A fragment-based docking simulation for investigating peptide–protein bindings
Abstract
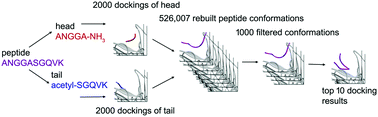
A fragment-based method was developed to investigate the binding conformations of peptide ligands. This method efficiently avoids the high degree of freedom (DOF) of peptide dockings by dividing a peptide into two half fragments. The fragments are separately docked on receptors and the results are used to rebuild a profile of massive possible docking conformations of the whole peptide. Through rapid scoring for filtering, the remaining peptide docking conformations are rigorously optimized by molecular dynamics (MD) and scored by molecular mechanics/generalized born surface area (MM/GBSA) method to predict the near-native binding conformations. This method has been tested on 17 cases of long peptide–protein interaction with known crystal structures, and also on 7 unbound protein receptors for which both the bound and unbound conformations are known. The resultant binding predictions fit very closely to the crystal structures.


 Please wait while we load your content...
Please wait while we load your content...