Accurate and cost-effective NMR chemical shift predictions for proteins using a molecules-in-molecules fragmentation-based method†
Abstract
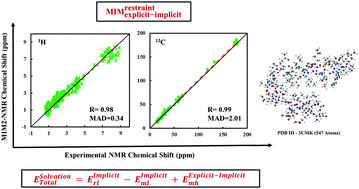
We have developed an efficient protocol using our two-layer Molecules-in-Molecules (MIM2) fragmentation-based quantum chemical method for the prediction of NMR chemical shifts of large biomolecules. To investigate the performance of our fragmentation approach and demonstrate its applicability, MIM-NMR calculations are first calibrated on a test set of six proteins. The MIM2-NMR method yields a mean absolute deviation (MAD) from unfragmented full molecule calculations of 0.01 ppm for 1H and 0.06 ppm for 13C chemical shifts. Thus, the errors from fragmentation are only about 3% of our target accuracy of ∼0.3 ppm for 1H and 2–3 ppm for 13C chemical shifts. To compare with experimental chemical shifts, a standard protocol is first derived using two smaller proteins 2LHY (176 atoms) and 2LI1 (146 atoms) for obtaining an appropriate protein structure for NMR chemical shift calculations. The effect of the solvent environment on the calculated NMR chemical shifts is incorporated through implicit, explicit, or explicit–implicit solvation models. The expensive first solvation shell calculations are replaced by a micro-solvation model in which only the immediate interaction between the protein and the explicit solvation environment is considered. A single explicit water molecule for each amine and amide proton is found to be sufficient to yield accurate results for 1H chemical shifts. The 1H and 13C NMR chemical shifts calculated using our protocol give excellent agreement with experiments for two larger proteins, 2MC5 (the helical part with 265 atoms) and 3UMK (33 residue slice with 547 atoms). Overall, our target accuracy of ∼0.3 ppm for 1H and ∼2–3 ppm for 13C has been achieved for the larger proteins. The proposed MIM-NMR method is accurate and computationally cost-effective and should be applicable to study a wide range of large proteins.


 Please wait while we load your content...
Please wait while we load your content...