Transcriptome-wide identification of sucrose synthase genes in Ornithogalum caudatum†
Abstract
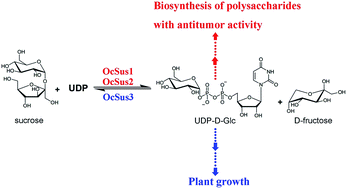
OCAP-2-1, OCAP-2-2, OCAP-3-1 and OCAP-3-3, four glucose-containing polysaccharides from Ornithogalum caudatum, exhibit antitumor activity, suggesting their potential application as natural antitumor drugs. Although the incorporation of glucose into these polysaccharides from UDP-D-glucose is reasonably well understood, the cDNA isolation and functional characterization of genes responsible for UDP-D-glucose biosynthesis from O. caudatum has not been identified. Here, we present a full characterization of the sucrose synthase family, a Leloir glycosyltransferase responsible for UDP-D-glucose biosynthesis from O. caudatum. Specifically, a transcriptome-wide search for Sus genes in O. caudatum was first performed in the present study. A total of 5 unigenes sharing high sequence identity with Sus were retrieved from transcriptome sequencing. Three full-length Sus-like candidates derived from this unigene assembly were then obtained and isolated by reverse transcription polymerase chain reaction (RT-PCR) from O. caudatum. Additional analysis showed two conserved domains (sucrose synthase and glycosyl transferase domains) were present in this family. Phylogenetic analysis indicated that the OcSus1 and OcSus2 could be clustered together into a monocots specific clade, while OcSus3 could be classified into M & D1 category with members from the monocots and dicots species, displaying an evolutionary consistency with other plant species. These candidate isoenzymes were screened by functional expression in E. coli individually as standalone enzymes. All three cDNAs were identified to be bona fide genes and encoded sucrose synthase with varied kinetic properties. To further explore the possible role of these Sus proteins in polysaccharide biosynthesis, transcript profiles of the three genes were subsequently examined by real-time quantitative PCR in various tissues. OcSus1 and OcSus2 were therefore assumed to be responsible for the biosynthesis of the four glucose-containing polysaccharides due to their expression profiles in O. caudatum. Taken together, these data provide further comprehensive knowledge for polysaccharide biosynthesis in O. caudatum and broaden the potential application of Sus in metabolic engineering or synthetic biology as a potential gene part.

 Please wait while we load your content...
Please wait while we load your content...