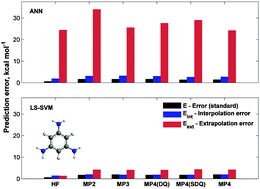
A multilayer feed-forward artificial neural network (MLP-ANN) with a single, hidden layer that contains a finite number of neurons can be regarded as a universal non-linear approximator. Today, the ANN method and linear regression (MLR) model are widely used for quantum chemistry (QC) data analysis (e.g., thermochemistry) to improve their accuracy (e.g., Gaussian G2–G4, B3LYP/B3-LYP, X1, or W1 theoretical methods). In this study, an alternative approach based on support vector machines (SVMs) is used, the least squares support vector machine (LS-SVM) regression. It has been applied to ab initio (first principle) and density functional theory (DFT) quantum chemistry data. So, QC + SVM methodology is an alternative to QC + ANN one. The task of the study was to estimate the Møller–Plesset (MPn) or DFT (B3LYP, BLYP, BMK) energies calculated with large basis sets (e.g., 6-311G(3df,3pd)) using smaller ones (6-311G, 6-311G*, 6-311G**) plus molecular descriptors. A molecular set (BRM-208) containing a total of 208 organic molecules was constructed and used for the LS-SVM training, cross-validation, and testing. MP2, MP3, MP4(DQ), MP4(SDQ), and MP4/MP4(SDTQ) ab initio methods were tested. Hartree–Fock (HF/SCF) results were also reported for comparison. Furthermore, constitutional (CD: total number of atoms and mole fractions of different atoms) and quantum-chemical (QD: HOMO–LUMO gap, dipole moment, average polarizability, and quadrupole moment) molecular descriptors were used for the building of the LS-SVM calibration model. Prediction accuracies (MADs) of 1.62 ± 0.51 and 0.85 ± 0.24 kcal mol−1 (1 kcal mol−1 = 4.184 kJ mol−1) were reached for SVM-based approximations of ab initio and DFT energies, respectively. The LS-SVM model was more accurate than the MLR model. A comparison with the artificial neural network approach shows that the accuracy of the LS-SVM method is similar to the accuracy of ANN. The extrapolation and interpolation results show that LS-SVM is superior by almost an order of magnitude over the ANN method in terms of the stability, generality, and robustness of the final model. The LS-SVM model needs a much smaller numbers of samples (a much smaller sample set) to make accurate prediction results. Potential energy surface (PES) approximations for molecular dynamics (MD) studies are discussed as a promising application for the LS-SVM calibration approach.

 Please wait while we load your content...
Please wait while we load your content...