Predicting the conformational variability of oncogenic GTP-bound G12D mutated KRas-4B proteins at zwitterionic model cell membranes†
Abstract
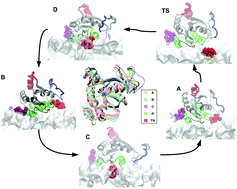
KRas proteins are the largest family of mutated Ras isoforms, participating in a wide variety of cancers. Due to their importance, large effort is being carried out on drug development by small-molecule inhibitors. However, understanding protein conformational variability remains a challenge in drug discovery. In the case of the Ras family, their multiple conformational states can affect the binding of potential drug inhibitors. To overcome this challenge, we propose a computational framework based on combined all-atom Molecular Dynamics and Metadynamics simulations in order to accurately access conformational variants of the target protein. We tested the methodology using a G12D mutated GTP bound oncogenic KRas-4B protein located at the interface of a DOPC/DOPS/cholesterol model anionic cell membrane. Two main orientations of KRas-4B at the anionic membrane have been determined. The corresponding torsional angles are taken as reliable reaction coordinates so that free-energy landscapes are obtained by well-tempered metadynamics simulations, revealing local and global minima of the free-energy hypersurface and unveiling reactive paths of the system between the two preferential orientations. We have observed that GTP-binding to KRas-4B has huge influence on the stabilisation of the protein and it can potentially help to open Switch I/II druggable pockets, lowering energy barriers between stable states and resulting in cumulative conformers of KRas-4B. This may highlight new opportunities for targeting the unique meta-stable states through the design of new efficient drugs.


 Please wait while we load your content...
Please wait while we load your content...