Multiscale landscape of molecular mechanism of SIRT1 activation by STACs†
Abstract
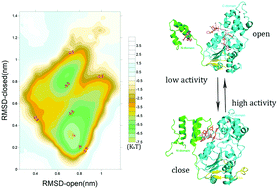
Sirtuins are a family of highly conserved NAD-dependent deacetylase that are involved in multiple biological processes in both prokaryotes and eukaryotes. Many sirtuin-activating compounds (STACs) have been reported for SIRT1, which is the best-characterized sirtuin. However, the molecular mechanism of SIRT1 activation by STACs remains controversial. Here, we developed a multiscale simulation model to explore this mechanism. By quantifying the free energy landscape for the closed conformation of a SIRT1–FdL peptide–resveratrol complex, we found a positive correlation between the barrier height of the active free energy basin and the experimentally determined fluctuations in the rate of SIRT1 deacetylation by resveratrol. In addition, by monitoring dynamics, we found that the open conformation of a SIRT1–p53–STAC-1 complex had a faster rate of conformational change than the closed structure. We also determined the structural properties of each thermodynamic or dynamic state and found that two potential activating factors, the stability of FdL peptide (the p53 peptide substrate including an AMC fluorophore group) binding and the stability of the SIRT1 conformation, were weakly correlated under certain conditions. These results address the controversial question of whether the AMC fluorophore group and native hydrophobic residues have similar roles in the SIRT1 activation process. Finally, we captured the global landscape of the transition, including less stable and more stable states, and proposed a global physical landscape for the mechanism of SIRT1 activation by STACs.


 Please wait while we load your content...
Please wait while we load your content...