NMR mapping of protein conformational landscapes using coordinated behavior of chemical shifts upon ligand binding†
Abstract
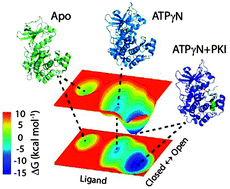
Proteins exist as an ensemble of conformers that are distributed on free energy landscapes resembling folding funnels. While the most stable conformers populate low energy basins, protein function is often carried out through low-populated conformational states that occupy high energy basins. Ligand binding shifts the populations of these states, changing the distribution of these conformers. Understanding how the equilibrium among the states is altered upon ligand binding, interaction with other binding partners, and/or mutations and post-translational modifications is of critical importance for explaining allosteric signaling in proteins. Here, we propose a statistical analysis of the linear trajectories of NMR chemical shifts (CONCISE, COordiNated ChemIcal Shifts bEhavior) for the interpretation of protein conformational equilibria. CONCISE enables one to quantitatively measure the population shifts associated with ligand titrations and estimate the degree of collectiveness of the protein residues' response to ligand binding, giving a concise view of the structural transitions. The combination of CONCISE with thermocalorimetric and kinetic data allows one to depict a protein's approximate conformational energy landscape. We tested this method with the catalytic subunit of cAMP-dependent protein kinase A, a ubiquitous enzyme that undergoes conformational transitions upon both nucleotide and pseudo-substrate binding. When complemented with chemical shift covariance analysis (CHESCA), this new method offers both collective response and residue-specific correlations for ligand binding to proteins.
- This article is part of the themed collection: The free energy landscape: from folding to cellular function

 Please wait while we load your content...
Please wait while we load your content...