Structural codons: linearity/helicity interconversion by pyridine/pyrimidine exchange in molecular strands†
Ibon
Odriozola
a,
Nathalie
Kyritsakas
b and
Jean-Marie
Lehn
*a
aInstitut de Science et d'Ingénierie Supramoléculaires (ISIS), 8 allée Gaspard Monge, BP 70028 67083 Strasbourg cedex, France. E-mail: lehn@isis.u-strasbg.fr
bService Commun de Rayons X, 4 rue Blaise Pascal, F-67070 Strasbourg cedex, France. E-mail: sercomrx@chimie.u-strasbg.fr
First published on 12th November 2003
Abstract
Pyridine and pyrimidine groups connected through amide functions can be combined into specific sequences that self-organize into either helical or linear structures enforced by the formation of intramolecular hydrogen bonds.
The control of the folding of molecular strands into well-defined architectures has been a subject of great interest in recent years in view of both its importance in molecular design and its significance for the understanding of the conformational features of biomolecules.1 Various intramolecular non-bonded interactional effects can be combined into specifically designed structure enforcing units, “structural codons”, capable of inducing into molecular strands well-defined shapes spanning the domain between helicity and linearity. In our laboratory, we have in particular made use of “helicity codons” involving specific combinations of heterocycles2 and pyridine carboxamide sequences3 to enforce helicity through respectively internal rotational preferences and hydrogen bond formation. Thus, molecular strands composed of alternating 2,6-diaminopyridine and 2,6-pyridinedicarbonyl units have been shown to adopt a helical conformation both in solution and in the solid state.3
We now report three new oligoamide model sequences showing that the exchange of pyridine by pyrimidine units results in a modification of the conformation of molecular strands, resulting in linearity/helicity interconversion.
The four pyridine and pyrimidine derived diamino and dicarbonyl groups shown in Fig. 1 may be classified as “linear” or “curved” units, according to the relative orientation of their termini in the conformation imposed by their hydrogen bonding pattern. These units can be combined in four different manners to give four different structural motifs. Thus, alternating linear units result in the formation of a linear strand; the alternating curved units lead to undulating forms; and finally, the two possible combinations of one linear and one curved unit generate helical oligoamide strands.
 | ||
| Fig. 1 Schematic representation of: (a) the four pyridine and pyrimidine-based monomeric units, and (b) four structurally different molecular strands composed of the combination of these monomeric units. The broken lines are meant to represent favourable electrostatic interactions. | ||
The corresponding four types of trimers4 shown in Fig. 2 are a representative example of these sequences. In each case the strand is represented in the conformation in which a maximum number of favourable electrostatic interactions (akin to markedly bent hydrogen bonds) can be established. The amide protons are in all the cases oriented so as to be surrounded by two aromatic nitrogen atoms, and the carbonyl oxygens are located at the same time between two aromatic protons, presenting respectively two favourable N–H⋯N and (weaker) C–H⋯O interactions.
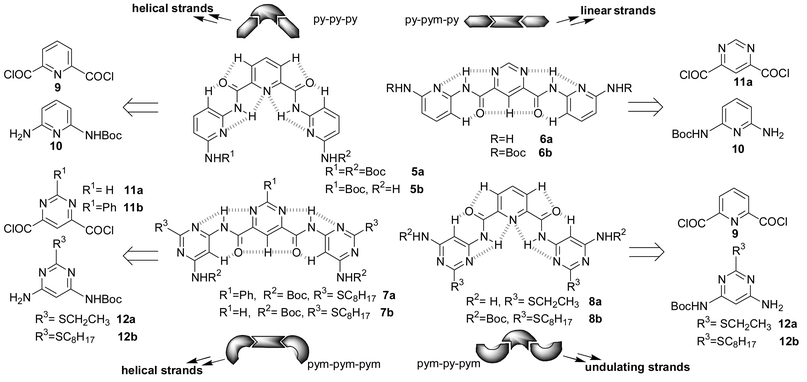 | ||
| Fig. 2 Schematic representation, pattern of favourable electrostatic interactions and retrosynthetic pathway for the four types of trimers. | ||
These oligomers (5–8) were prepared from the corresponding diacid5 chloride and Boc-monoprotected diamine,6 following the procedure described for compounds 5a,b (Fig. 2) in ref. 3b. Their analytical and spectroscopic data (NMR, mass) are in agreement with the assigned structure.
The conformations of compounds 5–8 were found to be as represented in Fig. 2 both in solution and in the solid state. Analysis of their 1H NMR and NOESY spectra clearly indicated, as expected, the absence of any short range H,H interactions. The conformations were confirmed by determination of the crystal structures of compounds 6b,77a and 8a8 (Fig. 3).‡ As seen, in all the cases the relative orientation of the heterocycles and the amide linkages is as depicted in Fig. 2.
 | ||
| Fig. 3 Views of the structure of one of the two independent molecules in the crystals of 6b (a), of 7a (b) and 8a (c). Some non-bonded N⋯H and O⋯H distances are given in Å. | ||
These spectroscopic and structural data indicate that the patterns of favourable electrostatic interactions enforce the predicted orientation of the different residues and that the replacement of a pyridine ring by a pyrimidine indeed interconverts the conformation of the tris-heterocyclic entities between the linear and the helical ones.
It is worth noting that the previously developed py–py strands3 fold so that the carbonyl oxygens are directed outside the helix whereas in the pym–pym strands these oxygens are oriented towards the inside, which makes them suitable for metal ion coordination and promising building blocks for the construction of ion channels.
In conclusion, three new sequences of oligoamides have been designed, which, by the appropriate choice of monomeric units, open up the possibility of constructing molecular strands designed to generate linear, undulating or helical shapes. The results obtained further extend the ability to direct the folding of molecular strands for both chemical and biological purposes.
A postdoctoral fellowship from Eusko Jaurlaritza/Govierno Vasco for I. O. is gratefully acknowledged.
Notes and references
- (a) D. J. Hill, M. J. Mio, R. B. Prince, T. S. Hughes and J. S. Moore, Chem. Rev., 2001, 101, 3893 CrossRef CAS; (b) S. H. Gellman, Acc. Chem. Res., 1998, 31, 173 CrossRef CAS; (c) A. E. Rowan and R. J. M. Nolte, Angew. Chem., Int. Ed., 1998, 37, 63 CrossRef CAS.
- (a) G. S. Hanan, J.-M. Lehn, N. Kyritsakas and J. Fischer, J. Chem. Soc., Chem. Commun., 1995, 765 RSC; (b) D. M. Bassani, J.-M. Lehn, G. Baum and D. Fenske, Angew. Chem., Int. Ed. Engl., 1997, 36, 1845 CrossRef CAS; (c) M. Ohkita, J.-M. Lehn, G. Baum and D. Fenske, Chem. Eur. J., 1999, 5, 3471 CrossRef CAS; (d) L. A. Cuccia, E. Ruiz, J.-M. Lehn, J.-C. Homo and M. Schmutz, Chem. Eur. J., 2002, 8, 3448 CrossRef CAS; (e) A. Petitjean, L. A. Cuccia, J.-M. Lehn, H. Nierengarten and M. Schmutz, Angew. Chem., Int. Ed., 2002, 41, 1195 CrossRef CAS; (f) J.-L. Schmitt, M. Stadler, N. Kyritsakas and J.-M. Lehn, Helv. Chim. Acta, 2003, 86, 1598 CrossRef CAS.
- (a) V. Berl, I. Huc, R. Khoury, M. J. Krische and J.-M. Lehn, Nature, 2000, 407, 720 CrossRef CAS; (b) V. Berl, I. Huc, R. Khoury and J.-M. Lehn, Chem. Eur. J., 2001, 7, 2798 CrossRef CAS; (c) V. Berl, I. Huc, R. Khoury and J.-M. Lehn, Chem. Eur. J., 2001, 7, 2810 CrossRef CAS.
- Synthesis and structural characterization of longer oligomers are in progress.
- The acid chlorides 11a and 11b were formed by boiling the corresponding diacids in SOCl2 for 24 h and recrystallized from hexane. For the preparation of 4,6-pyrimidinedicarboxylic acid see: R. R. Hunt, J. F. W. McOmie and E. R. Sayer, J. Chem. Soc., 1959, 525 Search PubMed . The 2-phenyl substituted diacid was prepared analogously from 4,6-dimethyl-2-phenylpyrimidine.
- Compounds 12a,b were prepared from 4,6-diamino-2-mercaptopyrimidine and the corresponding alkyl bromide using a procedure described by: R. A. Nugent, S. T. Schlachter, M. J. Murphy, G. J. Cleek, T. J. Poel, D. G. Wishka, D. R. Graber, Y. Yagi, B. J. Keiser, R. A. Olmsted, L. A. Kopta, S. M. Swaney, S. M. Poppe, J. Morris, W. G. Tarpley and R. C. Thomas, J. Med. Chem., 1998, 41(20), 3793 Search PubMed . The Boc group was inserted using the same procedure described in ref. 3b for 5a.
- Two independent molecules of 6b were found in the asymmetric unit, both with crystallographic 2-fold imposed symmetry and both with similar conformations.
- Compound 8a presented crystallographic 2-fold imposed symmetry.
Footnotes |
| † Electronic supplementary information (ESI) available: spectroscopic (1H,13C-NMR, mass) data for compounds 6a,b; 7a,b and 8a,b. See http://www.rsc.org/suppdata/cc/b3/b311045a/ |
| ‡ Crystallographic data: 6b C26H30N8O6·0.375CH2Cl2·0.125H2O, M = 584.67, orthorhombic, a = 25.0498(1), b = 20.9704(3), c = 11.7186(3) Å, V = 6155.8(2) Å3, space group P21212, Z = 8, µ = 0.141 mm−1, 18009 data measurements, 8581 data measurements with I > 3σ(I), R = 0.087, Rw = 0.111; 7a C46H64N10O6S2·CH2Cl2, M = 1002.15, orthorhombic, a = 13.5362(2), b = 18.8739(2), c = 41.3548(7) Å, V = 10565.4(3) Å3, space group Pbca, Z = 8, µ = 0.257 mm−1, 15327 data measurements, 4705 data measurements with I > 3σ(I), R = 0.086, Rw = 0.103; 8a C19H21N9O2S2·H2O, M = 489.58, monoclinic, a = 11.4672(3), b = 29.3393(8), c = 7.8434(3) Å, V = 2280.4(1) Å3, space group C12/c1, Z = 4, µ = 0.276 mm−1, 5616 data measurements, 1426 data measurements with I > 3σ(I), R = 0.043, Rw = 0.068. CCDC 219231–219233. See http://www.rsc.org/suppdata/cc/b3/b311045a/ for crystallographic data in .cif or other electronic format. |
| This journal is © The Royal Society of Chemistry 2004 |
