Renewable pencil electrodes for highly sensitive stripping potentiometric measurements of DNA and RNA
Joseph Wang*, Abdel-Nasser Kawde and Eskil Sahlin
Department of Chemistry and Biochemistry, New Mexico State University, Las Cruces, NM 88003, USA
First published on UnassignedUnassigned7th January 2000
Abstract
Renewable graphite pencil electrodes are demonstrated to be excellent materials for adsorptive stripping measurements of trace nucleic acids. While displaying an attractive stripping performance, comparable to that of conventional carbon paste electrodes, the pencil electrode offers a convenient (mechanical) renewal, with each stripping potentiogram recorded at a fresh surface. Various pencil lead materials and lengths have been examined and experimental variables of the pretreatment and measurement procedures have been explored and optimized. The extremely low detection limits (e.g., 3 μg l−1 tRNA with 10 min accumulation) are coupled to a good surface-to-surface reproducibility (RSD of 6.4% for 14 repetitive measurements of 1 mg l−1 ssDNA).
Introduction
Highly sensitive methods for quantifying nucleic acids are required in many aspects of molecular biology and biotechnology. Electroanalytical techniques have proved very useful for the analysis of DNA and RNA.1 In particular, the interfacial accumulation and reduction of the nucleobases at mercury electrodes have led to the development of highly sensitive adsorptive stripping voltammetric procedures for trace measurements of DNA and RNA.2 Analogous stripping measurements of nucleic acids can be performed at carbon-paste3 or microfabricated carbon-film4 electrodes by coupling the electrooxidation of the guanine moiety with the surface activity of nucleic acids, and a constant-current potentiometric stripping operation. Such replacement of the classical mercury drop electrode with solid-state electrochemical devices paves the way for increasing the role of electroanalysis in modern DNA diagnostics and nucleic-acid research.Here we report on the advantages of using pencil lead (graphite) electrodes for adsorptive stripping measurements of trace DNA and RNA. Pencil electrodes have been used successfully for anodic5,6 and abrasive7 stripping measurements of trace metals, as well as transducers for electrochemical immunosensors.8 In the following sections we will illustrate that the use of inexpensive pencil electrode materials results in highly sensitive potentiometric stripping measurements of nucleic acids (comparable to the commonly used carbon paste electrode), while offering a convenient surface renewal (through mechanical extrusion of the lead). Such surface renewal is simpler and faster than polishing procedures, common with solid electrodes, and results in good reproducibility for the individual surfaces. Factors influencing trace measurements of DNA and RNA at pencil electrodes are assessed below, and the attractive stripping performance is characterized.
Experimental
Instrumentation
Stripping potentiometric measurements were performed with a PSU 20 potentiometric stripping unit (Radiometer, Westlake, OH, USA), controlled by a PC, using TAP2 software (Radiometer). The electrode system consisted of a pencil lead working electrode, an Ag/AgCl (3M NaCl) reference electrode (Model RE-1, BAS) and a platinum wire counter electrode.A Pentel pencil, Model P205 (Japan), was used as a holder for the pencil lead. Electrical contact with the lead was achieved by soldering a metallic wire to the metallic part that holds the lead in place inside the pencil. Unless stated otherwise, the pencil was fixed vertically with 6 mm of the pencil lead exposed outside and 3 mm of the pencil lead immersed into the solution. Measurements were performed in a glass cell containing 2 ml of solution. Stirring was achieved with a magnetic stirring bar.
Working electrode materials
The pencil leads tested were from Pentel and named Hi-polymer Super C505 (black lead) of types B, F, HB, H, 4H and 6H. All leads had a total length of 60 mm and a diameter of 0.5 mm. Immersing 3 mm of the pencil lead into a solution resulted in an active electrode area of 4.91 mm2. The pencil leads were used as received.The preparation of the carbon paste disk electrode (2.5 mm diameter) was described earlier.3 The 3 mm diameter glassy carbon disk electrode (BAS) was polished with 0.05 μm alumina. All pipette tips and tubes for preparation of solutions were autoclaved prior to use.
Chemicals
The following chemicals and biochemicals were obtained from Sigma (St. Louis, MO, USA): transfer RNA (tRNA, from baker’s yeast; catalog number R8759), single stranded DNA (ssDNA; catalog number D8899), sodium acetate buffer solution (3M), Tris-EDTA buffer (1.0 M Tris HCl–0.10M EDTA) and diethyl pyrocarbonate (DEPC). Oligo(dG)20 was obtained from Life Technologies (Grand Island, NY, USA). The preparation of the tRNA stock solutions (1000 mg l−1) was described earlier.4 Stock solutions (1000 mg l−1) of ssDNA and oligo(dG)20 were prepared in a 10 mM Tris HCl–1 mM EDTA solution (pH 8.0). Other solutions were prepared in acetate buffer (0.2M, pH 5.0). All solutions were prepared using deionized and autoclaved water.Procedure
Unless mentioned otherwise, all measurements were performed by treating the surface at +1.40 V for 30s followed by accumulation at +0.50 V for 60s in a stirred acetate buffer solution (0.2 M, pH 5.0). The stripping step was carried out without stirring, after a 5s rest period, using a constant current of +5.0 μA. Each measurement was performed using a new pencil surface. The carbon paste and glassy carbon surfaces were polished prior to every measurement. Before evaluation, the stripping curve data were treated using the baseline-correction and filter commands of the TAP2 software.Results and discussion
Anodic measurements of trace nucleic acids are based primarily on measurements of the oxidation peak of the guanine residue.1,3Fig. 1 compares adsorptive stripping potentiograms for tRNA at the pencil (a), carbon-paste (b), and glassy-carbon (c) electrodes, following a 1 min adsorptive collection. Examination of the raw data (A, top) indicates that the pencil electrode results in a larger guanine peak, as well as solvent decomposition background, than the commonly used carbon paste electrode. A poorly defined peak, obscured by the background response, is observed at the glassy carbon electrode. The background contribution is corrected following the advanced data processing inherent to the computerized stripping potentiometric operation (B, bottom). As a result, the pencil electrode offers the most favorable detection of the tRNA guanine peak (Ep= +0.96 V). Note the appearance of an additional small peak, at both carbon paste and pencil electrodes (Ep= +1.10 V). A similar peak, observed for oligo(dG)20 (not shown), indicates that this is not an adenine response. Besides its favorable response characteristics, the pencil electrode offers a unique surface renewal (in connection with mechanical extrusion of the pencil lead), low cost, and rigidity. Such surface renewal is actually the closest solid-electrode analogue of mercury drop electrodes. | ||
| Fig. 1 Stripping potentiometric signals for 1 mg l−1 tRNA using the pencil (a), carbon paste (b) and glassy carbon (c) electrodes, before (A) and after (B) baseline correction. Pencil type and length (a), HB and 3 mm, respectively. Measurements were performed in a acetate buffer (0.2 M, pH 5.0) solution. Pretreatment at +1.40V for 30s followed by accumulation at +0.50V for 60s under stirred conditions. Stripping was performed in a quiescent solution using a constant current of +5.0μA. | ||
We screened different types of pencil leads for their stripping response to nucleic acids. Fig. 2 compares stripping chronopotentiograms for 2 μg l−1 tRNA, following 1 min accumulation, using six different pencil leads. Substantial differences in the stripping response and background signal are observed at the various types of pencil leads, with the HB one (a) displaying the most favorable signal-to-background characteristics. The F and H leads display a large guanine peak, along with a large background contribution (c,d). Lower backgrounds, and guanine signals, are observed with the B, 4H, and 6H leads (b,e and f, respectively). Such different response characteristics are expected considering the different compositions and roughness of the various leads (that contain various polymers and clays, besides graphite). Blank experiments (not shown) indicate that these non-graphite constituents do not yield a background peak. In view of the composite nature of the leads, and the complexity of the processes involved, it is not clear, at this stage, whether the different response characteristics reflect differences in the interfacial (adsorption) properties of the nucleic acids, in the kinetics of the guanine oxidation process, or in the effective surface area (roughness). Larger guanine signals, coupled to a large background (F and H leads), may indicate both a reactive graphite surface and a graphite-rich one. The HB pencil lead is advantageous as it offers a relatively large guanine peak in connection with a relatively low background signal and noise level (a). Different ferrocyanide voltammetric signals (and background profiles) were observed previously using different pencil leads.5 The HB pencil lead was found to be an excellent electrode in anodic and cathodic stripping voltammetric measurements of trace metals.6 Based on the profiles of Fig. 2, this lead was selected also for all subsequent DNA and RNA work.
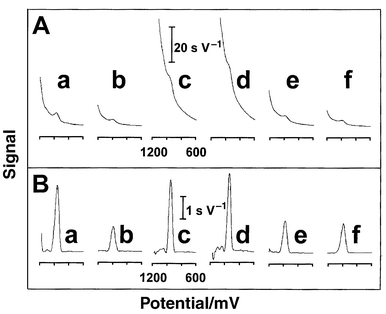 | ||
| Fig. 2 Stripping potentiograms for 2 mg l−1 tRNA using different pencil leads. Pencil type: HB (a), B (b), F (c), H (d), 4H (e) and 6H (f), before (A) and after(B) baseline correction. Other conditions as in Fig. 1(a). | ||
As is common for trace measurements of nucleic acids at other carbon electrodes,3,4,9 a short electrochemical activation is essential for obtaining favorable stripping signals. Fig. 3A shows the effect of the pretreatment potential on the response to 1 mg l−1 of tRNA. The guanine signal increases slowly upon changing the potential between +1.0 and +1.2 V, rises sharply between +1.2 and +1.5 V, and then levels off. A pretreatment potential of +1.4 V yielded the best tradeoff between sensitivity, background contribution, and corresponding noise, and was thus used for all subsequent work. A large noise level was observed without the electrochemical activation. The effect of the accumulation time is displayed in Fig. 3B. The response of tRNA increases rapidly upon raising the pretreatment time between 15 s and 3 min, than more slowly (up to 8 min), and subsequently more rapidly.
 | ||
| Fig. 3 Influence of the pretreatment potential (A) and accumulation time (B) on the stripping time for 1 mg l−1 tRNA using a pencil lead (type HB). Accumulation for 1 min (A) and pretreatment at +1.40 V (B). Other conditions as in Fig. 1(a). Each point is a mean value of two measurements. | ||
The pencil lead can be extruded to different lengths, to yield different surface areas. As expected, the length of the pencil lead (exposed to the sample) has a profound effect upon the response. The stripping signal for 1 mg l−1 tRNA increased linearly with the length over the 1–8 mm range (slope, 40 ms mm−1; correlation coefficient, 0.991; not shown; conditions, as in Fig. 1a). Due to a corresponding increase in the background, all subsequent work employed 3-mm pencil leads (with an active area of 4.91mm2). Of particular importance for renewable graphite electrodes is the surface-to-surface reproducibility. A series of 14 repetitive measurements of 1 mg l−1 ssDNA, using newly exposed leads, yielded a mean signal of 28.1 ms, with a relative standard deviation of 6.4% (conditions as in Fig. 1a). Note that such renewable graphite electrodes can be reproducibly used as received.
The optimal conditions offer convenient quantitation of mg l−1 concentrations of nucleic acids following short adsorption times. Fig. 4 displays calibration plots for oligo(dG)20 (A) and ssDNA (B) using a 1 min accumulation. These plots display a curvature characteristic of adsorptive stripping measurements. The peak of the shorter oligonucleotide increases rapidly with the concentration at first and starts to level off above 1 mg l−1. The response for ssDNA increases nearly linearly up to 2 mg l−1, with a curvature thereafter. Lower concentrations can be readily detected in combination with longer accumulation periods. For example, a detection limit of 3 μg l−1 tRNA was estimated based on the signal-to-noise characteristics (S/N=3) of the response for 10 μg l−1 tRNA following a 10 min adsorption (not shown).
 | ||
| Fig. 4 Calibration curves for oligo(dG)20 (A) and ssDNA (B) using a pencil lead (type HB). Conditions as in Fig. 1(a). Each point is a mean value of two measurements. | ||
In conclusion, the pencil electrode has been demonstrated to be an excellent electrode material for adsorptive stripping measurements of trace nucleic acids. Such suitability is attributed to the composite (mixed graphite–insulator) character of pencil leads, which is analogous to that of other composite carbons used for the analysis of nucleic acids.3,4 While offering very favorable signal-to-background characteristics, the main advantage of the pencil DNA electrode appears to be its readily renewable surface. Other bioanalytical applications should benefit from the facile detection of nucleic acids at pencil electrodes. For example, the strong adsorptive accumulation of DNA could lead to renewable, low-cost, DNA modified electrodes for detecting DNA hybridization (with the mechanical extrusion obviating the need for regenerating the ssDNA probe). Similarly, the well-defined guanine response should offer convenient flow detection of nucleic acids following HPLC or CZE separations. The later should address the selectivity challenge expected in assays of biological samples. Alternatively, potential interferences could be minimized in connection with a proper sample pretreatment (e.g., extraction, PCR amplification).
Acknowledgements
This work was supported by NIH Grant Number RR14549. E. S. and A. K. acknowledge fellowships from STINT (Sweden) and the Egyptian government, respectively.References
- E. Palecek, Electroanalysis, 1996, 8, 7 CAS.
- E. Palecek and M. Fojta, Anal. Chem., 1994, 66, 1566 CrossRef CAS.
- J. Wang, X. Cai, J. Wang, C. Jonsson and E. Palecek, Anal. Chem., 1995, 67, 4065 CrossRef CAS.
- J. Wang, X. Cai, B. Tian and H. Shiraishi, Analyst, 1996, 121, 965 RSC.
- A. M. Bond, P. J. Mahon, J. Schiewe and V. Vicente-Beckett, Anal. Chim. Acta, 1994, 345, 67 CrossRef CAS.
- T. Nagunuma, F. Anuda and J. Jin, Aichi Kyoiku Daigaku Kenkyu Hokoku, Shizen Kagaku, 1995, 44, 25 Search PubMed.
- D. Blum, W. Leyffer and R. Holze, Electroanalysis, 1996, 8, 296 CAS.
- L. Engel and W. Baumann, Fresenius J. Anal. Chem., 1993, 346, 745 CrossRef CAS.
- X. Cai, G. Rivas, P.A. Farias, H. Shiraishi, J. Wang and E. Palecek, Electroanalysis, 1996, 8, 753 CAS.
| This journal is © The Royal Society of Chemistry 2000 |
