Pyrene–neomycin conjugate: dual recognition of a DNA triple helix†
Liang
Xue
,
I.
Charles
and
Dev P.
Arya
*
Laboratory of Medicinal Chemistry, Department of Chemistry, Clemson University, SC 29634, USA. E-mail: dparya@clemson.edu; Fax: +1 864-656-6613; Tel: +1 864-656-1106
First published on 5th December 2001
Abstract
We report the synthesis of pyrene–neomycin conjugate and its ability to stabilize DNA/RNA triple helices.
Oligonucleotides directed to a precise DNA sequence provide a promising approach to artificially control the transcription process.1,2 Triplex DNA has attracted attention because of the potential application of triplex forming oligonucleotides (TFOs) as therapeutic agents, for applications such as intracellular gene targeting, and as rational chemical solutions to sequence specific recognition of a DNA duplex.1,3
A popular strategy to improve triplex stability involves the use of intercalators and groove binding ligands.4–7 Most groove binders prefer duplex grooves or even destabilize triplex DNA.4–7 There is little information available for antibiotics that selectively bind DNA triplex grooves or RNA triplex grooves. The stabilization of the poly(dA)·2poly(dT), a mixed base DNA 22 mer, RNA and hybrid triple helices by neomycin has recently been reported by us.6–8 Neomycin was shown to be the most effective triplex groove binder that remarkably stabilized DNA and RNA triple helices with little effect on DNA double helix. We hypothesized that with a combination of neomycin’s strong ionic/groove recognition and an intercalating moiety, new classes of molecules with high triplex affinity can be identified. A pyrene–neomycin conjugate has therefore been synthesized by forming an amide linkage between neomycin B and 1-pyrenebutyric acid N-hydroxysuccinimide ester (Fig. 1). We report that pyrene–neomycin is more potent in stabilizing DNA triple helices than neomycin B or pyrene or a combination of both. It increases the melting temperature of a DNA triple helix by >10 °C compared to neomycin B at 4 μM concentration {base triplet–drug ratio (rdb) = 0.26}.
![UV melting profile of poly(dA)·2poly(dT) at 260 nm in the presence of 150 mM KCl, 10 mM sodium cacodylate, 0.5 mM EDTA, pH 7.2. (a) No ligand, (b) 4 μM-neomycin, (c) 4 μM neomycin + 4 μM 1-aminopyrene, (d) 4 μM pyrene–neomycin, (e) 4 μM 1-aminopyrene. [DNA] = 15 μM per base triplet, Tm = ±1 °C. (The y-axis has been artificially offset to differentiate the melting curves).](/image/article/2002/CC/b108171c/b108171c-f1.gif) | ||
| Fig. 1 UV melting profile of poly(dA)·2poly(dT) at 260 nm in the presence of 150 mM KCl, 10 mM sodium cacodylate, 0.5 mM EDTA, pH 7.2. (a) No ligand, (b) 4 μM-neomycin, (c) 4 μM neomycin + 4 μM 1-aminopyrene, (d) 4 μM pyrene–neomycin, (e) 4 μM 1-aminopyrene. [DNA] = 15 μM per base triplet, Tm = ±1 °C. (The y-axis has been artificially offset to differentiate the melting curves). | ||
The following assumptions were made in the design of the conjugate. (i) The amino groups on rings I, II and IV are necessary in stabilizing and in recognizing the Watson–Hoogsteen groove (aminoglycosides without any of these amines do not stabilize triplexes as efficiently).7 (ii) For triplex stabilizing structures based on neomycin, these amines must be retained. The 5″-OH on ring III was thus chosen to provide the linkage to the intercalating unit.
The intercalator (Scheme 1, pyrene) was linked to neomycin amine 2 (prepared in 3 steps from neomycin) using its activated acid. A successful example of coupling of pyrene succinimide ester with neomycin amine 2 is illustrated in Scheme 1. The synthesis rests on the selective conversion of ring III 5″-OH of neomycin into a good leaving group, 2,4,6-triisopropylbenzenesulfonyl (TIBS), as previously reported by Tor.9–10 The displacement of the TIBS leaving group by aminothiol, followed by coupling with the activated ester and deprotection with HCl gives the target conjugate 3 in good yield (Scheme 1).
 | ||
| Scheme 1 Reagents and conditions a–e: (a) (Boc)2O, DMF, H2O, Et3N, 60 °C, 5h, 60%; (b) 2,4,6-triisopropylbenzenesulfonyl chloride, pyridine, rt, 40 h, 50%; (c) HCl·H2NCH2CH2SH, NaOEt–EtOH, rt, 18h, 50%; (d) 1-pyrenebutyric acid N-hydroxysuccinimide ester, CH2Cl2, DMAP, rt, overnight, 80%; (e) 4 M HCl–dioxane, HSCH2CH2SH, rt, 5 min, 80%. | ||
The UV melting studies of triplexes formed from poly(dA)·poly(dT) and poly(dT) were carried out using UV spectroscopy at 260 and 284 nm. Melting curves of poly(dA)·2poly(dT) with different ligands, pyrene–neomycin, neomycin and 1-aminopyrene at 4μM concentration are shown in Fig. 1. The melting profiles are clearly biphasic, as typically observed when polydA·2polydT triplex is formed. The low temperature transition represents the conversion from the triplex to duplex and a single-strand of poly(dT). The high temperature transition represents the dissociation of the duplex, poly(dA)·poly(dT) into single strands. Without any ligand, triplex melts at 32 °C. In the presence of 4 μM neomycin and 4 μM pyrene–neomycin, triplex Tm rises to 43 and 58 °C, respectively. These results show that pyrene–neomycin can stabilize the triplex much more effectively than neomycin at these low concentrations (rdb = 0.26). This is quite remarkable since pyrene itself is not a good triplex stabilizer. Addition of 4 μM 1-aminopyrene and 4 μM neomycin to a DNA triplex leads to a small ΔTm3→2 increase (a biphasic transition, Fig. 1c, Fig. 2). Covalent attachment of the two yields a single transition with a ΔTm about 10 °C higher than the sum of the two (added together). With all three ligands, duplex melting points were unchanged (71°C), signifying that both neomycin and pyrene–neomycin (at these concentrations) have no effect on the stabilization of this DNA duplex. At higher pyrene–neomycin concentrations, a larger increase in Tm is observed, but the ΔΔTm values (from neomycin) do not increase significantly, suggesting saturation of the drug binding site at rdb ∼0.13.
 | ||
| Fig. 2 Plot of variation of Tm3→2 and Tm2→1 of poly(dA)·2poly(dT) as a function of increasing pyrene–neomycin and neomycin concentrations. Solution conditions: same as Fig. 1. | ||
A slightly different picture emerges with the RNA triplex. ΔTm plots of poly(rA)·2poly(rU) with different ligands, 1-aminopyrene, pyrene–neomycin, and neomycin from 0–4 μM concentration are shown in Fig. 3. Without any ligand, Tm3→2 is 32 °C; while in the presence of 4 μM pyrene–neomycin, Tm3→2 rises to 41 °C and Tm2→1 rises from 49 to 55 °C. Pyrene–neomycin then stabilizes RNA triplex as well as the duplex, but the ΔTm3→2 values are slightly less than those induced by neomycin. Duplex stabilization by both ligands is, however, quite comparable. The inability of pyrene to intercalate/stabilize an RNA triplex is perhaps responsible for this lower stabilization observed with the conjugate (1-aminopyrene has no effect on the RNA duplex/triplex, Fig. 3). Appropriately designed conjugates could then, in principle, be made to differentiate such higher order DNA/RNA nucleic acid structures, and help us explore the structural variations responsible for their recognition.
![Plot of variation of Tm3→2 and Tm2→1 of poly(rA)·2poly(rU) as a function of pyrene–neomycin and neomycin. 10 mM sodium cacodylate, 0.1 mM EDTA, pH 6.8. (a) Triplex + neomycin, (b) triplex + pyrene–neomycin, (c) duplex + pyrene–neomycin, (d) duplex + neomycin, (e) triplex + 1-aminopyrene, (f) duplex + 1-aminopyrene. [RNA] = 30 μM base triplet.](/image/article/2002/CC/b108171c/b108171c-f3.gif) | ||
| Fig. 3 Plot of variation of Tm3→2 and Tm2→1 of poly(rA)·2poly(rU) as a function of pyrene–neomycin and neomycin. 10 mM sodium cacodylate, 0.1 mM EDTA, pH 6.8. (a) Triplex + neomycin, (b) triplex + pyrene–neomycin, (c) duplex + pyrene–neomycin, (d) duplex + neomycin, (e) triplex + 1-aminopyrene, (f) duplex + 1-aminopyrene. [RNA] = 30 μM base triplet. | ||
Computer modeling of pyrene–neomycin docked in a TAT triplex suggests that pyrene can intercalate between the base pairs while neomycin stays bound to the Watson–Hoogsteen groove (Fig. 4). Ring I may be embedded inside the groove with amino groups H-bonded to the anti-parallel T strand, whereas the ribose can provide the optimum conformation and size for a good fit into the triplex groove. Rings I and IV are responsible for bringing the two pyrimidine strands together. Because of this possible dual binding mode of triplex recognition, pyrene–neomycin conjugate shows significant ability to stabilize DNA triple helices. Such chromophores should then be effective model ligands for surveying the groove recognition properties of a triplex bound neomycin. These results are now being extended to the stabilization of mixed sequences/hybrid structures and will be reported in due course.
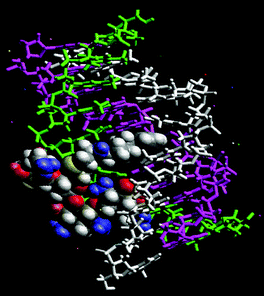 | ||
| Fig. 4 Pyrene–neomycin docked in the Watson–Hoogsteen groove of a TAT DNA triplex. (Atom colors: pyrene–neomycin; purple: poly(dA); white: poly(dT); green: poly(dT)—the third strand.) | ||
Financial support for the work was provided by Clemson University and Greenville Hospital System. The support of NMR facility by NSF grant (CHE-9700278) is gratefully acknowledged. We thank Dr Ljiljana Micovic for help with modeling studies.
Notes and references
- L. J. Maher III, Cancer Invest., 1996, 14, 66–82 CAS.
- H. E. Moser and P. B. Dervan, Science, 1987, 238, 645–650 CrossRef CAS.
- N. T. Thuong and C. Helene, Angew. Chem., Int. Ed. Engl., 1993, 32, 666–690 CrossRef.
- G. Felsenfeld, D. Davies and A. Rich, J. Am. Chem. Soc., 1957, 79, 2023–2024 CrossRef CAS.
- T. C. Jenkins, Curr. Med. Chem., 2001, 7, 99–115 Search PubMed (for a comprehensive list of references; also see reference 7).
- D. P. Arya and R. L. Coffee, Jr, Bioorg. Med. Chem. Lett., 2000, 10, 1897–1899 CrossRef CAS.
- D. P. Arya, R. L. Coffee, Jr, B. Willis and A. I. Abramovitch, J. Am. Chem. Soc., 2001, 123, 4385–4396.
- D. P. Arya, R. L. Coffee, Jr and I. Charles, J. Am. Chem. Soc., 2001, 123, 11093–11094 CrossRef CAS.
- K. Michael, H. Wang and Y. Tor, Bioorg. Med. Chem. Lett., 1999, 7, 1361–1371 CrossRef CAS.
- S. R. Kirk, N. W. Luedtke and Y. Tor, J. Am. Chem. Soc., 2000, 122, 980–981 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: NMR spectra, UV spectra, extinction coefficients, melting curves of pyrene–neomycin conjugate, details of modeling studies. See http://www.rsc.org/suppdata/cc/b1/b108171c/ |
| This journal is © The Royal Society of Chemistry 2002 |
