Computational modeling of cytokine signaling in microglia†
Abstract
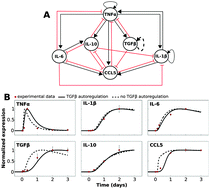
Neuroinflammation due to glial activation has been linked to many CNS diseases. We developed a computational model of a microglial cytokine interaction network to study the regulatory mechanisms of microglia-mediated neuroinflammation. We established a literature-based cytokine network, including TNFα, TGFβ, and IL-10, and fitted a mathematical model to published data from LPS-treated microglia. The addition of a previously unreported TGFβ autoregulation loop to our model was required to account for experimental data. Global sensitivity analysis revealed that TGFβ- and IL-10-mediated inhibition of TNFα was critical for regulating network behavior. We assessed the sensitivity of the LPS-induced TNFα response profile to the initial TGFβ and IL-10 levels. The analysis showed two relatively shifted TNFα response profiles within separate domains of initial condition space. Further analysis revealed that TNFα exhibited adaptation to sustained LPS stimulation. We simulated the effects of functionally inhibiting TGFβ and IL-10 on TNFα adaptation. Our analysis showed that TGFβ and IL-10 knockouts (TGFβ KO and IL-10 KO) exert divergent effects on adaptation. TFGβ KO attenuated TNFα adaptation whereas IL-10 KO enhanced TNFα adaptation. We experimentally tested the hypothesis that IL-10 KO enhances TNFα adaptation in murine macrophages and found supporting evidence. These opposing effects could be explained by differential kinetics of negative feedback. Inhibition of IL-10 reduced early negative feedback that results in enhanced TNFα-mediated TGFβ expression. We propose that differential kinetics in parallel negative feedback loops constitute a novel mechanism underlying the complex and non-intuitive pro- versus anti-inflammatory effects of individual cytokine perturbations.

 Please wait while we load your content...
Please wait while we load your content...