In vitro selection and evaluation of rna aptamers that recognize arginine-rich-motif model peptide on a quartz-crystal microbalance
Shinobu Fukusho, Hiroyuki Furusawa and Yoshio Okahata*
Department of Biomolecular Engineering, Tokyo Institute of Technology, 4259 Nagatsuda, Midori-ku, Yokohama, 226–8501, Japan. E-mail: yokahata@bio.titech.ac.jp
First published on 21st December 2001
Abstract
To study RNA–peptide interactions, we performed an in vitro selection of RNA on a simple α-helical peptide-immobilized quartz-crystal microbalance (QCM) and evaluated the association constants (107 M−1) of the selected RNA to the model peptide on the same QCM plate.
RNA-binding proteins play a key role in fundamental cellular processes such as translation, mRNA processing and in viral processes on infection by RNA viruses.1 Understanding RNA–protein interactions is important in the study of how RNA-binding proteins specifically recognize the target RNA and for the design of drugs to inhibit infection of RNA viruses. RNA-binding domains of these proteins can be grouped into families such as a ribonucleoprotein motif, a double-strand RNA binding domain2 and an arginine-rich motif.3 A simple arginine-rich Rev peptide (17 amino acids) as a part of Rev protein (regulator of expression of virion proteins) from HIV-I has been studied to bind to an internal loop of a target RNA (RRE RNA) without a large protein unit.4 This indicates that the arginine-rich α-helical peptide can be expected to bind to the specific RNA sequence (see Fig. 1).
 | ||
| Fig. 1 Schematic illustrations of specific interaction between RRE RNA and Rev peptide and design of a simple α-helix model peptide. | ||
In this study, we designed a simple α-helical peptide as a model of the Rev peptide. The model peptide retains only five arginine residues that interact with phosphate anions of the target RNA and the other amino acids are replaced simply by alanines (Fig. 1). In vitro selection of RNA was studied by using the model peptide-immobilized quartz-crystal microbalance (QCM). QCM is a highly sensitive mass measuring device whose resonance frequency decreases linearly with the increase of a mass on the QCM electrode at the nanogram level in aqueous solutions.5,6 We were able to monitor a selection process from a random RNA pool as a mass change and evaluate an association constant of the selected RNA quantitatively on the same QCM plate, without recourse to radioactive labeling or fluorescent probes.
A schematic illustration of experimental setup is shown in Fig. 2. One side of the QCM plate is sealed with a rubber casing, maintaining it in air environment to avoid contact with the ionic aqueous solution.5a–c The model peptide was immobilized through a Cys-SH group with a long poly(ethylene oxide) spacer on the other side of the Au electrode (area: 5 mm2) of a 27 MHz QCM.5d The model peptide-immobilized QCM plate was soaked in the buffer solution and the frequency decrease (mass increase) responding to the addition of random ssRNA was monitored in the aqueous solution. After monitoring the selection process as frequency decrease (mass increase), the QCM plate was washed with the selection buffer (0.01 M HEPES, pH 7.5, 0.1 M NaCl) and the selected ssRNA was recovered with the elution buffer (0.01 M HEPES, pH 7.5, 1 M NaCl). The ssRNA was reverse transcribed to DNA and amplified with PCR. Then the dsDNA was transcribed to RNA and used for the next selections.7–10 Selection processes were repeated for 1–7 cycles.
 | ||
| Fig. 2 Experimental procedures of in vitro selection of RNA that shows a high affinity for the model peptide on a 27 MHz QCM plate. | ||
Selection processes were monitored as mass changes by using a QCM and results are summarized in Fig. 3. On the 1st cycle selection, random RNA was hardly bound to the model peptide on the QCM plate. With increasing selection cycles, the selected RNA was observed to bind reasonably to the model peptide. Since the frequency changes of the 6th to the 7th cycle of selections were constant, the selection was finished at the 7th cycle.
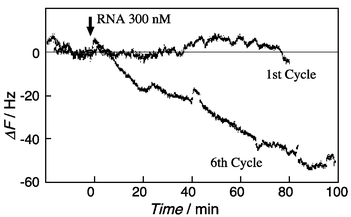 | ||
| Fig. 3 Monitoring of selection processes of RNA bound to the model peptide-immobilized QCM (0.01 M HEPES, pH 7.5, 0.1 M NaCl, at 20 °C). | ||
The selected RNA was reverse transcribed to DNA and was PCR-amplified, cloned by T/A cloning method, and the sequence for the determined by a standard dideoxynucleotide method using a ABI Prism 310 genetic analyzer (Applied Biosystems). The obtained sequences for the random region of 30 mer after the 7th cycle are summarized in Table 1 and the calculated secondary structure of Group I RNA is also shown.
| a Symbols of –, △, and bold letters show conserved, deletion sequences, and verified sequences, respectively. |
|---|
 |
 |
Binding behaviors of cloned RNAs to the model peptide-immobilized QCM plate are evaluated as shown in Fig. 4. A-3 RNA of Group I bound strongly to the model peptide, and saturation binding behavior was observed with increasing injection concentrations. From the reciprocal plots,5a–c the association constant of the selected A-3 RNA to the model peptide was found to be 7.2 × 107 M−1. This value is equal to or higher than association constants of Rev peptide to RRE RNA (2.3 × 107 M−1) or to the aptamers (2.7–5.3 × 107 M−1).7–10 We expected that the Arg-Ala model peptide would recognize an internal loop similar to the RRE RNA that is the recognition sequence of the Rev peptide (see Fig. 1). Contrary to expectation, there was no correlation between the selected Group I RNA and RRE RNA, even though the Group I RNA has a high affinity to the model peptide. It is likely that only five arginines are not enough to recognize particular internal loops, nevertheless the five arginines play an important role in the recognition of the specific RNA.
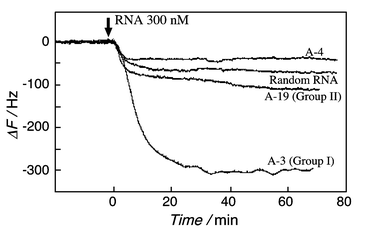 | ||
| Fig. 4 Evaluation of binding behavior of the RNA selected clones on the model peptide-immobilized QCM plate (0.01 M HEPES, pH 7.5, 0.1 M NaCl, at 20 °C). | ||
The QCM system will become a useful tool for in vitro selection of RNAs, because it can monitor in situ both selection and evaluation processes.
Notes and references
- C. Jain and J. G. Belasco, Cell, 1996, 87, 115–125 CrossRef CAS.
- H. Siomi and G. Dreyfuss, Curr. Opin. Genet. Dev., 1997, 7, 345–353 CrossRef CAS.
- M. A. Weiss and N. Narayana, Biopolymers, 1998, 48, 167–180 CrossRef CAS.
- J. L. Battiste, H. Mao, N. S. Rao, R. Tan, D. R. Muhandiram, L. E. Kay, A. D. Frankel and J. R. Williamson, Science, 1996, 273, 1547–1551 CAS.
- (a) H. Matsuno, K. Niikura and Y. Okahata, Chem. Eur. J., 2001, 7, 3305–3312 CrossRef CAS; (b) Y. Okahata, K. Niikura, H. Furusawa and H. Matsuno, Anal. Sci., 2000, 16, 1113–1119 Search PubMed; (c) H. Matsuno, K. Niikura and Y. Okahata, Biochemistry, 2001, 40, 3615–3622 CrossRef CAS; (d) A. Watanabe, H. Furusawa and Y. Okahata, Nucleic Acids Symp., 1999, 42, 193–194 Search PubMed.
- G. Sauerbrey, Z. Physik., 1956, 155, 206–222 Search PubMed.
- W. Xu and A. D. Ellington, Proc. Natl. Acad. Sci. USA, 1996, 93, 7475–7480 CrossRef CAS.
- C. Tnerk and S. MacDougal-Waugh, Gene, 1993, 137, 33–39 CrossRef.
- L. Giver, D. Bartel, M. Zapp, A. Pawul, M. Green and A. D. Ellington, Nucleic Acids Res., 1993, 21, 5509–5516 CAS.
- D. Nieuwlandt, M. Weoker and L. Gold, Biochemistry, 1995, 34, 5651–5659 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2002 |
