during compression. S. cerevisiae has an improved piezo-resistance after being exposed to a mild stress, including heat-shock, cold-shock, ethanol-shock and hydrogen peroxide-shock. However, yeast cells subjected to a mild pressure do not acquire resistance to any subsequent severe pressure increase, unless they are incubated at room pressure between the two pressure treatments. This is likely related to the specific problems induced by pressure on the cells, that is reduction of membrane fluidity and impaired protein synthesis. When the stress is applied, the up-regulated genes are induced but cannot be translated. Once the cells return to ambient conditions, the biosynthesis can take place and protects the cells against further pressure increase for a relatively long period of time, compared to the duration observed for heat-shock.
Pressure changes the fluidity of the membrane in a similar way to low temperature, by enhancing the order of the phospholipid bilayers, and causing the fatty acid to pack more tightly. The fluidity of a membrane at 100 MPa and 2 °C (typical deep-sea conditions) is similar to that at atmospheric pressure and −18 °C. This is compensated by an increase in unsaturated fatty acids, which leads to highly disordered phospholipid bilayers that are less permeable to water molecules. Hence, this maintains the membrane in a functional liquid crystalline state despite the effect of pressure. The increased proportion of unsaturated fatty acids is common among deep-sea organisms. As stresses that both decrease membrane fluidity, the application of cold and pressure to yeast shows a slight induction of the ERG25 gene, whereas heat-shock down-regulates the ERG25 gene. ERG25 codes for the protein ERG25p, which is a sterol desaturase enzyme involved in ergosterol biosynthesis. Ergosterol is a sterol group with an unsaturated side chain, while cholesterol has a saturated one. Membranes enriched with ergosterol seem to be more resistant to ethanol and probably to temperature than cells enriched with cholesterol, whereas those containing cholesterol have proved to be more resistant to pressure than cholesterol-free ones.
Pressure alters the structure of the cell wall and cytoskeleton. Those effects are counterbalanced by the up-regulation by pressure of the gene HSP12, which codes for HSP12p. HSP12p is a small hydrophilic protein located in the cell wall. It improves the flexibility of the cell wall, by disrupting interactions between adjacent polysaccharide layers that would else build a rigid structure. Once more, there is a common feature between pressure and low-temperature stress: they do not induce the majority of HSPs but small HSPs (like HSP12 and HSP26) related to membrane destabilization. In particular, the larger HSPs induced by heat-shock, and related to chaperone activity that prevents protein folding, are not induced by pressure.
Another effect of pressure on yeast cell is the acidification of the cytoplasm, and of the vacuole. At atmospheric pressure, cytoplasm and vacuole have a constant pH of 7.0 and 6.0, respectively. Increasing pressure to 50 MPa decreases the cytoplasm pH by 0.3 units, and the vacuole pH by 0.3 to 0.5 units. The increased acidity is due to the increased solubility of CO2 and greater dissociation of carbonic acid at high pressure. Intracellular acidification is also observed in the case of heat-shock, ethanol or osmotic stresses. The higher acidification of the vacuole has its origin in the induction of the gene HSP30, which codes for a down-regulator of the H+-ATPase activity in yeast plasma membrane. H+-ATPases pump out protons accumulated in the cytoplasm into the vacuole. As pressure increases, the yeast vacuole is assumed to serve as proton sequestrant to maintain favourable cytoplasmic pH.
5. High pressure effects in molecular bioscience
In the preceding sections, the relevance of high hydrostatic pressure (HHP) for the development of life has been discussed. Besides its relevance for biology, interest in pressure as a thermodynamic and kinetic variable has been growing also in physico-chemical studies and in biotechnological applications of biological materials in recent years.33–36 The fundamental reasons are: i) Changing temperature of a biochemical system at atmospheric pressure produces a simultaneous change in thermal energy and volume; therefore, to separate thermal and volume effects, one must carry out high pressure experiments. ii) Because noncovalent interactions play a primary role in the stabilization of biochemical systems, the use of pressure allows one to change, in a controlled way, the intermolecular interactions without the major perturbations produced by changes in temperature or co-solvent concentration. iii) Pressure affects chemical equilibria and reaction rates, depending on the reaction (ΔV) and activation (ΔV≠) volumes involved. The behavior of all systems under high pressure is governed by Le Châtelier's principle, which predicts that the application of pressure shifts an equilibrium towards the state that occupies a smaller volume, and accelerates processes for which the transition state has a smaller volume than the ground state. For example, if a reaction is accompanied by a ΔV≠ value of −50 ml mol−1, it is enhanced more than 3000-fold by applying a pressure of 400 MPa at ambient temperature. With the knowledge of ΔV and ΔV≠ values, one can draw valuable conclusions about the nature of the reaction and its mechanism. iv) Pressure-dependent studies often lead to the discovery of new phases and processes. v) One can extend the range of temperature conditions and carry out experiments at subzero °C temperatures and in the supercritical state. Therefore, protein solutions can be measured at subzero Celsius temperatures to investigate their cold-denaturation behavior.Pressures used to investigate biochemical systems usually range from 0.1 MPa to about 1 GPa. Such pressures only change intermolecular distances and affect conformations, but do not change covalent bond distances or bond angles. The covalent structure of low molecular mass biomolecules (peptides, lipids, saccharides), as well as the primary structure of macromolecules (proteins, nucleic acids and polysaccharides), is not perturbed by pressures up to about 2 GPa. Pressure acts predominantly on the conformation and supramolecular structures of biomolecular systems. High pressure studies generally call for unique methods, which have been developed in recent years.33–40 Here, some basic concepts and results are discussed.
Nucleic acids
Due to the stabilizing effect of HHP on DNA hydrogen bonds, the duplex to single strand transition temperature (known as the melting temperature, TM) increases under pressure.41,42 Stacking interactions, which have been shown to produce a negative volume change, stabilize the double helix at high pressure and, consequently, increase TM. Dubins et al. pointed out that the effect of pressure on the stability of nucleic acid complexes strongly depends on its melting temperature TM and must always be defined in the context of the solution ionic strength and in a specific pressure-temperature domain.43 Nucleic acid duplexes with TM values above ∼50 °C are stabilized by pressure.Lipid membranes
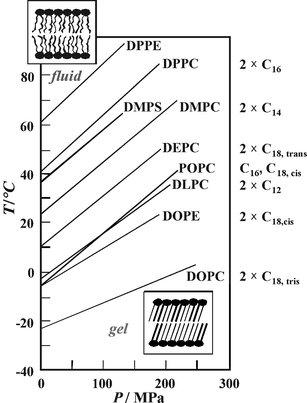 | ||
| Fig. 4 T,P-phase diagram for the main (chain-melting) transition of different phospholipid bilayer systems. The fluid (liquid-crystalline) Lα-phase is observed in the low-pressure, high-temperature region of the phase diagram; the gel phase regions appear at low temperatures and high pressures, respectively. The acyl chains of the various phospholipids are denoted on the right hand side of the figure. | ||
Upon compression, the lipids adapt to volume restriction by changing their conformation and packing. A common slope of ∼220 °C GPa−1 has been observed for the gel–fluid phase boundary of the saturated phosphatidylcholines (Fig. 4). Using the Clapeyron relation, dTm/dP = TmΔVm/ΔHm, the positive slope can be explained by an endothermic enthalpy change, ΔHm, and a volume increase, ΔVm, for the gel–fluid transition that have been found in direct measurements of these thermodynamic properties. Similar transition slopes have been found for most phospholipid bilayer systems, such the mono-cis-unsaturated lipid POPC, the phosphatidylserine DMPS, and the phosphatidylethanolamine DPPE. Only the slopes of the di-cis-unsaturated lipids DOPC and DOPE are markedly smaller. The two cis-double bonds of DOPC and DOPE lead to very low transition temperatures and slopes, as they impose kinks in the linear conformations of the lipid acyl chains, thus creating significant free volume in the bilayer so that the ordering effect of high pressure is reduced.
It has been noted that applying high pressure can lead to the formation of additional gel phases that are not observed under ambient conditions, such as the interdigitated high pressure gel phase Lβi found for phospholipid bilayers with long acyl chain lengths. To illustrate this polymorph, the results of a detailed study of the P,T-phase diagram of DPPC in excess water are shown in Fig. 5a. At much higher pressures, further gel phases appear.34,35 The data demonstrate that biological organisms could modulate the physical state of their membranes in response to changes in the external environment by regulating fractions of the lipid components in the cell membrane that vary in chain length, chain unsaturation or headgroup structure via “homeoviscous adaption”. In fact, several studies have demonstrated that membranes are significantly more fluid in barophilic and/or psychrophilic species, which is principally a consequence of an increase in the unsaturated/saturated lipid ratio, as noted in the previous section.
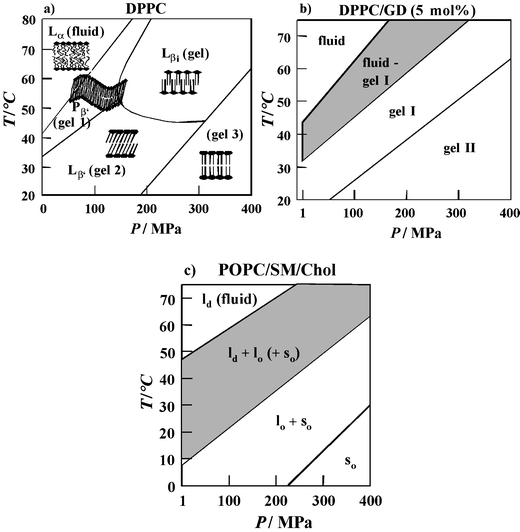 | ||
| Fig. 5 (a) T,P-phase diagram of DPPC bilayers in excess water (besides the Gel 1 (Pβ′), Gel 2 (Lβ′) and Gel 3 phase, an additional crystalline gel phase (Lc) can be induced in the low-temperature regime after prolonged cooling). (b) Phase diagram of DPPC-GD (5 mol%) in excess water as obtained from SAXS and FT-IR spectroscopy data. (c) Tentative P,T-phase diagram of the model raft mixture POPC/SM/Cholesterol (1 : 1 : 1) as obtained from spectroscopic and SAXS data. The lo + ld (+so) fluid/ordered domain coexistence region is hatched. | ||
However, Nature has further means to regulate the membrane fluidity. Biological membranes consist of lipid bilayers, which typically comprise a complex mixture of phospholipids and sterol, along with embedded or surface associated proteins. The sterol cholesterol is an important component of animal cell membranes that contain up to 50 mol%. Cholesterol thickens a liquid-crystalline bilayer and increases the packing density of lipid acyl chains in the plane of the bilayer in a way that has been termed a “condensing effect”. Measurements of the acyl chain orientational order of the lipid bilayer system demonstrated the ability of sterols to efficiently regulate their structure, motional freedom and hydrophobicity.34,35,44,45 Addition of increasing amounts of cholesterol leads to a drastic increase of the chain order parameter S in the lower pressure region. For concentrations above about 30–50 mol% cholesterol, the conformational order is almost independent of pressure and the fluid-to-gel phase transition can hardly be detected any more. Hence, sterols have the ability to regulate the structure, motional freedom and hydrophobicity of biomembranes, so that they can withstand drastic changes in environmental conditions, such as temperature and external pressure.
Recent studies have been carried out on more complex models for biomembrane systems, such as cholesterol-containing ternary mixtures that contain an unsaturated lipid like phosphatidylcholine and a saturated lipid like sphingomyelin. Such lipid systems are supposed to mimic distinct liquid-ordered (lo) lipid regions, called “rafts”, which coexist with liquid-disordered (ld), fluid-like domains. Rafts are also present in cell membranes and are thought to be important for cellular functions such as signal transduction and the sorting and transport of lipids and proteins. FT-IR spectroscopy in combination with calorimetry, fluorescence spectroscopy and synchrotron X-ray scattering has been used to characterize T- and P- dependent changes in the conformation, hydration, structure and phase behavior of the canonical lipid raft mixture POPC/SM/Cholesterol, and to establish a P,T-phase diagram of the system over an extended temperature and pressure range (Fig. 5c). The lo/ld phase coexistence region of the model raft mixture extends over a rather wide temperature range of about 40 °C. An overall fluid phase is reached at rather high temperatures (above ∼50 °C), only. At ambient temperature, a fully ordered lipid state is reached at 100–200 MPa. Interestingly, ceasing of membrane protein function in natural membrane environments has been observed for a variety of systems in this pressure range.34,35 This might be correlated with the membrane matrix reaching a physiologically unacceptable overall ordered state at these pressures.
Little is known about pressure effects on the motions of lipid bilayers at elevated pressures.34,35,37–39 Of particular interest is the effect of pressure on lateral diffusion, which is related to biological functions such as electron transport and some hormone-receptor interactions. Pressure effects on lateral diffusion of lipid molecules in relation to other membrane components have yet to be carefully studied, however. Pressure effects on the lateral self diffusion coefficient D of DPPC and POPC vesicles have been studied by Jonas.37 The lateral diffusion coefficient of DPPC in the liquid-crystalline phase decreases by about 30% from 1 to 30 MPa at 50 °C. A further 70% decrease in the D-value occurs at the pressure-induced Lα to gel phase transition. The effect of cholesterol incorporation into fluid lipid bilayers has a significant effect on the conformational order, but a less pronounced effect on the dynamic properties of the lipid membrane. Hence, lipid bilayers are able to regulate their structure and fluidity by an adjustment of their sterol composition as well as by a lateral redistribution of their various lipid components, and saturated (ordered) and unsaturated (fluid) domains. An increase in the sterol level in a membrane generally reduces the effect of variations in pressure.
Temperature and pressure effects on proteins
Also with respect to the kinetics of the protein folding reaction, pressure studies are of particular use, as they allow us to evaluate the volume profile during the folding process and to characterize the nature of the barrier to folding or unfolding and the corresponding transition state. Moreover, pressure studies present an important advantage due to the generally observed positive activation volume for folding, the result of which is to slow down the folding reaction substantially, in turn allowing for relatively straightforward measurements of structural order parameters characteristic for folding intermediate states, that are difficult or even impossible to quantify on much faster timescales corresponding to ambient pressure conditions.
As an example, we show results of a study of the pressure-induced unfolding and refolding of staphylococcal nuclease (SNase), a small protein of 149 amino acids, consisting of about 26% α-helices and 25% β-sheets.46,47 The high pressure SAXS data at 25 °C revealed that over a pressure range from atmospheric to ∼300 MPa, the radius of gyration Rg of the protein doubles from roughly 17 Å for native SNase two-fold to nearly 35 Å (Fig. 6a). The scattering curves reveal a transition from a globular to an ellipsoidal structure. The FT-IR amide I′ absorption band reveals a pressure-induced denaturation process that is evidenced by an increase in disordered and turn structures and a drastic decrease in the content of β-sheets and α-helices. Contrary to the temperature-induced unfolded state, the pressure-induced denatured state retains some degree of β-like secondary structure and the protein molecules cannot be described as fully extended random polypeptide coil. Temperature-induced denaturation involves further unfolding of the protein molecule that is indicated by a larger Rg-value of 45 Å. There are many indications now that the conformation of a protein denatured by pressure is more compact than that of a protein denatured by temperature or chemical agents, and often resembles “molten globule”-type structures. This does not seem too surprising, as pressure is known to favour the formation of hydrogen bonds, which maintain the secondary-structure network, but is unfavourable for hydrophobic interactions, which are predominantly responsible for maintaining the tertiary structure of a protein. The idea is supported by theoretical results that suggest water penetration into the protein interior as a likely mechanism for pressure-denaturation of proteins due to a weakening of hydrophobic interactions, as opposed to the temperature-induced unfolding process. Assuming the pressure-induced unfolding transition of SNase to occur essentially as a two-state process, a standard Gibbs free energy change for unfolding of ΔG0 = 17 kJ mol−1 and a volume change for unfolding of ΔV = −80 ml mol−1 is obtained. Generally, proteins are 5–10 times less compressible than water. As a result, pressure-induced volume changes in proteins are quite small, typically <1%. For monomeric proteins such as SNase, the difference corresponds approximately to the volume of 4–5 water molecules (∼18 ml mol−1).
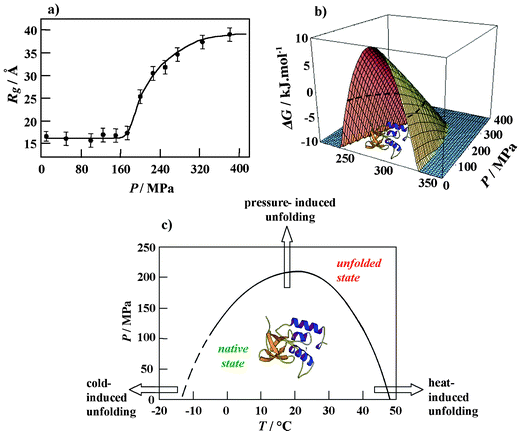 | ||
| Fig. 6 (a) Radius of gyration Rg of SNase as a function of pressure at T = 25 °C. (b) Calculation of the three-dimensional free energy landscape of SNase (pH 5.5) using experimentally determined thermodynamic parameters. The Gibbs free energy of unfolding, ΔG, is plotted as a function of temperature and pressure. The slice of the three-dimensional free energy landscape for ΔG = 0 (dashed line) yields the P,T stability diagram of the protein. (c) P,T-stability diagram of SNase at pH 5.5 as obtained by SAXS, FT-IR spectroscopic and DSC measurements. | ||
The pressure midpoints at several temperatures obtained from the FT-IR spectroscopy and SAXS profiles are plotted as a P,T-phase diagram in Fig. 6b. Such a partially elliptic-like phase diagram is typical for monomeric proteins.33–36 Knowing experimentally obtained thermodynamic parameters, such as the changes in heat capacity, expansivity, compressibility, enthalpy and volume at the unfolding transition, allows the calculation of the three-dimensional free-energy landscape.34,35 The corresponding plot for the protein SNase (Fig. 6c) clearly shows that the protein is stable only (Gibbs free energy of unfolding ΔG > 0) within a limited P,T-phase-space. The good agreement between the experimental data points and the theoretical curve for ΔG = 0 justifies the two-state assumption for the unfolding transition of SNase. Certainly, the temperature and pressure dependence of the thermodynamic parameters involved must be considered to obtain quantitative agreement with the experimental data. Moreover, only if the denatured state has a rather well-defined average free energy, an effective two-state model may be a reasonable approximation. Generally, the unfolding process is best described by a funnel-like energy landscape picture.48 Certainly, the shape of the stability diagram depends on the individual protein structural composition and it may be more complicated, in particular for larger proteins. Also, additional regions in the phase diagram may appear, such as an extended region at high temperatures where aggregation occurs. We also note that the unfolded state ensemble in the P,T-plane can be of considerably different structure, and that long-lived metastable states may occur. Whereas monomeric proteins generally unfold at pressures above 200 MPa (at 400–800 MPa in most cases), oligomeric proteins dissociate at much lower pressures (mostly at ∼100–200 MPa).33–36
The perplexing effect of high pressure on protein aggregation consists in, on the one hand, inducing aggregation-prone intermediate states, and on the other hand the ability of high-pressure to prevent aggregation and to dissociate aggregates.51–54 The susceptibility of protein aggregates to pressure largely depends on the degree of the structural order of an aggregate. Fresh, amorphous aggregates are more sensitive to pressure and prone to refolding to the native state than mature amyloid fibrils. In the latter case, effectiveness of pressure-induced dissociation depends on the particular mode of polypeptide backbone and side chain packing that allows reducing remaining void volumes. The pressure-sensitivity of fresh aggregates and the virtual insensitivity of mature fibrils allows us not only to differentiate between various stages of the amyloid-formation, but also to obtain reliable thermodynamic data, such as Gibbs' free energy and volume changes, of the early stage of the protein transformation. This is only possible due to the reversibility of the process under high-pressure conditions. Such an approach has been successfully employed in studies on lysozyme, insulin, PrP and TTR amyloidogenesis.51–54 Our work on insulin fibrillation at high pressure conveyed an astonishing example of how studies employing HHP may shed new light on aggregation pathways and subfibrillar structure of amyloid. Though, as high pressure disfavors insulin aggregation, in fact it permits amyloidogenesis through an alternative, less effective pathway that brings about a negligible volume expansion, finally leading to insulin amyloid of a unique morphology. Concerning the free energy landscape of proteins, probably a more generalized protein landscape picture including an additional “aggregation funnel”—eventually consisting of different deep minima for different strains—must be envisaged. One of the most interesting prospects for application of high pressure in protein aggregation research was the idea of destroying prion infectivity through pressure treatment.55
6. Is high-pressure water the cradle of life?
If one takes into account the earliest of life traces identified on Earth (−3.8 Ga), from isotopically light carbon traces, one has to assume that this carbon was reduced from CO2 to organic carbon by a mechanism that incorporates more isotopically light (12C) than heavy carbon (13C). In addition, due to the age and pressure and temperature history of the rock, one has to assume that the original δ13C of the Archean carbon was much lower than that measured today (−10‰). It is thus likely that this original δ13C value ranged from −25 to −40‰. On todays Earth, only a few metabolic processes can create such a significant isotopic fractionation of carbon : photosynthesis (δ13C ranging from −15 to −35‰) and methanogenesis (δ13C ranging from −40 to −60‰). Both require complex energy harvesting and transfer machineries, even in the simplest of organisms. Inventing these pathways obviously needed time.How long did it take life to emerge from the limbs, invent the cell, the genetic information and the proteins, to finally reach the photosynthetic or methanogene outcome? Nobody knows, and the question is highly debated amongst the scientific community. However, as discussed in the introduction, catastrophic meteoritic events with the capability to vaporize the totality of the Earth' ocean waters occurred at least to −4.2 Ga, and most probably up to 3.9 Ga. Therefore, if life emerged on Earth, its path from nothingness to its almost full complexity must have taken between 100–400 million years. Several lines of evidence support the hypothesis that life could have originated under pressure.
One of the bottlenecks for the emergence of life is the synthesis of its first building blocks. On one hand, the thermal and physico-chemical conditions favorable to their “spontaneous” synthesis in the primordial soup are also those that favor their chemical instability. On the other hand, water in deep hydrothermal systems is under pressure (20–35 MPa) temperature (350–450 °C) conditions that correspond to the thermodynamic supercritical state. Under such conditions, experiments show that physicochemical properties like the dielectric constant ε, the viscosity η, the density ρ and the ionic hydration decrease in supercritical water (see review by Bassez56). Consequently, the solubility of ionic and polar compounds diminishes, while that of simple apolar molecules is enhanced. Hence, the apolar supercritical water in hydrothermal vents could concentrate prebiotic molecules, which would react more efficiently.56
High pressure allows the synthesis of a set of molecules that cannot be synthesized at ambient pressures, or at the expense of enzymatic activities. High pressure can shift the temperature requirement for a given chemical reaction towards lower temperature, lowering the ΔG0, allowing for prebiotic synthesis at lower temperatures. Furthermore, high pressure can stabilize several essential biological macro molecules such as DNA and RNA. Therefore, it could be possible for the proto-life to “invent” RNA, and make use of its catalytic properties, something that could not occur in the temperature conditions required for the spontaneous synthesis of RNA at surface conditions.
Perhaps, the most crucial indication about whether life originated from the deep waters of the proto-ocean is given to us by the study of the current life forms on Earth. When we compare the adaptations to different physico-chemical conditions that can be observed within the three domains of life, we are overwhelmed by the adaptation ability of life. However, only a few specific families of organisms for each environment can live in these extreme environments, whether hot, acidic, halophilic, etc. In fact, most organisms today can live in moderate physico-chemical conditions, from which only moderate variations can be tolerated. In contrast, most, if not all, organisms can live under a large range of hydrostatic pressures. In fact, surface organisms can withstand pressures as high as 20 MPa, e.g. the pressure equivalent to 2 km of water, without consequence on its life cycle or metabolism. Indeed, several will be able to live under much higher pressure before hydrostatic pressure is perceived as a stress. Tolerance to high hydrostatic pressure is the only physical or chemical parameter found in all organisms in which it was sought for, and may well indeed represent one of the most ancestral physical conditions under which life had to emerge.
Abbreviations
| GD | gramicidin D |
| HHP | high hydrostatic pressure |
| DMPC | 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (di-C14:0) |
| DPPC | 1,2-dipalmitoyl-sn-glycero-3-phosphatidylcholine (di-C16:0) |
| DPPE | 1,2-dipalmitoyl-sn-glycero-3-phosphatidylethanolamine (di-C16:0) |
| DMPS | 1,2-dimyristoyl-sn-glycero-3-phosphatidylserine (di-C14:0) |
| DSPC | 1,2-distearoyl-sn-glycero-3-phosphatidylcholine (di-C18:0) |
| DOPC | 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (di-C18:1,cis) |
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine (di-C18:1,cis) |
| DEPC | 1,2-dielaidoyl-sn-glycero-3-phosphatidylcholine (di-C18:1,trans) |
| POPC | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (C16:0,C18:1,cis) |
| SNase | staphylococcal nuclease |
| DSC | differential scanning calorimetry |
| SAXS | (WAXS) small(wide)-angle X-ray scattering |
| SANS | small-angle neutron scattering |
| TTR | transthyretin |
| PrP | prion protein |
| FT-IR | Fourier-transform infrared |
| ΔV and ΔG0 | standard volume and free energy change |
| ΔV≠ | activation volume of reaction |
Acknowledgements
Financial support from the French GEOMEX program of the CNRS to ID and PO, from the French MRCT of the CNRS to PO, and from the Deutsche Forschungsgemeinschaft (DFG) and the Fonds der Chemischen Industrie to RW is gratefully acknowledged. Thanks to P. F. McMillan for improving the English writing of the manuscript.References
- E. G. Nisbet and N. H. Sleep, Nature, 2001, 409, 1083 CrossRef CAS.
- M. J. Russell and N. T. Arndt, Biogeosciences, 2005, 2, 97 Search PubMed.
- K. Zahnle, N. Arndt, C. Cockell, A. Halliday, E. Nisbet, F. Selsis and N. H. Sleep, in Geology and Habitability of Terrestrial Planets, ed. K. Fishbaugh, D. des Marais, O. Korablev, P. Lognonne and F. Raulin, Space Sciences Series of ISSI, Springer, Amsterdam, 2006 Search PubMed.
- Y. Amelin, A. N. Krot, I. D. Hutcheon and A. A. Ulyanov, Science, 2002, 297, 1678–1683 CrossRef CAS.
- S. A. Wilde, J. W. Valley, W. H. peck and C. M. Graham, Nature, 2001, 409, 175 CrossRef CAS.
- J. E. Chambers, Earth Planet. Sci. Lett., 2004, 223, 241 CrossRef CAS.
- M. Boyet and R. W. Carlson, Science, 2005, 309, 576 CrossRef CAS.
- J. D. Kramers, Precamb. Res., 2003, 126, 379 Search PubMed.
- A. J. Cavosie, S. A. Wilde, D. Liu, P. W. Weiblen and J. W. Valley, Precamb. Res., 2004, 135, 251 Search PubMed.
- A. J. Cavosie, J. W. Valley and S. A. Wilde, Earth Planet. Sci. Lett., 2005, 235, 663 CrossRef CAS.
- J. F. Kasting, Precamb. Res., 2005, 137, 119 Search PubMed.
- M. J. Burchell, J. R. Mann and A. W. Bunch, Mon. Notes R. Astronom. Soc., 2004, 352, 1273 Search PubMed.
- H. W. Jannasch and C. D. Taylor, Annu. Rev. Microbiol., 1984, 38, 487 CrossRef CAS.
- T. Gold, Proc. Natl. Acad. Sci. USA, 1992, 89, 6045 CrossRef CAS.
- J. B. Corliss, J. Dymond, L. I. Gordon, J. M. Edmond, R. P. V. Herzen, R. D. Ballard, K. Green, D. Williams, A. Bainbridge, K. Crane and T. H. Vanandel, Science, 1979, 203, 1073 CrossRef CAS.
- C. M. Cavanaugh, Biol. Soc. Wash. Bull., 1985, 6, 373 Search PubMed.
- G. Erauso, A.-L. Reysenbach, A. Godfroy, J. R. Meunier, B. Crump, F. Parensky, J. A. Baross, V. T. Marteinsson, G. Barbier, N. R. Pace and D. Prieur, Arch. Microbiol., 1993, 160, 338 CAS.
- J. T. Trevors, Res. Microbiol., 2002, 153, 487–491 CrossRef CAS.
- B. M. Jakosky and E. L. Shock, J. Geophys. Res., Planets, 1998, 103, 19359 Search PubMed.
- E. V. Koonin, Nat. Rev. Microbiol., 2003, 1, 127 Search PubMed.
- P. Forterre, Mol. Microbiol., 1999, 33, 457 CrossRef CAS.
- C. A. Ouzounis, V. Kunin, N. Darzentas and L. Goldovsky, Res. Microbiol., 2006, 157, 57 CrossRef CAS.
- J. I. Glass, N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison, H. O. Smith and J. C. Venter, Proc. Natl. Acad. Sci. USA, 2006, 103, 425 CrossRef CAS.
- C. Woese, Proc. Natl. Acad. Sci. USA, 1998, 95, 6854 CrossRef CAS.
- S. Gribaldo and H . Philippe, Theor. Population Biol., 2002, 61, 391 Search PubMed.
- D. C. Reanney, J. Theor. Biol., 1974, 48, 243 CrossRef CAS.
- J. T. Beatty, J. Overmann, M. T. Lince, A. K. Manske, A. S. Lang, R. E. Blankenship, C. L. Van Dover, T. A. Martinson and F. G. Plumley, Proc. Natl. Acad. Sci. USA, 2005, 102, 9306 CrossRef CAS.
- C. E. ZoBell and F. H. Johnson, J. Bacteriol., 1949, 57, 179 CAS.
- D. H. Bartlett, Biochim. Biophys. Acta, 2002, 1595, 367 CAS.
- F. Abe, Cell. Mol. Biol., 2004, 50, 437 Search PubMed.
- P. M. B. Fernandes, Braz. J. Med. Biol. Res., 2005, 38, 1239 Search PubMed.
- T. J. Welch, A. Farewell, F. C. Neidhardt and D. H. Bartlett, J. Bacteriol., 1993, 175, 7170 CAS.
- C. Balny, P. Masson and K. Heremans, Biochim. Biophys. Acta, 2002, 1595, 3 CrossRef CAS.
- R. Winter, Advances in High Pressure Bioscience and Biotechnology II, Springer-Verlag, Amsterdam, 2003 Search PubMed.
- R. Winter, in High pressure effects in molecular bioscience, ed. M. R. Manaa, Elsevier, Amsterdam, 2005 Search PubMed.
- R. Lange, Proteins Under High Pressure, Elsevier, Amsterdam, 2006 Search PubMed.
- J. Jonas, High pressure NMR, Springer-Verlag, Amsterdam, 1991 Search PubMed.
- M. R. Arnold, H. R. Kalbitzer and W. Kremer, J. Magn. Reson., 2003, 161, 127 CrossRef.
- T. N. Niraula, T. Konno, H. Li, H. Yamada, K. Akasaka and H. Tachibana, Proc. Natl. Acad. Sci. USA, 2004, 101, 4089 CrossRef CAS.
- R. Winter and R. Köhling, J. Phys.: Condens. Matter, 2004, 16, S327 CrossRef CAS.
- P. L. G. Chong, M. Zein, T. K. Khan and R. Winter, J. Phys. Chem. B, 2003, 107, 8694 CrossRef CAS.
- P. L. G. Chong, R. Ravindra, M. Khurana, V. English and R. Winter, Biophys. J., 2005, 89, 1841 CrossRef CAS.
- D. N. Dubins, A. Lee, R. B. Macgregor and T. V. Chalikian, J. Am. Chem. Soc., 2001, 123, 9254 CrossRef CAS.
- C. Bernsdorff, A. Wolf and R. Winter, Z. Phys. Chem., 1996, 151, 151.
- C. Bernsdorff, A. Wolf, R. Winter and E. Gratton, Biophys. J., 1997, 72, 1264 CrossRef CAS.
- G. Panick, R. Malessa, R. Winter, G. Rapp, K. J. Frye and C. A. Royer, J. Mol. Biol., 1998, 275, 389 CrossRef CAS.
- G. Panick, G. J. A. Vidugiris, R. Winter and C. A. Royer, Biochemistry, 1999, 38, 4157 CrossRef CAS.
- J. C. Onuchic and P. G. Wolynes, Curr. Opin. Struct. Biol., 2004, 14, 70 CrossRef CAS.
- L. Mitra, N. Smolin, R. Ravindra, C. A. Royer and R. Winter, Phys. Chem. Chem. Phys., 2006, 8, 1249 RSC.
- H. Herberhold, C. A. Royer and R. Winter, Biochemistry, 2004, 43, 3336 CrossRef CAS.
- J. L. Silva, D. Foguel and C. A. Royer, Trends Biochem. Sci., 2001, 26, 612 CrossRef CAS.
- Y. Cordeiro, J. Kraineva, M. P. B. Gomes, M. H. Lopes, V. R. Martins, L. M. T. R. Lima, D. Foguel, R. Winter and J. L. Silva, Biophys. J., 2005, 89, 2667 CrossRef CAS.
- R. Jansen, S. Grudzielanek, W. Dzwolak and R. Winter, J. Mol. Biol., 2004, 338, 203 CrossRef CAS.
- S. Grudzielanek, V. Smirnovas and R. Winter, J. Mol. Biol., 2006, 356, 497 CrossRef CAS.
- J. M. Zhou, L. Zhu, C. Balny and S. Perrett, Biochem. Biophys. Res. Commun., 2001, 287, 147 CrossRef CAS.
- M.-P. Bassez, J. Phys.: Condens. Matter, 2003, 15, L353 CrossRef CAS.
- R. Margesin and Y. Nogi, Cell. Mol. Biol., 2004, 50, 429 Search PubMed.
| This journal is © The Royal Society of Chemistry 2006 |
