Differential receptor arrays and assays for solution-based molecular recognition
Aaron T.
Wright
and
Eric V.
Anslyn
*
1 University Station A5300, Austin, TX, USA. E-mail: Anslyn@ccwf.cc.utexas.edu; Fax: 512-471-7791; Tel: 512-471-1669
First published on 7th November 2005
Abstract
Nature has inspired an emergent supramolecular field of synthetic receptor arrays and assays for the pattern-based recognition of various bioanalytes and metal species. The synthetic receptors are not necessarily selective for a particular analyte, but the combined signal response from the array is diagnostic for the analyte. This tutorial review describes recent work in the literature for this emerging supramolecular field and details basic array and assay design principles. We review the analytes targeted, signaling types used, and pattern recognition.
Developing specific receptors for the solution-based analysis of complex analytes and mixtures is a daunting task. A solution to this difficult task has been inspired by nature's use of arrays of receptors in the senses of taste and smell. An emerging field within supramolecular chemistry is the use of synthetic and readily available receptors in array formats for the detection of analytes in solution. Each receptor in a differential array does not necessarily have selectivity for a particular analyte, but the combined fingerprint response can be extracted as a diagnostic pattern visually, or using chemometric tools. This new genre of molecular recognition is advancing rapidly with several groups developing novel array platforms and receptors.
 Aaron T. Wright | Aaron T. Wright earned his B.S. in Chemistry in 2001 from George Fox University in Newberg, OR. He is currently a graduate student at the University of Texas at Austin working under the guidance of Eric V. Anslyn. His Ph.D. research has focused on the development of synthetic receptors for the selective detection of analytes in complex solutions, and the synthesis of synthetic receptor libraries for use in differential receptor arrays for pattern-based detection of bioanalytes. Next summer he will begin a post-doctoral position with Benjamin Cravatt at the Scripps Research Institute. |
 Eric V. Anslyn | Eric V. Anslyn received his Ph.D. in Chemistry from the California Institute of Technology in 1987 under the direction of Robert H. Grubbs. After a two year stay at Columbia University as a National Science Foundation Post-Doctoral Fellow with Ronald Breslow, he joined the chemistry faculty at the University of Texas at Austin in 1989. He was named a Sloan Scholar, Dreyfus Teacher Scholar, Searle Scholar, and a Presidential Young Investigator and was promoted to Associate Professor in 1994. In 1997 he was named Full Professor, and in 2004 he was awarded the Norman Hackerman Professorship in Chemistry. He is also a 2005 Cope Scholar Awardee. His research encompasses physical organic and bioorganic chemistry, and his research generally explores the use of synthetic receptors for sensing and catalysis applications. |
1 Introduction
Nature has provided inspiration for the field of molecular recognition, but attempting to rival the affinity and selectivity of enzymes and antibodies has proved to be a difficult task. Advances have been made in the design and synthesis of selective molecular receptors for small molecule targets and metal ions. Despite this progress, it continues to be difficult to design selective receptors for larger and more complex bioanalytes such as proteins, nucleic acids, and complex carbohydrates.Recently, chemists have investigated the application of arrays of molecular receptors for the analysis of small and medium sized analytes, and large bioanalytes. The inspiration for this work has been nature's use of “differential” receptors in the mammalian tongue and nose for the senses of taste and smell. Rather than specifically designed binding interactions between receptors and analytes, each of the arrayed receptors responds to a differing degree to each analyte. Composite fingerprints made up from multiple differential binding interactions provide unique diagnostic patterns for the individual analytes or mixtures thereof.
Arrays of synthetic differential receptors transcend the relative lack of specificity for particular analytes by organizing multiple binding events into single diagnostic patterns for each analyte. Differential arrays do not have to rely on specific binding sites, but multiple interactions may be pertinent. This is to be expected when targeting large and conformationally dynamic analytes such as proteins, where several surface interactions may be possible. Synthetic receptor arrays are generating exciting developments and leading to applications in the detection of bioanalytes, pathogens in solution, environmental analysis, and medical diagnostics.
To visualize binding events a signalling assay, such as a colorimetric or fluorescent response, is included. A method for data acquisition and interpretation of patterns using a variety of chemometric tools for pattern-recognition is often necessary as well.1
Herein we present a tutorial review on differential receptor arrays for solution-based assays. This review will discuss several array platforms, and evaluate the progress of this field.
There is already a wealth of information available on differential vapor and solution phase sensor arrays composed of fiber optic sensors, microelectrodes, conductive polymers, metal oxide field effect transistors, surface acoustic wave devices, and electrochemical sensors.2–7 Therefore, we will only provide minimal reference to these arrays and the results that have been obtained.
2 Micromachined Si/SiN chip
The mammalian tongue has a number of receptors within pores that respond differentially to sweet, sour, salty, and bitter tastes. The combined fingerprint signals from the tongue's receptors result in the diagnostic patterns for each tastant. As a mimic of the mammalian tongue, a team of researchers at the University of Texas developed a sensor methodology that allowed for analyte detection in solution.8 Receptors were created by synthetically derivatizing solid phase resin beads, and positioning them within micromachined pyramidal wells in Si/SiN wafers, one bead per well, to make individually addressable resin “taste buds.” Each bead signals a unique response to an analyte, and the combined fingerprint response gives the pattern for the analyte in solution.2.1 Early “electronic tongue” development
In a proof of concept experiment a 3 × 3 array of derivatized beads was created to simultaneously identify multiple analytes as a primitive simulation of the mammalian tongue.8 The array is contained within a transparent disc frame, with PEEK tubing for delivery and extrication of the analyte solution. The solution enters from the top of the array, and proceeds through the beads and out the bottom of the micromachined pyramidal wells. Solutions are pumped through the array using a fast protein liquid chromatograph. The transparent disc frame is sealed within further aluminium housing, and this is affixed to a stereoscope stand allowing both top and bottom illumination of the resin beads. A charge-coupled device (CCD) was fixed to the stereoscope for image capture (red, green, and blue (RGB) light density) and data acquisition. The CCD is a powerful tool allowing parallel acquisition of spectral data from multiple beads. Absorption properties of the beads are measured via the CCD, and patterns were obtained for various analytes and pH conditions by evaluation of the cumulative array response.In this initial study four sensors were placed within the array. The sensors were developed by derivatizing polymeric resins with the following: fluorescein (FLU) for pH, o-cresolphthalein complexone (CRP) for Ca2+ and pH, alizarin complexone (ALZ) for Ce3+, Ca2+, and pH, and a boronic ester (BOH) of resorufin-derivatized galactose for fructose and pH.
Evaluation of the RGB relative absorbance values for 0.1 M solutions of the metals, as well as mixtures of the metals, at pH values of 3, 5, 7, 9, and 11 gave distinctive colorimetric patterns. This sensor array model provides rapid generation of responses (less than one minute), and clear quantifiable colorimetric patterns. Illustrated in Fig. 1 are the responses of the derivatized resins at multiple pHs to Ca2+. As shown, the patterns are composed of various levels of RGB absorbance. This work led to more diverse and complex arrays for more challenging and dynamic analytes.
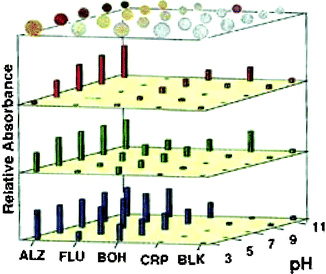 | ||
| Fig. 1 The illustration shows the derivatized beads on top of the colorimetric bar graph charts. The extent of color attenuation (linear scale) obtained from the CCD in response to Ca2+ (0.1 M Ca(NO3)2) is shown for red (λ = 700 nm), green (λ = 550 nm), and blue (λ = 435 nm). Reprinted with permission from Neikirk et al.,8J. Am. Chem. Soc., 1998, 120, 6429. © 1998 American Chemical Society. | ||
2.2 Enhancement of the Si/SiN chip array
Refinement of the sensor array8 by McDevitt led to new instrumentation with increased concentration thresholds, reproducibility, and reversibility.9 The instrumentation was enhanced by addition of a video capture card for recording multiple images over a defined time period in order to monitor the kinetics of binding.In the refined system each resin bead within the pyramidal pits on the Si/SiN wafer acts as a microreactor and microanalyzer. Drawing an area of interest (AOI) for each resin bead in the array permits extraction of data from each individual resin-bound receptor. Data can be extracted from multiple images to observe chemical changes as analtyes bind to the receptors. The extracted data was defined as an effective absorbance (AR = −log [IR/IB]), in which the absorbance of a certain RGB channel defined as “R” is obtained by measuring the −log value of the intensity of each receptor bead, divided by the intensity of an acylated blank bead.
In the new system the receptors can be “recharged” and used multiple times for reproducible results. As a microreaction chamber it was found that increased diversity between the beads enhanced the wide range of analyte detection possible. In fact, a single receptor in the array might not give a detectable signal for a certain analyte, but the cumulative response from all the receptors leads to a unique pattern. Therefore, diversity leads to arrays that can sense different classes of analytes. It was also found that using agarose beads in addition to the polymeric beads allowed for enzyme and immunological assays.10,11
This refined system has several advantages including optical collection at multiple sites, uniform flow characteristics, real-time data acquisition, and rapid adaptation of the array for detecting different classes of analytes. Indeed, the authors cite that their system can now be thought of as a “programmable taste chip.”9
In accordance with the “programmable taste chip” several resin-bound synthetic receptors have been developed and employed to detect many classes of analytes. By synthesizing unique receptors with functionalities targeted toward certain analyte classes, customizable differential sensor arrays were created.
2.3 Differential sensor arrays with synthetic receptors
Combinatorial chemistry is an excellent tool for rapidly generating large numbers of synthetic receptors.12,13 Our group has exploited the array discussed above for three differential array studies involving the recognition and discrimination of nucleotide phosphates,14 proteins and glycoproteins,15 and tripeptides and tripeptide mixtures.16 We initially generated core host structures that provide binding functionalities and preorganization for binding particular analyte classes. The differential binding element is introduced by incorporating peptide arms onto the core structures of the receptors using split-and-pool combinatorial chemistry. This generates a large receptor library from which a few receptors are randomly chosen and placed into the differential array.To observe binding events occurring at each receptor in the array an indicator-displacement assay (IDA) was employed.17 Fluorescein, an anionic chromophore, was initially passed through the array resulting in receptor–indicator associations that “color” the beads. The effective absorbance values were calculated for the displacement assay when a 2 mL injection of a 20 mM nucleotide phosphate solution was injected through the array. This was completed by capturing several images with the CCD during the displacement assay. By combining the effective absorbance values for just a single transmission (i.e. only the red channel) over the entire displacement time period, graphs were made for each receptor in the array corresponding to the rate of indicator-displacement at that site. Fig. 2 shows the displacement profiles for two separate receptors in the array in response to AMP. The unique pattern for each nucleotide phosphate is composed of the cumulative collection of displacement rates from all the receptors in the array.
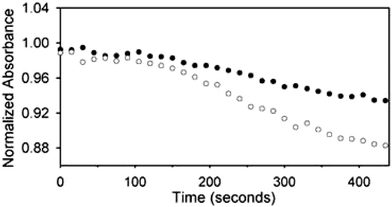 | ||
| Fig. 2 Displacement profiles for two separate receptors in the array in response to AMP (20 mM in 25 mM HEPES) pH 7.5 buffered aqueous solution. Effective absorbance is equivalent to normalized absorbance. Reprinted with permission from Anslyn et al.,14J. Am. Chem. Soc., 2003, 125, 1114. © 2003 American Chemical Society. | ||
Clearly, if all 30 displacement profiles were plotted side by side it would be implausible to visually detect noticeable differences between the analytes. Principal component analysis1 (PCA) was employed to reduce the dimensionality of the data set. In this mathematical model the 30 dimension patterns are reduced to single data points on a new principal component (PC) axis. The first PC axis lies along the line of maximum variance; subsequent PC axes define diminishing levels of variance. Separation between data points on a PC plot describe how unlike they are from one another. Ideally, multiple trials of the same analyte should cluster very closely, and other analytes should be spatially separated from them. As seen in Fig. 3, the three nucleotide phosphates are spatially separated indicating the ability of the array to discriminate the three analytes from one another. Sequencing of the peptide arms on eight of the receptors showed that several of them incorporated aromatic residues and serine, or the structurally similar threonine. It was found earlier in a screening study of the same library that ATP was bound strongly by library members containing hydroxyl and aromatic residues.18
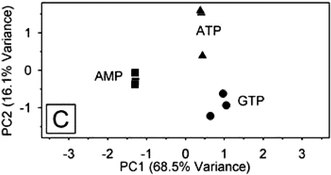 | ||
| Fig. 3 A PCA score plot indicating the separation and discrimination of the three nucleotide phosphates (▲ ATP, ■ AMP, ● GTP) by 30 receptors from library 1. Reprinted with permission from Anslyn et al.,14J. Am. Chem. Soc., 2003, 125, 1115. © 2003 American Chemical Society. | ||
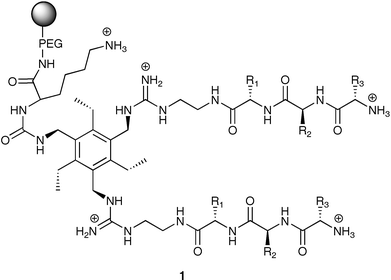
Employing a hexasubstituted benzene scaffold, we developed library 2 for the pattern recognition and discrimination of various proteins and glycoproteins.15 Library 2 was created with two combinatorially synthesized tripeptide arms giving differential binding variance for separation within the two classes of proteins, and boronic acid moieties for assisted differentiation between proteins and glycoproteins. The library was created using all natural amino acids except Cys resulting in a library with 6,859 unique members. Twenty-nine members of 2 were randomly selected and placed in a 7 × 5 array with 6 acetylated amine blanks.
The previous research using an array for nucleotide phosphates utilized an IDA. In this study it was determined that the required concentrations of the analytes could be significantly reduced by using a staining, or indicator-uptake assay. In this assay the proteins and glycoproteins (355 µM) were initially added to the synthetic sensor array. Next, the indicator bromopyrogallol red (3.0 µM) was added. During the indicator-uptake 215 images were captured by the RGB CCD, and the kinetics of indicator-uptake for the green channels of each receptor were obtained. Using PCA, the 29 slopes were reduced to single data points on a PC chart, as seen in Fig. 4.
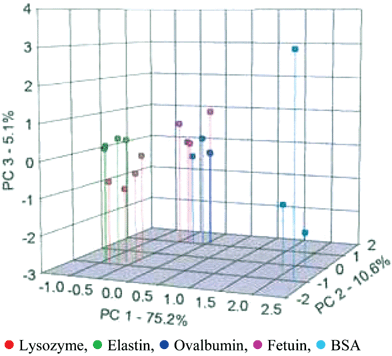 | ||
| Fig. 4 A PCA score plot indicating the recognition and separation of four trials of five different proteins and glycoproteins by 2. Percentages on the axes detail the amount of variance defined within the PC axes. Reprinted with permission from Anslyn et al.,15Angew. Chem., Int. Ed., 2005, 44, 6375. © 2005 Wiley-VCH. | ||
The glycoproteins (ovalbumin and fetuin) somewhat separated from one another and clearly separated from the three proteins (lysozyme, elastin, and bovine serum albumin (BSA)). Furthermore, lysozyme and elastin did have some minimal spatial separation, and were clearly separated from BSA. It was found that the separations were not solely based upon pI or molecular weight, but resulted from specific contacts between the receptors and surfaces of the proteins.
Seven of the receptors from the array were sequenced and shown to have the greatest contribution to the formation of PC axis 1. In this case no observable similarities were found and we concluded that sensor arrays composed of chemically diverse receptors can provide good discrimination and recognition of analytes.
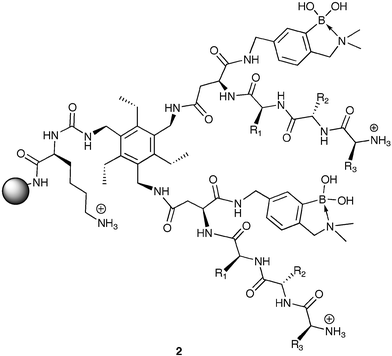
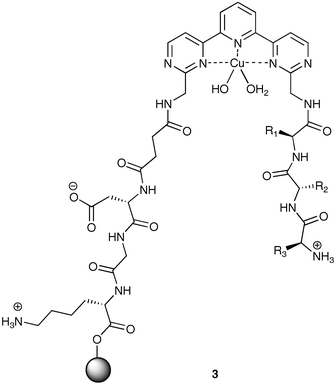
In a recent study, we developed library 3 incorporating a Cu(II) ligand backbone for preorganization of the binding cavity,16 as well as enhanced affinity for analtyes containing Cu(II) ligands.20 Emanating outward from the core of 3 are two tripeptide arms. The arm originating from the resin was fixed as Asp-Gly-Lys, with a succinic acid group as a linker between the tripeptide and the core. The second tripeptide arm was derivatized using split-and-pool combinatorial chemistry to create library 3 with 6,859 members.
Akin to our previous study,15 an indicator-uptake assay was used with Orange G as the indicator. Again 215 images were captured by the CCD and indicator-uptake slopes were evaluated from each of 30 receptors in the array to create diagnostic patterns using PCA for each of four tripeptides, and three mixtures of the tripeptides.
Four tripeptides were discriminated, three terminating in the Cu(II) ligand His, and one in Gly. As is evident in Fig. 5, the array of synthetic receptors effectively separated the individual tripeptides (13 µM), as well as the mixtures of tripeptides (13 µM). As expected, the mixtures fell in between the individual PCA score points for the tripeptides comprising the mixture illustrating the 50/50 nature of the mixtures. In this case, metal coordination to 3 acts as an initial source of binding affinity toward the analytes. The tripeptide arms provide the discriminating force resulting in differential binding between the tripeptides and the 30 receptors. Had the Cu(II) center been solely responsible for the binding interaction, it would have resulted in a PCA plot with all the analytes with N-terminal His overlapping one another.
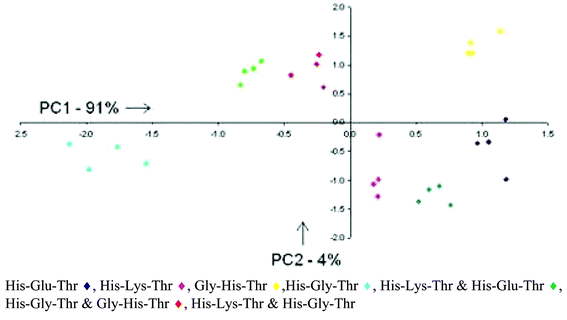 | ||
| Fig. 5 Two-dimensional PCA plot describing 95% of the variance. Clustering of the analytes illustrates the ability of the differential array of resin-bound receptors to discriminate various tripeptides and mixtures of tripeptides. Reprinted with permission from Anslyn et al.,16J. Am. Chem. Soc., 2005, submitted. © 2005 American Chemical Society. | ||
2.4 Differential chromatographic sensor arrays
Rather than differentiating the receptor on each resin bead within an array, McDevitt21 derivatized resin beads with a colorimetric indicator at the core of the bead, and binding functionalities on the periphery of the beads for analyzing a number of metals. By derivatizing the resin periphery with metal ligands as well as relatively “inert” functional groups, the time elapsed for penetration of the metal into the core of the bead resulted in a colorimetric modulation dependent on the resin-derivatization and the metal. This work demonstrates a nice solution-based approach to metal sensing using synthetically derivatized resin beads in a microchromatographic sensing assay.The researchers chose the indicator alizarin complexone to derivatize the core of the resin as it responds to a wide variety of metal cations. The periphery of the beads was derivatized with either acetic anhydride (“inert”) or EDTA anhydride. Beads from both derivatization batches were subsequently placed into the pyramidal wells of the Si/SiN wafer. As the metal cations were delivered to the receptors, the effective absorbance of each bead was monitored versus time. To determine patterns for each metal cation, multiple variables were monitored: 1) color change of a bead was calculated as the difference between the initial and final effective absorbances, 2) tD is the time from the beginning of a bead's color change to the halfway point, 3) tL was defined as the elapsed time before a color change was observed, and 4) the red, green, and blue transmission data showing bead color changes were collected via CCD images. Fig. 6 shows the pattern responses for the metal cations Zn2+ (10 mM), Ni2+ (10 mM), Pb2+ (5 mM), and Pb2+ (10 mM).
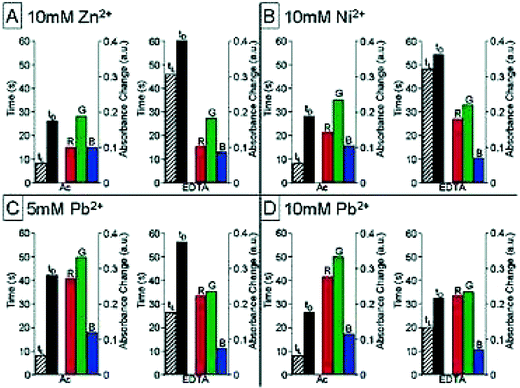 | ||
| Fig. 6 Pattern responses to various metal cations by an array of microchromatographical receptors. A) Zn2+ (10 mM), B) Ni2+ (10 mM), C) Pb2+ (5 mM), and D) Pb2+ (10 mM). The bars on the left side indicate the tL and tD values (hatched and black bars respectively). The red, green, and blue channels are shown on the right indicating bead color changes. In each quadrant two beads are shown for each metal cation (Ac = acetylated, and EDTA). Reprinted with permission from McDevitt et al.,21J. Am. Chem. Soc., 2003, 125, 2870. © 2003 American Chemical Society. | ||
Interestingly, in the case of Pb2+ the colorimetric signal is not enough to distinguish between the two concentrations, but the temporal data provides the necessary discrimination. However, as seen in Fig. 6 between panels A and B, the discrimination between the two metals is based almost entirely on the colorimetric response. Also, the tL values for the acetylated beads vary minimally between the four cases indicating the importance of the derivatization using EDTA for chromatographic purposes. This work demonstrates that a minimal sensor approach can still be complementary to discriminating multiple analytes.
3 Molecularly imprinted polymers
Molecularly imprinted polymers (MIPs) are highly crosslinked polymer matrices developed in the presence of a template analyte molecule. After templating, the analyte molecule is removed leaving a binding cavity functionally suitable to the target analyte. MIPs possess high chemical and thermal stability, and can be fashioned to selectively bind a number of different analytes. They can also be rapidly and inexpensively generated making them amenable to an array setting.The drawbacks associated with MIPs, low overall affinities and high levels of cross-reactivity, can actually be advantageous for differential sensor arrays. As long as signals are generated for an analyte from one or more MIP sensors a unique fingerprint can be obtained. Pattern recognition can be completed by mathematical multivariate analyses such as PCA and linear discriminant analysis (LDA).1
3.1 Molecularly structured monolayers—spreader bar technology
As an initial demonstration of MIPs within sensor arrays, Mirsky22 used a simple spreader-bar approach for the development of differential sensors. In this case thiol modified purines and pyrimidines (spreader-bar molecules) were co-adsorbed onto a gold surface with dodecanethiol (matrix molecule). Self-assembled monolayers formed having significant differential reactivity towards a number of nucleic acids: adenine, cytosine, thymine, uracil, caffeine, and uric acid. The primary detection method was measurement of the capacitive current.Five different spreader-bar molecules were employed in the development of the array: 6-mercaptopurine (ASH), 2-amino-6-purinethiol (GSH), 4-amino-2-mercaptopyrimidine (CSH), 4-hydroxy-5-methyl-2-mercaptopyrimidine (TSH), and 4-hydroxy-2-mercaptopyrimidine (USH). The change in the capacitive current was measured for each electrode in the array upon addition of an analyte. This meant that five separate values were calculated for each of the analytes; resulting in unique patterns. PCA (Fig. 7) showed discrimination of the analytes. Interestingly, rather than analyzing six trials of each analyte, six different concentrations were analyzed (20–470 µM), and these were clustered using PCA.
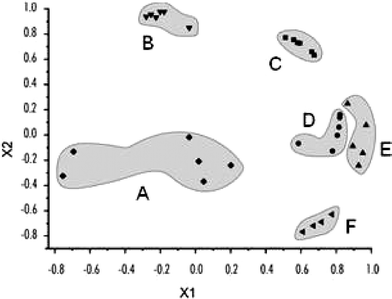 | ||
| Fig. 7 PCA shows patterns for the five different analytes (caffeine (A), uracil (B), adenine (C), cytosine (D), thymine (E), and uric acid (F)) generated using an electrode array of artificial receptors developed with spreader-bar technology. X1 and X2 are the first and second principal components. These two principal components define 75% of the variance. Reprinted with permission from Mirsky et al.,22Chem. Commun. 2003, 432. © 2003 The Royal Society of Chemistry. | ||
The utility of this technology lies in the relative simplicity of artificial receptor development and the large variety of chemoreceptors with differing selectivities prepared. Technically, it may be possible to develop near limitless numbers of sensors using this technology.
3.2 Development of MIP sensor arrays
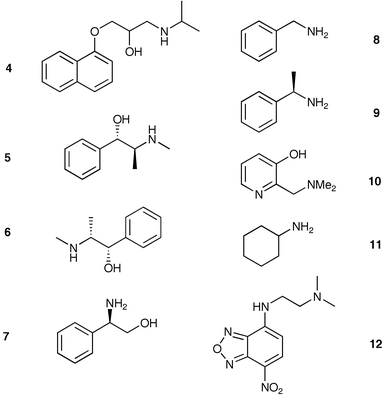
An initial demonstration of the utility of MIPs in sensor arrays was completed by Shimizu.23 In this case an eight-member sensor array was used to discriminate six different aryl amines. The eight polymers were synthesized using seven different aryl amines as templates, and one sensor was not templated. Some of the aryl amines have biological activity (4 – propanolol) and others were diastereomers (5 – ephedrine, 6 – pseudoephedrine).A constant mass of each polymer was mixed with a 3 mM solution of each analyte in acetonitrile. The response was measured as the ratio of absorbances (258 nm, (A0 − Ai)/A0) of the solutions before and after equilibration with each of the eight MIPs in the array. Thus the unique analyte patterns were composed of the eight different absorbance values. Five trials were run for each analyte resulting in an enormous amount of data that could not be visually interpreted as patterns. Therefore, LDA was employed to deconvolute the data into recognizable clusters. The axes of the LDA plot are formed from linear combinations of the original data set weighted by coefficients that produce the greatest analyte differentiation. A single point on a LDA plot is representative of the original eight responses from the MIP sensor array, much like PCA.
As demonstrated in Fig. 8, the two-dimensional LDA plot shows individual clustering of six analytes (10 was not tested because it could not be evaluated under the same conditions as the other aryl amines), and spacing between the different aryl amines. Using a ‘jack-knife’ analysis, the MIP array accurately classified 94% of the measured samples. A drawback of this method was the necessity of employing analytes with an optical handle for evaluation of the binding between the MIPs and analytes. In a subsequent analysis, a colorimetric IDA was employed to circumvent this limitation.24
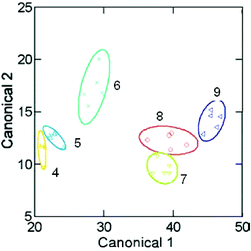 | ||
| Fig. 8 An LDA shows the patterns and clustering of the six analytes measured with the MIP array. Reprinted with permission from Shimizu et al.,23Chem. Commun., 2004, 1172. © 2004 The Royal Society of Chemistry. | ||
To test a MIP array with an IDA assay, Shimizu24 templated six analytes (4–9) and included a blank MIP sensor. For a colorimetric signal modulation, indicator 12 was utilized in an IDA format. Furthermore, the authors showed that a nontemplated analyte, cyclohexylamine 11, could be identified by the array using a pattern recognition protocol with LDA. Thus, the IDA strategy allows the detection of analytes not originally used for generating the array. The general scheme for generating patterns with an MIP sensor array incorporating a colorimetric IDA for signal modulation is shown (Scheme 1).
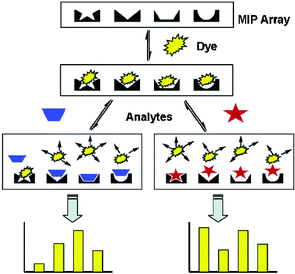 | ||
| Scheme 1 General scheme for a MIP sensor array using a colorimetric IDA. A unique colorimetric response from each sensor in the array results in diagnostic patterns for each of the analytes tested. Reprinted with permission from Shimizu et al.,24J. Am. Chem. Soc. 2005, 127, 5695. © 2005 American Chemical Society. | ||
A requirement of the array was that the individual sensors possess differential selectivity toward the various analytes so that fingerprint responses were generated. This is a general conclusion in the field of differential sensor arrays and is akin to the conclusions drawn from the ongoing work in our and McDevitt's laboratories. Shimizu also determined that an individual analyte would have the strongest interaction with the sensor specifically templated for that analyte. Therefore, the selectivity of the array arose from the templating process.
Identification of differences in the response patterns was done using LDA and the raw responses from the array as charted in Fig. 9. Tight clustering of the analytes shows both the recognition capabilities of the array, and the reproducibility of the responses. Furthermore, the nontemplated analyte, 11, is clearly separated from the other analytes. Using MIP chemistry to access analytes not originally templated is certainly a powerful advance. Certainly, the advancement of imprinting technologies may lead to exciting new developments in differential sensing with synthetic arrays.
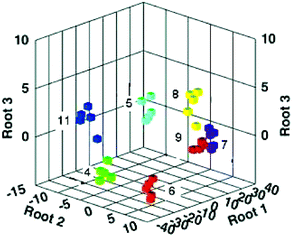 | ||
| Fig. 9 A three-dimensional LDA plot that describes 99.7% of the variance in the original data. Response patterns to the seven analytes from the MIP array are demonstrated. Reprinted with permission from Shimizu et al.,24J. Am. Chem. Soc. 2005, 127, 5695. © 2005 American Chemical Society. | ||
4 Porphyrin-incorporating differential arrays
Porphyrins and metalloporphyrins are excellent choices for use in aqueous recognition of analytes using differential sensor arrays. Both are readily modifiable, stable, and have large surface areas for binding. Furthermore, they can be used in both fluorescent and colorimetric signalling assays. These qualities have contributed to recent advances in sensor array technologies utilizing these adaptable synthetic receptors exclusively,19 and in conjunction with other pH and solvatochromic indicators.254.1 Porphyrin fluorescent surface protein receptors
Hamilton19 has created a sensor array for the detection of multiple proteins using tetraphenylporphyrins (TPP). These porphyrins have large hydrophobic surface areas that are excellent for protein recognition. Derivatization of the TPP periphery with various amino acids and dipeptides resulted in library 13 with receptors encompassing differing charges, size, hydrophobicity, and symmetry well-suited to the recognition of proteins with various surface characteristics. The TPP derivatives also are highly fluorescent making signal detection and pattern development facile.A “one-pot” synthesis (Scheme 2) resulted in 13 with 35 members isolated and purified. These 35 members were derivatized with four to eight hydrophobic groups and comprised all charge combinations from +8 to −8. Eight of the TPP derivatives were arranged in 8 rows of a 96 well quartz plate. Four proteins (15 µM) with varying pIs (ferredoxin (pI 2.75), cytochrome c551 (pI 4.7) myoglobin (pI 6.8), and cytochrome c (pI 10.6)) were tested against each receptor resulting in various distinguishable quenching patterns (irradiated with UV light at 302 nm) as seen in Fig. 10. The stronger the interaction between receptor and protein, the greater fluorescence quenching. However, this can not be related to the relative binding affinities as the mechanism of quenching may vary from receptor to receptor based upon the associated protein.
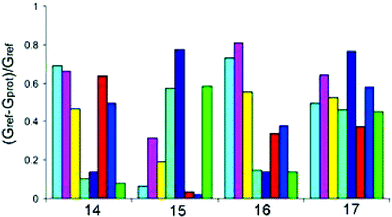 | ||
| Fig. 10 Patterns for cytochrome c (14), ferredoxin (15), cytochrome c551 (16), and myoglobin (17) based on the eight membered array of TPP derivatives. The bars quantify the extent of color attenuation measured as the quantity (Gref − Gprot)/Gref, where Gref is the average gray value for the blank wells and Gprot is the average gray value for the wells with proteins. Reprinted with permission from Hamilton et al.,19J. Am. Chem. Soc. 2004, 126, 5656. © 2004 American Chemical Society. | ||
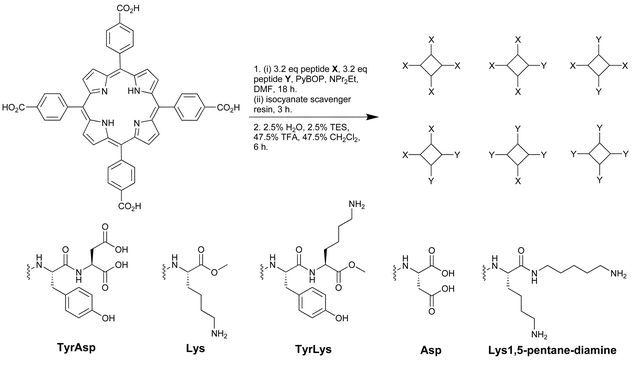 | ||
| Scheme 2 A mixed condensation synthesis resulted in 13 comprised of TPP derivatives with various peptidic components in the X and Y positions. Reprinted with permission from Hamilton et al.,19J. Am. Chem. Soc. 2004, 126, 5656. © 2004 American Chemical Society. | ||
The response of the array is directly correlated to the charge complementarity with the proteins. The more basic receptors showed increased fluorescence quenching with the acidic protein ferredoxin, and the more acidic receptors showed increased quenching with basic cytochrome c. The more neutral myoglobin showed interactions with nearly every protein in the array.
4.2 Colorimetric array for organics in water
Detecting organic compounds in an aqueous environment is inherently difficult due to the high concentration of water. Suslick25 printed a hydrophobic surface with the following: 1) metalloporphyrins which are metal-ion containing dyes that respond to Lewis basicity, 2) derivatives of and precursors to various pH indicators that respond to Brønsted acidity and basicity, and 3) conventional solvatochromic dyes with large permanent dipoles that respond to local environment polarity. These arrays were made of 36 various indicators and were used to detect organic compounds (concentrations as low as 1 µM) and a number of common soft drinks with colorimetric patterns.The array is initially saturated in an aqueous liquid that does not contain any analytes, and is then imaged by a flatbed scanner. Then an analyte is introduced, and following rapid colorimetric changes a second image is obtained by the scanner. Each printed receptor on the array is represented by a single spot that is the absolute value of the RGB color changes. As illustrated in Fig. 11, the color change profiles give diagnostic patterns for the organic analytes in aqueous solvents. It should also be mentioned that the pH was set at 7.0, so the pH indicators do not necessarily respond solely to pH, but also other interactions.
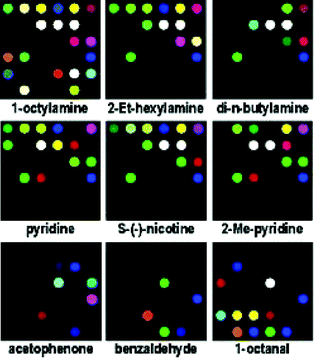 | ||
| Fig. 11 A base-sensitive array was used to obtain the color change profiles exhibited for various organic compounds in aqueous solvents (all amines 10 mM, all other analytes 50 mM, in pH 7 phoshate buffer). Not all patterns are shown from the original publication. Reprinted with permission from Suslick et al.,25J. Am. Chem. Soc. 2005, 127, 11548. © 2005 American Chemical Society. | ||
The authors used hierarchical cluster analysis (HCA) to identify familial characteristics in the patterns observed. They found that the array of printed receptors was capable of distinguishing such subtle differences as primary versus secondary amines and branched versus cyclic amines. They also determined that the HCA did not have any misclassifications out of 144 cases. Finally, it was determined that the limit of discrimination is very large with the possible number of distinct patterns being 34.8
As an extension to the identification of various organics in aqueous solutions, color change profiles (as in Fig. 11) were obtained for a number of soft drinks with an acid-sensitive array. Though this is not a mixture analysis that identifies the individual components, it does readily identify one complex mixture from another. Soft drinks show the ability to discriminate analytes in low concentration in the presence of a common high concentration species such as fructose. This is a nice real-world example for employing a sensor array, and facile discrimination of the soft drinks was exhibited.
This scientific example of a sensor array demonstrates a judicious use of readily available sensor components. This is a much simplified process that still yields excellent results. Formation of many arrays with similar usage of available components is likely to be seen in the future, and is a continued advancement of sensor technologies.
5 Microtiter plate arrays
Microtiter plate (MTP) technology is a readily available and disposable format for sensor arrays. It is an easily prepared flexible format that allows incorporation of many different types of receptors and assays such as IDAs. In this section two studies by Wolfbeis are reviewed, but other examples exist for using this technology including Hamilton's previously discussed work. Indeed, MTP arrays are a widely employed and accepted sensor format.5.1 Multi-ion imaging with fluorescent sensors
Wolfbeis26 employed a MTP array for the detection of several ions, and mixtures of ions. In this study a uniform analytical protocol was employed with fluorescent indicators dispersed into water soluble thin films of poly(ethylene glycol) on the bottom of MTP wells. A CCD for optical detection of fluorescence changes was also incorporated into the array.Fluorescent indicators were chosen that have very similar excitation (excite with blue LED at 470 nm) and emission wavelengths (>505 nm), as well as similar decay times (2–6 ns). Upon addition of one of six ions (Ca2+, Mg2+, Na+, Hg2+, SO42−, and Cl−), or mixtures thereof, at concentrations ranging from 0.1–25 mM a quantifiable fluorescence signal was captured using the CCD. However, after obtaining problematic fluorescence results they incorporated a new spectroscopic scheme called dual-lifetime referencing (DLR). This method converts fluorescence intensity information into a time dependent manner. In DLR, the decay profile of an indicator is referenced against the phosphorescence of beads added as reference dyes.
Due to the use of DLR, the quantitative information was not analyzed, and a simple “on/off” traffic light signalling system was developed for pattern recognition. In this case high or low luminescence intensities are distinguished by set threshold values. Therefore, if the luminescence is enhanced beyond the threshold limit for a particular dispersed indicator in an MTP well, that well is changed from red to green. If a strong response is actually a quenching mechanism, then that too will go from red to green if a low threshold limit is passed. Because the CCD was recording grayscale images, this technique significantly eased the analysis and made patterns readily identifiable by visual inspection. The traffic light patterns for ions and ion mixtures are illustrated in Fig. 12.
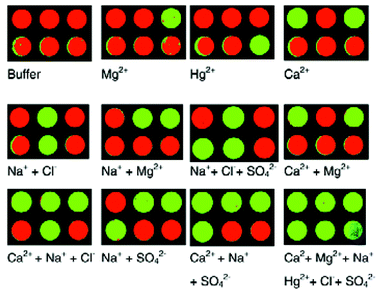 | ||
| Fig. 12 Traffic light patterns for various ions and ion mixtures. Green indicates the presence of an ion, and red indicates the lack of an ion. Reprinted with permission from Wolfbeis et al.,26Analyst, 2002, 127, 201. © 2002 The Royal Society of Chemistry. | ||
5.2 Metal ion sensor array
Wolfbeis27 refined the previously mentioned array system by improving the imaging setup for the recognition of five divalent metal cations. This improvement included developing a homogenous light field with a single blue LED for illumination of each well in the MTP. It also included a CCD image capture assay that did not require amplification, yet could detect ion concentrations as low as 10 µM. Furthermore, all 96 wells of a MTP could be read within a few microseconds. This enhanced assay led to greater imaging and recognition capabilities, and good pattern analysis using the chemometric tool of support vector machines, which is a learning tool for classification of unknowns.In this array study nine commercially available fluorescent indicators were dispersed in polymeric thin films. The excitation of the indicators occurred between 400–500 nm, and emissions at >500 nm. The decay times were in the nanosecond range. Again, DLR was employed such that the fluorescence decay of the indicator was referenced against the phosphorescence of an inert dye molecule bound to a bead that was placed in each MTP well. Also, as before, pseudo color changes were developed based upon exceeded threshold detection limits resulting in diagnostic patterns.
Using a leave-one-out accuracy classification method, the error rate of support vector machine classifications was evaluated. At a concentration greater than 10 µM, the error rates for the classification of the five divalent cations were never worse than 17.7% and were as little as 6.9%. It was found that higher classification errors were related to the number of indicators a particular analyte responded to, and more training patterns may improve the classifications.
The use of DLR and the improved imaging capabilities of this most recent differential recognition assay demonstrate both the power and capabilities of this method. Data can be rapidly acquired since multiple parameters can be measured simultaneously. There is great promise for methods such as this in water and environmental analysis, medical diagnostics, and high-throughput screening.
6 Synthetic biomolecules for array development
Tailoring of biomolecules through synthetic chemical modifications, mutations, and biochemical mismatches has also permitted the development of receptors. Following this lead, Landry28 has developed biomolecular receptors with nucleic-acid based three-way junctions that promote the formation of lipophilic cavities. These have been used in an array format for the characterization of hydrophobic molecules in solution.Three-way junctions are formed at the intersection of three or more double helices. At these junctions are three exposed aromatic surfaces of unstacked base pairs creating the lipophilic cavity. To achieve differential binding to multiple guest molecules, mutations and mismatched base pairs (bulges) could be introduced to the three-way junction structures leading to the creation of several different receptors. To identify binding events a fluorescent molecule was covalently bound to a phosphorothioate handle. Thus, the fluorescent molecule is initially within the lipophilic cavity, and upon analyte introduction it is displaced resulting in a signal modulation.
Incorporating a known selective three-way junction cocaine receptor, a modified variant was developed with the appended fluorescein (Fig. 13). This derivative was also tested against other hydrophobic steroid molecules that are potential targets in “mix and measure” assays for urine samples. Current assays for detecting these steroids are multistep and tedious. The single receptor was shown to differentially respond to various hydrophobic molecules. By placing several such receptors in an array, all with differential selectivity toward the analytes, the binding events can be incorporated into a diagnostic pattern.
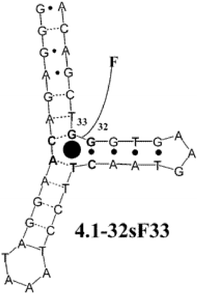 | ||
| Fig. 13 Structure of the modified cocaine variant (4.1–32sF33). Reprinted with permission from Landry et al.,28J. Am. Chem. Soc. 2003, 125, 6085. © 2003 American Chemical Society. | ||
The other three-way junction receptors were developed by changing the shape of the cavity by base-pair mismatches and through repositioning of the fluorescein at various positions around the cavity. Hundreds of possible variants can be developed using these methods. Eight structural variants were placed in an array, and their increase in fluorescence intensity determined upon introduction of a guest. Fig. 14 shows the bar graph patterns for the four different guest molecules. The power of a differential cross-reactive array is evident here as it is unlikely that a single receptor could have adequately discriminated the four hydrophobic guests. It was also shown that these arrays could be used for quantitative analysis.
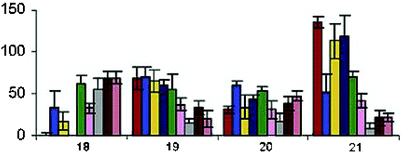 | ||
| Fig. 14 Patterns based upon the responses of eight three-way junction receptors toward the following hydrophobic guests: cocaine hydrochloride 18 (500 µM), deoxycorticosterone 21-glucoside 19 (32 µM), dehydroisoandrosterone 3-sulfate sodium 20 (125 µM), and sodium deoxycholate 21 (2 mM). Triplicate measurements were taken and standard deviations are shown. Concentrations were used for each guest that would result in a 50–70% fluorescence intensity response. Reprinted with permission from Landry et al.,28J. Am. Chem. Soc. 2003, 125, 6085. © 2003 American Chemical Society. | ||
As a last example of the power of their three-way junction arrays, the arrays were used to develop a characteristic pattern for a urine standard. By comparing the pattern of the standard to that of patterns of urine spiked with either dehydroisoandrosterone 3-sulfate or deoxycorticosterone 21-glucoside, the steroids were discriminated. This is a nice real-world example of synthetic solution-based differential arrays. As described earlier, it is imperative that differential arrays can be refined and improved to, at a minimum, qualitatively detect analytes in complex mixtures and solutions.
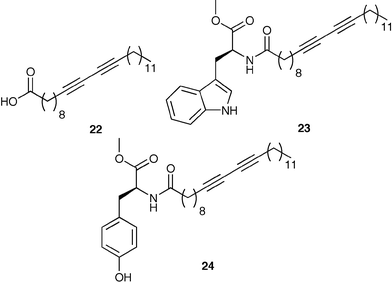
A second example of synthetic biomolecules in arrays was completed by Basu.29 In this study two liposomes were prepared by polymerization of 22 (95 mol%) and either 23 or 24 (5 mol%). These liposomes change color from red to blue when bound to an analyte. Using this rather simple two sensor array, lipopolysaccharides (LPS) from various Gram negative bacteria were discriminated. Lipopolysaccharides are complex glycolipids embedded within the outer membranes of Gram negative bacteria. A single cell contains as many as two million copies of the LPS.
The LPS have terminal sugars that are unique to each bacterial type. Therefore, identification of the terminal sugars directly correlates to the various Gram negative bacteria. The colorimetric responses (CR) from the synthetic liposomes form the patterns necessary for fingerprint identification of the bacterial types. The authors define the CR as the change in the ratio of absorbances at 640 and 550 nm in the presence and absence of LPS. The overall pattern was enhanced by using the two sensors in four different conditions. Therefore, a total of eight responses from only two differential sensors gave diagnostic bar-graph patterns.
The four conditions employed were: 1) room temperature (rt), 2) 35 °C, 3) with the detergent sodium dodecyl sulfate (SDS), and 4) with EDTA. The detergent likely affects the aggregation of LPS, and EDTA sequesters divalent cations interacting with phosphate groups in the LPS. Increased temperature enhances the CR. The CR values were obtained from interactions between the liposomes and five Gram negative bacterial LPS: Escherichia coli O26∶B6, Pseudomonas aeruginosa, Salmonella enteriditis, Salmonella minnesota, and Shigella flexneri.
The five patterns generated were unique enough for discrimination of the five bacterial LPS. The importance of the additives and increased temperatures was demonstrated, especially in discriminating between E. coli and P. aeruginosa where the EDTA experiment was needed.
This research provides a nice background for more intense research with greater numbers of analytes and receptors, and the use of chemometric tools. This is a novel assay for discriminating between strains of bacteria.
7 Other arrays and assays using IDAs
Several applications have been found for the use of IDAs, and as discussed earlier there has been good success using them in synthetic sensor arrays. In this last section three additional cases are reviewed in which the power of the arrays lie in the usage of an IDA. In these cases there are only one or two receptors used, but multiple indicators are used creating differential equilibria and multiple data. Chemometric tools were employed to determine the relationship between patterns.7.1 Identification of 20 natural amino acids
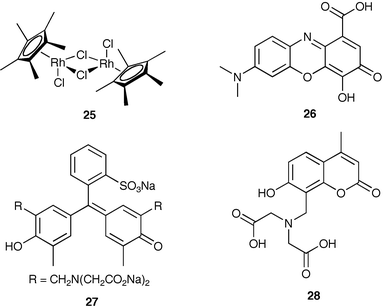
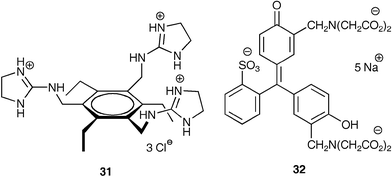
The sensitivity and selectivity of an assay for analytes can be modulated by changing the pH, or other conditions, while using an IDA. Sensing of amino acids, especially nonfunctionalized amino acids such as alanine and valine, is very difficult due to the lack of specific receptors. Due to this, Severin30 realized that a single synthetic sensor array incorporating commercially available units could be employed for identifying 20 natural amino acids.Severin used an organometallic Cp*Rh complex (25) as the amino acid receptor. The receptor has fast exchange kinetics for the three facial coordination sites opposite the Cp* ligand, is water soluble and air stable, and has a strong affinity for amino acids. Three indicators were used in the assay, gallocyanine (26), xylenol orange (27), and calcein blue (28). The indicators and amino acids have similar affinities for 25 resulting in a competition for at least one indicator–amino acid combination. Therefore, in this study they used multiple receptor–indicator combinations at various pH values which change the strength of associations resulting in multiple data points for each analyte. These points were analyzed with LDA and PCA to determine classification errors and separation of analytes.
In their study 20 natural amino acids were initially associated with a complex of 25 and 26 at pH 7.0. The amino acids with the strongest associations (as determined by near complete indicator-displacement) were placed in Group I, and all others into Group II. Following classifications each Group I member was analyzed by four different receptor/indicator combinations at multiple pH, and Group II members were analyzed by 25 and 26 at five pHs. All trials were completed 12 times. The predictive behavior of the assay was determined by a leave-one-out validation routine. After a single measurement is removed, the remaining data is used for developing a LDA. The error rate of misclassification in Group I was 0.6%, and 3.3% for Group II due to misclassifications of Val and Leu. A PCA of Group II amino acids shows excellent clustering of the individual amino acids, and nice separation in general, except minor overlaps between Val and Leu (Fig. 15). This work demonstrates the utility of an IDA assay with commercially available components to achieve high sensing capabilities. It is likely that this type of assay will be employed for the analysis of complex analytes such as proteins and complex mixtures.
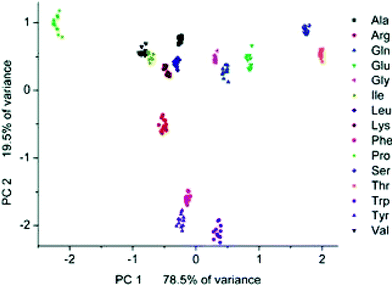 | ||
| Fig. 15 PCA score plot for Group II amino acids. The two principal component axes contain 98% of the variance in the original data set. Reprinted with permission from Severin et al.,30J. Am. Chem. Soc. 2005, 127, 3700. © 2005 American Chemical Society. | ||
7.2 Multicomponent sensing ensembles
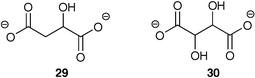
The electronic array utilizing a Si/SiN wafer was described earlier. Though it is a powerful tool for the recognition of analytes and mixtures, it is a relatively complex assay. Therefore, we have pursued simpler sensing ensembles using widely available UV/Vis spectrophotometers.As a first example, homogenous solutions of receptor and indicator combinations were used to report environmental responses to two similar analytes, malate (29) and tartrate (30).31 Two hexasubstituted benzene receptors, both incorporating boronic acids and guanidiniums, were placed in a single solution with two indicators, bromopyrogallol red and pyrocatechol violet. These indicators have differing affinities to the two receptors, and their absorbance spectrums span a large wavelength area. Therefore, several absorbance data points from each of multiple UV/Vis spectra can be used to generate patterns and quantify the tartrate and malate concentrations.
In practice, a four component solution was prepared: two different receptors (each 150 µM), pyrocatechol violet (60 µM), and bromopyrogallol red (30 µM). The concentrations of the receptors and indicators were held constant, and increasing concentrations of the two analytes were added simultaneously (0.2 mM increments). Forty-nine distinct spectra were obtained with 27 data points collected from each.
All the data was analyzed using the supervised learning method of classification called multilayer perceptron artificial neural networks (MLP-ANN). This chemometric tool has an input and output channel and a hidden layer where data is processed (learned). The network was initially trained using 27 data points from 45 of the 49 UV/Vis spectra. The four trials left out were then entered into the network to test its training. It was found that the network was able to predict the concentrations of both tartrate and malate in the four unknowns with a maximum error rate of only 6%. Additional experiments and further training of the network reduced the maximum error to 2%. This single vessel differential sensing assay effectively predicted the concentrations of two analytes using a pattern recognition algorithm. This array-less assay avoids the need for large numbers of receptors, and may be applied to the analysis of multiple analytes.
An additional study was carried out that was very similar to the previous one,32 except with two added dimensions of difficulty. First, a cross-reactive indicator was added that both associates with the receptor, and with one of the analytes. Therefore, the indicator is one of the differential components along with the receptor. Second, the complexity of the analysis was increased by evaluating vodka solutions, thus demonstrating a real-world analysis.
In this report the hexasubstituted benzene receptor, 29, and the cross-reactive indicator xylenol orange, 30, were placed in a single cuvette and used to measure the concentrations of citrate and Ca(II) in doped vodka solutions (25% v/v vodka in water). After the initial doped measurements, citrate and Ca(II) concentrations were determined from a neat vodka sample.
The single cuvette has multiple equilibria occurring simultaneously. Binding of citrate to 29 releases 30 so it can bind Ca(II). Binding of Ca(II) by 32 frees up citrate to bind 31. Though multiple equilibria are occurring, only the UV/Vis absorbance spectrum of 32 reports the activities within the solution. As with the previous study, the two analytes were mixed at various amounts (Ca(II), 0–400 µM, 50 µM increments; citrate, 0–800 µM, 100 µM) to obtain 80 spectra, from which 25 data points were extracted. As shown in Fig. 16, subtle differences in the absorbance spectrum of 32 upon exposure to varying concentrations of Ca(II) and citrate can generate enough data to use a chemometric tool to determine quantitative values.
![A UV/Vis absorbance spectrum of an IDA created from a solution (25% v/v vodka in water with 10 mM HEPES at pH 7.5) containing 31 (240 µM) and 32 (10 µM). Two spectra are shown demonstrating various concentrations of the two analytes. [(): Ca(ii) 400 µM, citrate 100 µM. (): Ca(ii) 50 µM, citrate 600 µM.] Reprinted with permission from Anslyn et al.,32Tetrahedron, 2003, 59, 10089. © 2003 Elsevier.](/image/article/2006/CS/b505518k/b505518k-f16.gif) | ||
Fig. 16 A UV/Vis absorbance spectrum of an IDA created from a solution (25% v/v vodka in water with 10 mM HEPES at pH 7.5) containing 31 (240 µM) and 32 (10 µM). Two spectra are shown demonstrating various concentrations of the two analytes. [(![[thick line, graph caption]](https://www.rsc.org/images/entities/char_e117.gif) ): Ca(II) 400 µM, citrate 100 µM. ( ): Ca(II) 400 µM, citrate 100 µM. (![[dash dash, graph caption]](https://www.rsc.org/images/entities/char_e091.gif) ): Ca(II) 50 µM, citrate 600 µM.] Reprinted with permission from Anslyn et al.,32Tetrahedron, 2003, 59, 10089. © 2003 Elsevier. ): Ca(II) 50 µM, citrate 600 µM.] Reprinted with permission from Anslyn et al.,32Tetrahedron, 2003, 59, 10089. © 2003 Elsevier. | ||
Again a MLP-ANN was used for determining the reliability of the assay to quantify the concentrations of the two analytes. In this study, 75 of the 80 spectra were used for training. The five left out were then put into the network and the predicted values were correlated with the actual values. This time the error ranged from 3.4–30%. Despite this, studies continued to analyze just the citrate concentrations in five flavored vodkas. Comparing the predicted citrate concentrations from the ANN to NMR-determined concentrations resulted in differences ranging from 1.7–33%. Though this study generated greater error than the previous, it demonstrates a real-world application using a simple single vessel assay that can be replicated using readily available UV/Vis spectrophotometers.
As a final example of a single-molecule multianalyte sensor, Suzuki33 developed a jewel pendant ligand for the colorimetric UV/Vis detection of metal cations. Three dyes were linked to a metal ligand comprised of three heteroatoms (N, O, S) having differential hard–soft acid–base (HSAB) characteristics. The multichromogenic receptor absorbs with two peaks (340 and 710 nm) and a shoulder (460 nm). Upon binding to a metal cation (Mg(II), Ca(II), Mn(II), Co(II), Ni(II), Zn(II), Ag(I), Hg(II), Cu(II), Fe(III), Al(III), Cr(III), and Pb(II); 10 eq. of each added), the ligand's absorbance changes. The change in absorbance at the three key wavelengths represented the signature pattern for each cation. In some cases only a single wavelength changed reflecting the HSAB nature of the receptor. The authors went on to measure 25 combinations of Cu(II) and Fe(III) in the 3–50 µM range in acetonitrile. Using a back-propagation ANN method the concentrations of the cation mixtures were simultaneously estimated. They found a root mean-square of prediction of R = 0.94 for Cu(II) and R = 0.9 for Fe(III) indicating the ability of the single-molecule metal cation sensor.
8 Fluorescent self-assembled monolayers
As a final example, Crego-Calama34 utilized a glass substrate functionalized with a combinatorially derived set of fluorescent self-assembled monolayers for the aqueous pattern recognition of metal cations. Two binding molecules and three fluorophores were sequentially reacted with an 11-aminoundecyltrichlorosilane self-assembled monolayer to give a nine-membered receptor library. The functionalized glass slides were placed at a 45 degree angle in a cuvette and solutions of the chloride salts of Hg2+, Cu2+, Ca2+, and Co2+ (10−4 M) were added. Fluorescence emissions were measured for each of the nine receptors, which corresponded to the pattern for each cation. The nature of the fluorophore and binding groups influenced the observed differential response. This was the first report of ion sensing in water by fluorescence and a self-assembled monolayer on a glass slide.9 Conclusions
Several arrays and assays exist for differential sensing of various analytes using synthetic and commercially available components. Differential sensing permits the recognition and discrimination of complex analytes and mixtures of analytes. They have also been applied to some real-world situations. The arrays can be readily tailored to accommodate a wide swath of analytes and will likely see increased usage and applications as the technology is refined and enhanced. Though differential sensing does not replace the standard lock-and-key recognition protocol, it does allow the analysis of complex analytes and mixtures, a task that is laborious for standard protocols. Differential arrays will likely be used in new technologies for detecting bioterrorism agents, environmental analysis, medical diagnostics, and other societal needs.References
- P. C. Jurs, G. A. Bakken and H. E. McClelland, Chem. Rev., 2000, 100, 2649 CrossRef CAS.
- K. J. Albert, N. S. Lewis, C. L. Schauer, G. A. Sotzing, S. E. Stitzel, T. P. Vaid and D. R. Walt, Chem. Rev., 2000, 100, 2595 CrossRef CAS.
- J. P. Epstein and D. R. Walt, Chem. Soc. Rev., 2003, 32, 203 RSC.
- K. Toko, Biosens. Bioelectron., 1998, 13, 701 CrossRef CAS.
- K. Toko, Meas. Sci. Technol., 1998, 9, 1919 CrossRef CAS.
- D. R. Walt, Acc. Chem. Res., 1998, 31, 267 CrossRef CAS.
- I. R. Laukis, Acc. Chem. Res., 1998, 31, 317 CrossRef CAS.
- J. J. Lavigne, S. Savoy, M. B. Clevenger, J. E. Ritchie, B. McDoniel, S.-J. Yoo, E. V. Anslyn, J. T. McDevitt, J. B. Shear and D. Neikirk, J. Am. Chem. Soc., 1998, 120, 6429 CrossRef CAS.
- A. Goodey, J. J. Lavigne, S. M. Savoy, M. D. Rodriguez, T. Curey, A. Tsao, G. Simmons, J. Wright, S.-J. Yoo, Y. Sohn, E. V. Anslyn, J. B. Shear, D. P. Neikirk and J. T. McDevitt, J. Am. Chem. Soc., 2001, 123, 2559 CrossRef CAS.
- N. Christodoulides, M. Tran, P. N. Floriano, M. Rodriguez, A. Goodey, M. Ali, D. Neikirk and J. T. McDevitt, Anal. Chem., 2002, 74, 3030 CrossRef CAS.
- T. Curey, A. Goodey, A. Tsao, J. Lavigne, Y. Sohn, J. T. McDevitt, E. V. Anslyn, D. Neikirk and J. B. Shear, Anal. Biochem., 2001, 293, 178 CrossRef CAS.
- J. J. Lavigne and E. V. Anslyn, Angew. Chem., Int. Ed., 2001, 40, 3118 CrossRef.
- A. Furka, F. Sebestyen, M. Asgedom and G. Dibo, Int. J. Pept. Protein Res., 1991, 37, 487 CAS.
- S. C. McCleskey, M. J. Griffin, S. E. Schneider, J. T. McDevitt and E. V. Anslyn, J. Am. Chem. Soc., 2003, 125, 1114 CrossRef CAS.
- A. T. Wright, M. J. Griffin, Z. Zhong, S. C. McCleskey, E. V. Anslyn and J. T. McDevitt, Angew. Chem., Int. Ed., 2005, 44, 6375 CrossRef CAS.
- A. T. Wright, E. V. Anslyn and J. T. McDevitt, J. Am. Chem. Soc., 2005 Search PubMed submitted.
- For a detailed review see: S. L. Wiskur, H. Ait-Haddou, J. J. Lavigne and E. V. Anslyn, Acc. Chem. Res., 2001, 34, 963 Search PubMed.
- S. Schneider, S. O'Neil and E. V. Anslyn, J. Am. Chem. Soc., 2000, 122, 542 CrossRef CAS.
- L. Baldini, A. J. Wilson, J. Hong and A. D. Hamilton, J. Am. Chem. Soc., 2004, 126, 5656 CrossRef CAS.
- A. T. Wright and E. V. Anslyn, Org. Lett., 2004, 6, 1341 CrossRef CAS.
- A. P. Goodey and J. T. McDevitt, J. Am. Chem. Soc., 2003, 125, 2870 CrossRef CAS.
- T. Hirsch, H. Kettenberger, O. S. Wolfbeis and V. M. Mirsky, Chem. Commun., 2003, 432 RSC.
- N. T. Greene, S. L. Morgan and K. D. Shimizu, Chem. Commun., 2004, 1172 RSC.
- N. T. Greene and K. D. Shimizu, J. Am. Chem. Soc., 2005, 127, 5695 CrossRef CAS.
- C. Zhang and K. S. Suslick, J. Am. Chem. Soc., 2005, 127, 11548 CrossRef CAS.
- T. Mayr, G. Liebsch, I. Klimant and O. S. Wolfbeis, Analyst, 2002, 127, 203 Search PubMed.
- T. Mayr, C. Igel, G. Liebsch, I. Klimant and O. S. Wolfbeis, Anal. Chem., 2003, 75, 4389 CrossRef CAS.
- M. N. Stojanović, E. G. Green, S. Semova, D. B. Nikić and D. W. Landry, J. Am. Chem. Soc., 2003, 125, 6085 CrossRef CAS.
- M. Rangin and A. Basu, J. Am. Chem. Soc., 2004, 126, 5038 CrossRef CAS.
- A. Buryak and K. Severin, J. Am. Chem. Soc., 2005, 127, 3700 CrossRef CAS.
- S. L. Wiskur, P. N. Floriano, E. V. Anslyn and J. T. McDevitt, Angew. Chem., Int. Ed., 2003, 42, 2070 CrossRef CAS.
- S. C. McCleskey, P. N. Floriano, S. L. Wiskur, E. V. Anslyn and J. T. McDevitt, Tetrahedron, 2003, 59, 10089 CrossRef CAS.
- H. Komatsu, D. Citterio, Y. Fujiwara, K. Minamihashi, Y. Araki, M. Hagiwara and K. Suzuki, Org. Lett., 2005, 7, 2857 CrossRef CAS.
- R. Zimmerman, L. Basabe-Desmonts, F. van der Baan, D. N. Reinhoudt and M. Crego-Calama, J. Mater. Chem., 2005, 15, 2772 RSC.
| This journal is © The Royal Society of Chemistry 2006 |