Deep learning model predicts water interaction sites on the surface of proteins using limited-resolution data†
Abstract
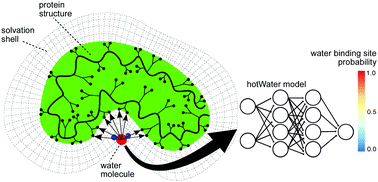
We develop a residual deep learning model, hotWater (https://pypi.org/project/hotWater/), to identify key water interaction sites on proteins for binding models and drug discovery. This is tested on new crystal structures, as well as cryo-EM and NMR structures from the PDB and in crystallographic refinement with promising results.


 Please wait while we load your content...
Please wait while we load your content...