Fluidic switching in nanochannels for the control of Inchworm: a synthetic biomolecular motor with a power stroke†
Abstract
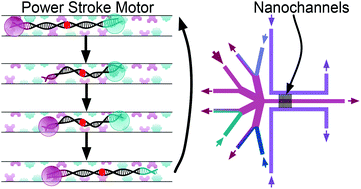
Synthetic molecular motors typically take nanometer-scale steps through rectification of thermal motion. Here we propose Inchworm, a DNA-based motor that employs a pronounced power stroke to take micrometer-scale steps on a time scale of seconds, and we design, fabricate, and analyze the nanofluidic device needed to operate the motor. Inchworm is a kbp-long, double-stranded DNA confined inside a nanochannel in a stretched configuration. Motor stepping is achieved through externally controlled changes in salt concentration (changing the DNA's extension), coordinated with ligand-gated binding of the DNA's ends to the functionalized nanochannel surface. Brownian dynamics simulations predict that Inchworm's stall force is determined by its entropic spring constant and is ∼0.1 pN. Operation of the motor requires periodic cycling of four different buffers surrounding the DNA inside a nanochannel, while keeping constant the hydrodynamic load force on the DNA. We present a two-layer fluidic device incorporating 100 nm-radius nanochannels that are connected through a few-nm-wide slit to a microfluidic system used for in situ buffer exchanges, either diffusionally (zero flow) or with controlled hydrodynamic flow. Combining experiment with finite-element modeling, we demonstrate the device's key performance features and experimentally establish achievable Inchworm stepping times of the order of seconds or faster.


 Please wait while we load your content...
Please wait while we load your content...