Probing protein aggregation at buried interfaces: distinguishing between adsorbed protein monomers, dimers, and a monomer–dimer mixture in situ†
Abstract
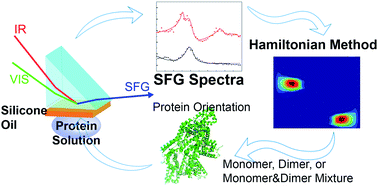
Protein adsorption on surfaces greatly impacts many applications such as biomedical materials, anti-biofouling coatings, bio-separation membranes, biosensors, antibody protein drugs etc. For example, protein drug adsorption on the widely used lubricant silicone oil surface may induce protein aggregation and thus affect the protein drug efficacy. It is therefore important to investigate the molecular behavior of proteins at the silicone oil/solution interface. Such an interfacial study is challenging because the targeted interface is buried. By using sum frequency generation vibrational spectroscopy (SFG) with Hamiltonian local mode approximation method analysis, we studied protein adsorption at the silicone oil/protein solution interface in situ in real time, using bovine serum albumin (BSA) as a model. The results showed that the interface was mainly covered by BSA dimers. The deduced BSA dimer orientation on the silicone oil surface from the SFG study can be explained by the surface distribution of certain amino acids. To confirm the BSA dimer adsorption, we treated adsorbed BSA dimer molecules with dithiothreitol (DTT) to dissociate these dimers. SFG studies on adsorbed BSA after the DTT treatment indicated that the silicone oil surface is covered by BSA dimers and BSA monomers in an approximate 6 : 4 ratio. That is to say, about 25% of the adsorbed BSA dimers were converted to monomers after the DTT treatment. Extensive research has been reported in the literature to determine adsorbed protein dimer formation using ex situ experiments, e.g., by washing off the adsorbed proteins from the surface then analyzing the washed-off proteins, which may induce substantial errors in the washing process. Dimerization is a crucial initial step for protein aggregation. This research developed a new methodology to investigate protein aggregation at a solid/liquid (or liquid/liquid) interface in situ in real time using BSA dimer as an example, which will greatly impact many research fields and applications involving interfacial biological molecules.


 Please wait while we load your content...
Please wait while we load your content...