Successful batch and continuous lyophilization of mRNA LNP formulations depend on cryoprotectants and ionizable lipids†
Alexander
Lamoot‡
ab,
Joris
Lammens‡
c,
Emily
De Lombaerde
ab,
Zifu
Zhong
ab,
Mark
Gontsarik
ab,
Yong
Chen
ab,
Thomas R. M.
De Beer
*c and
Bruno G.
De Geest
 *ab
*ab
aDepartment of Pharmaceutics, Ghent University, Ghent, Belgium. E-mail: br.degeest@ugent.be
bCancer Research Institute Ghent (CRIG), Ghent University, Ottergemsesteenweg 460, B-9000 Ghent, Belgium
cLaboratory of Pharmaceutical Process Analytical Technology, Department of Pharmaceutics, Ghent University, Ottergemsesteenweg 460, B-9000 Ghent, Belgium. E-mail: Thomas.DeBeer@UGent.be
First published on 4th April 2023
Abstract
The limited thermostability and need for ultracold storage conditions are the major drawbacks of the currently used nucleoside-modified lipid nanoparticle (LNP)-formulated messenger RNA (mRNA) vaccines, which hamper the distribution of these vaccines in low-resource regions. The LNP core contains, besides mRNA and lipids, a large fraction of water. Therefore, encapsulated mRNA, or at least a part of it, is subjected to hydrolysis mechanisms similar to unformulated mRNA in an aqueous solution. It is likely that the hydrolysis of mRNA and colloidal destabilization are critical factors that decrease the biological activity of mRNA LNPs upon storage under ambient conditions. Hence, lyophilization as a drying technique is a logical and appealing method to improve the thermostability of these vaccines. In this study, we demonstrate that mRNA LNP formulations comprising a reduction-sensitive ionizable lipid can be successfully lyophilized, in the presence of 20% w/v sucrose, both by conventional batch freeze-drying and by an innovative continuous spin lyophilization process. While the chemical structure of the ionizable lipid did not affect the colloidal stability of the LNP after lyophilization and redispersion in an aqueous medium, we found that the ability of LNPs to retain the mRNA payload stably encapsulated, and mediate in vivo and in vitro mRNA translation into protein, post lyophilization strongly depended on the ionizable lipid in the LNP formulation.
Introduction
The nucleoside-modified mRNA lipid nanoparticle (LNP)-formulated COVID-19 vaccines developed by Pfizer-BioNTech and Moderna have greatly contributed to controlling the COVID-19 pandemic.1,2 However, the limited thermostability and need for ultracold storage of this class of vaccines remain important challenges, which impede the global deployment of mRNA vaccines, particularly in resource-poor countries.2–5 Notably, poor global supply of COVID-19 vaccines may lead to the emergence of novel variants.5–7 Arteta et al. reported that the LNP core, besides mRNA and lipids, comprises over 20% (v/v) of water.8 LNP-encapsulated mRNA is, by consequence, at least partly, exposed to an aqueous environment and prone to hydrolysis of the phosphodiester bonds in the mRNA backbone. Hence, the limited shelf life of mRNA LNP formulations might be, at least in part, due to mRNA hydrolysis occurring inside the LNP core.5,7Thermostable mRNA LNP formulations and manufacturing strategies are hence of great relevance. Lyophilization (freeze-drying) is an appealing method to extend the half-life of mRNA LNPs, by removing water from the formulation.9,10 The lyophilization of pharmaceutical unit doses is currently performed via batch-wise production (i.e., conventional batch freeze-drying). However, innovative continuous manufacturing drying techniques are successfully emerging.11,12
During conventional batch freeze-drying (Fig. 1), vials filled with a liquid formulation are put on heat-controlled shelves in the drying chamber. During the freezing step, the shelves are cooled to a temperature below the glass transition temperature of the maximally freeze concentrated solute (T′g). When the product is fully frozen, primary drying is initiated by lowering the pressure in the chamber, allowing the ice to sublimate. Concomitantly, the shelf temperature is increased to accelerate the drying process. Notably, during this step, care is taken to ensure that the product temperature remains below its collapse temperature. After sublimation of all ice, secondary drying starts by gradually raising the shelf temperature, leading to desorption of residual dissolved and bound water.13 Although batch freeze-drying is commonly used, it is expensive, and time- and energy-consuming, and does not allow the production of flexible batch sizes. In addition, differences in the quality aspects of the product (e.g., cake structure and residual moisture) might occur, depending on the location of the vial in the chamber due to non-homogeneous heat transfer and nucleation moments across the entire shelf.11,13,14
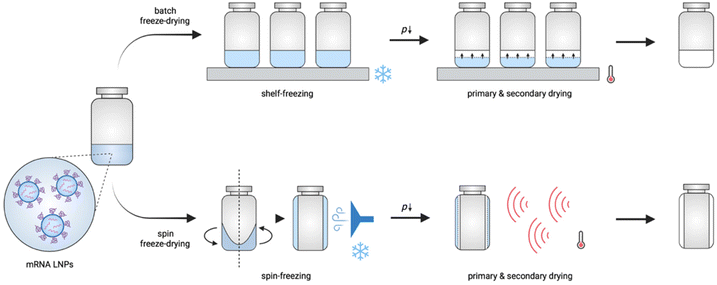 | ||
| Fig. 1 Schematic illustration of conventional batch freeze-drying and continuous spin freeze-drying. | ||
Novel and innovative continuous technologies exist, including continuous spin freeze-drying developed by Corver et al. (RheaVita, Ghent, Belgium), which overcome these limitations.11–15 This innovative technology integrates all freeze-drying process steps in a continuous production line and drastically reduces the production time and cost, and avoids scale-up issues. In this manufacturing technique, a vial filled with a liquid formulation is first spin-frozen by rapid rotation along its longitudinal axis while lowering the temperature by a flow of cold inert gas. This step generates a thin cylindrical frozen product layer that is uniformly spread across the inner wall of the vial. By reducing the product layer thickness and increasing the (macroscopic) surface area via spin-freezing, a significantly higher sublimation rate and, up to 40 folds, shorter total drying time can be achieved.11–15 Subsequently, the spin-frozen product is transferred via a load-lock to a drying chamber to initiate the primary drying. This load-lock system enables fast transfer of the vial from the atmospheric pressure (during spin-freezing) to vacuum pressure conditions (during primary drying) without the risk of any pressure increase in the drying chamber. Several infrared (IR) heaters are positioned in series in the drying chamber to provide energy for sublimation. Continuous spin freeze-drying also enables straightforward up-scaling and offers the possibility for in-line monitoring and controlling critical process parameters and critical quality attributes of each vial separately.11,15 Hence, identical quality can be assured for all processed vials.
In this work, we explored batch and continuous lyophilization technologies to produce thermostable nucleoside-modified mRNA LNP formulations. We selected two reduction-sensitive ionizable lipids and tested whether the mRNA LNP can be lyophilized in the presence of sucrose as a lyoprotectant. We tested the influence of lyophilization on the colloidal stability, mRNA payload encapsulation and transfection efficiency in vitro and in vivo, post lyophilization and redispersion in an aqueous medium.
Experimental section
Materials
Unless otherwise stated, all chemicals were purchased from Sigma Aldrich. Cell culture media and supplements, DPBS (1×), TBS (1×), Opti-MEM, Tryple Select and Quanti-iT RiboGreen RNA assay kit were purchased from Thermo Fischer. DPSC, DSPE-Cy5 and DMG-PEG2000, were purchased from Avanti Polar Lipids. MC38, CT26 and HEK293 cell lines were obtained from ATCC.mRNA-LNP formulations
mRNA-LNPs were fabricated by mixing an aqueous mRNA solution and an ethanolic lipid solution using a NanoAssemblr Benchtop device (Precision NanoSystems, Vancouver, Canada), which contained a staggered herringbone microfluidic mixer. Ethanol solutions (0.667 mL) contained S-Ac7-Dog or S-Ac7-DHDa, respectively, DSPC, cholesterol and DMG-PEG (mol% ratio 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5). Aqueous mRNA solutions (1.333 mL) were made by dissolving eGFP- or Fluc-encoding mRNA, respectively, in 5 mM acetate buffer at pH 4 at an mRNA concentration of 0.15 mg mL−1. Of note, eGFP mRNA LNP formulations were supplemented with 0.1 mol% fluorescently labeled DSPE-Cy5 (S-Ac7-Dog/S-Ac7-DHDa, DSPC, DSPE-Cy5, cholesterol, DMG-PEG2000 at a molar ratio of 50
1.5). Aqueous mRNA solutions (1.333 mL) were made by dissolving eGFP- or Fluc-encoding mRNA, respectively, in 5 mM acetate buffer at pH 4 at an mRNA concentration of 0.15 mg mL−1. Of note, eGFP mRNA LNP formulations were supplemented with 0.1 mol% fluorescently labeled DSPE-Cy5 (S-Ac7-Dog/S-Ac7-DHDa, DSPC, DSPE-Cy5, cholesterol, DMG-PEG2000 at a molar ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.9
9.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1
0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5). To remove ethanol, the LNP formulations were dialyzed against RNase-free water or Tris buffered saline (TBS), respectively, in Slide-A-Lyzer cassettes (cut-off 3.5 kDa) (Thermo Fischer, USA). LNP suspensions dialyzed against TBS were stored at 2–8 °C. LNP suspensions dialyzed against RNase-free water were immediately supplemented with 20% (w/v) sucrose and lyophilized. Control LNP suspensions in RNase-free water without sucrose were also lyophilized. Prior to lyophilization or storage, all dialyzed LNP suspensions were concentrated in Amicon Ultra 10K centrifugal filters (MilliporeSigma, USA), resulting in an mRNA concentration of 100 μg mL−1.
1.5). To remove ethanol, the LNP formulations were dialyzed against RNase-free water or Tris buffered saline (TBS), respectively, in Slide-A-Lyzer cassettes (cut-off 3.5 kDa) (Thermo Fischer, USA). LNP suspensions dialyzed against TBS were stored at 2–8 °C. LNP suspensions dialyzed against RNase-free water were immediately supplemented with 20% (w/v) sucrose and lyophilized. Control LNP suspensions in RNase-free water without sucrose were also lyophilized. Prior to lyophilization or storage, all dialyzed LNP suspensions were concentrated in Amicon Ultra 10K centrifugal filters (MilliporeSigma, USA), resulting in an mRNA concentration of 100 μg mL−1.
Batch freeze-drying
First, 2R glass vials (Schott, Müllheim, Germany) were filled with 350 μL of each mRNA-LNP formulation. Then, all vials were placed on a precooled shelf of an Amsco FINNAQUA GT4 freeze-dryer (GEA, Koln, Germany) at −40 °C. The shelf was kept at −40 °C for 2 h to ensure equal freezing of all vials. Subsequently, during the primary drying step, the pressure was reduced to 10 Pa and the temperature of the shelf was increased to −35 °C to let the samples dry for 24 h. To remove the residual moisture, the shelf temperature was further increased to 25 °C at a rate of 0.05 °C min−1. After 5 h, the drying procedure was ended by aerating the drying chamber with inert nitrogen gas and stoppering the vials. All freeze-dried vials were capped and eventually stored at 2–8 °C before use. Prior to physicochemical characterization, in vitro and in vivo transfection experiments samples were reconstituted in 875 μL 1× TBS resulting in an mRNA concentration of 40 μg mL−1 and a sucrose concentration of 8% w/v.Spin freezing and continuous single-vial lyophilization
First, 2R glass vials (Schott, Müllheim, Germany) were filled with 350 μL of each mRNA-LNP formulation. Then, each vial was separately placed vertically inside a single-vial spin freeze-dryer (RheaVita, Zwijnaarde, Belgium). Subsequently, the vial was rapidly rotated around its longitudinal axis at 3500 rpm. The temperature of the vial was lowered by a flow of cold compressed air, cooled using a heat exchanger. The heat exchanger consisted of a stainless steel container filled with liquid nitrogen, where a polyurethane tubing (with an internal diameter of 5 mm and a total length of 3 m) was submersed. The temperature of the vial was monitored via thermal imaging (FLIR A655sc, Thermal focus, Ravels, Belgium). In addition, the gas temperature was monitored using a thin gauge type-K thermocouple (Labfacility, Leeds, UK), which was positioned between the vial and the gas outlet. The vial and gas temperature were used as the input for an in-house scripted LABVIEW 2019 (National Instruments, Austin, TX, USA) application to control the freezing step. A cooling rate of 25 °C min−1 was used during the freezing step. In the single-vial drying chamber, vacuum was applied as soon as the vial obtained a temperature of −50 °C. Each vial was dried at 10 Pa without the use of an infrared heater, while rotating at 5 rpm (i.e., drying was based on the radiation from the surroundings) to ensure conservative drying conditions. After 5 h, the spin freeze-dried vial was stoppered under vacuum. Subsequently, the single vial drying chamber was aerated with inert nitrogen gas to end the drying procedure. All freeze-dried vials were capped and eventually stored at 2–8 °C before use. Prior to physicochemical characterization, in vitro and in vivo transfection experiment samples were reconstituted in 875 μL 1× TBS resulting in an mRNA concentration of 40 μg mL−1 and a sucrose concentration of 8% w/v.Dynamic light scattering (DLS) and electrophoretic light scattering (ELS)
DLS and ELS measurements were performed using a Zetasizer Nano ZS (Malvern Instruments Ltd, Malvern, UK) equipped with a HeNe laser (λ = 633 nm) and detection was done at a scattering angle of 173°. The samples were diluted 10 folds in 5 mM HEPES buffer (pH 7.4) and measured in triplicate by DLS. Cumulant analysis of the data yielded the z-average and polydispersity index (data provided as average). For ELS, the samples were also diluted 10 folds in 5 mM HEPES buffer (pH 7.4) and measured in triplicate. The zeta potential values were calculated based on the Smoluchowski equation and are expressed as average ± standard deviation.RiboGreen assay: mRNA encapsulation efficiency
LNP samples were diluted in a TE (Tris-EDTA) buffer to obtain a (theoretical) concentration of 1 μg mL−1 mRNA (working solutions). Then, 50 μL working solution was diluted two folds in a black 96-well plate with the TE buffer. In parallel, samples were prepared in the TE buffer supplemented with 2% Triton X-100 to dissolve the LNPs. Next, 100 μL of RiboGreen solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted RiboGreen reagent in TE buffer) was added to each well. Fluorescence was measured using an Ensight Multimode Microplate reader (PerkinElmer, USA) (λex = 485 nm, λem = 528 nm). The resulting fluorescence values were subtracted by the fluorescence value of the reagent blank. The encapsulation efficiency (EE%) was calculated according to the equation
100 diluted RiboGreen reagent in TE buffer) was added to each well. Fluorescence was measured using an Ensight Multimode Microplate reader (PerkinElmer, USA) (λex = 485 nm, λem = 528 nm). The resulting fluorescence values were subtracted by the fluorescence value of the reagent blank. The encapsulation efficiency (EE%) was calculated according to the equation 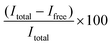 , where Itotal is the fluorescence intensity of samples measured with total mRNA in the presence of 2% Triton X-100 and Ifree is the fluorescence intensity of samples measured with free or unencapsulated mRNA in the absence of 2% Triton X-100.
, where Itotal is the fluorescence intensity of samples measured with total mRNA in the presence of 2% Triton X-100 and Ifree is the fluorescence intensity of samples measured with free or unencapsulated mRNA in the absence of 2% Triton X-100.
Small-angle X-ray scattering (SAXS)
SAXS experiments were performed using a Xeuss 3.0 XRS system (Xenocs) with an Eiger2R 1M detector (Dectris). An X-ray beam with a wavelength of 1.54 Å was generated by a Cu-source (Genix 3D; Xenocs) at 50 kV and 0.60 mA. LNPs were concentrated to a total lipid content of about 10 mg mL−1 with centrifugal filters (Amicon Ultra 10K centrifugal filters) and filled into 1.5 mm-thick borosilicate capillaries (wall thickness 0.01 mm; WJM-Glas). The measurements were done at room temperature, with an exposure time of 3 h and a sample-to-detector distance of 748 mm, providing a scattering vector magnitude, q, in the range of 0.1–3.4 nm−1 (q = 4π/λ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin
sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, where λ is the wavelength and 2θ is the scattering angle). Collected scattering patterns on the detector were azimuthally averaged to provide the 1-D curves with the experimental uncertainties, and scattering from PBS was subtracted as background (XSACT software; Xenocs). Additional SAXS exposures were carried out on the same samples to check for the radiation damage. No radiation damage was observed. Correlation spacing, d, was estimated from the q value of the correlation peak maxima as d = 2π/qmax.
θ, where λ is the wavelength and 2θ is the scattering angle). Collected scattering patterns on the detector were azimuthally averaged to provide the 1-D curves with the experimental uncertainties, and scattering from PBS was subtracted as background (XSACT software; Xenocs). Additional SAXS exposures were carried out on the same samples to check for the radiation damage. No radiation damage was observed. Correlation spacing, d, was estimated from the q value of the correlation peak maxima as d = 2π/qmax.
In vitro mRNA transfection
MC38, CT26 and HEK293 cells were seeded in 96-well plates at a density of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 100 μL complete medium and allowed to adhere and reach 80% confluency for 24 h at 37 °C (5% CO2). Lyophilized LNP were reconstituted in TBS. Next, each well was pulsed in triplicate with 10 μL (200 ng of mRNA) of Cy5 labeled eGFP mRNA LNP formulations (diluted in Opti-MEM). Plates were subsequently incubated for 24 h at 37 °C (5% CO2). After 24 h, cells were first centrifuged (350g, 5 min), the supernatant was removed and 30 μL prewarmed TrypleSelect was added to each well and incubated for 5 min at 37 °C (5% CO2) to detach cells. Next, cells were suspended with 200 μL FACS buffer and analyzed using a BD Accuri flow cytometer (BD Bioscience, USA). Data were processed using the FlowJo software package (BD Bioscience, USA).
000 cells per well in 100 μL complete medium and allowed to adhere and reach 80% confluency for 24 h at 37 °C (5% CO2). Lyophilized LNP were reconstituted in TBS. Next, each well was pulsed in triplicate with 10 μL (200 ng of mRNA) of Cy5 labeled eGFP mRNA LNP formulations (diluted in Opti-MEM). Plates were subsequently incubated for 24 h at 37 °C (5% CO2). After 24 h, cells were first centrifuged (350g, 5 min), the supernatant was removed and 30 μL prewarmed TrypleSelect was added to each well and incubated for 5 min at 37 °C (5% CO2) to detach cells. Next, cells were suspended with 200 μL FACS buffer and analyzed using a BD Accuri flow cytometer (BD Bioscience, USA). Data were processed using the FlowJo software package (BD Bioscience, USA).
In vivo mRNA transfection
All animal procedures were performed in accordance with the ethical guidelines for the welfare of laboratory animals of Ghent University and approved by the ethical committee of the Faculty of Veterinary Sciences (Ghent University, Ghent, Belgium, EC approval 2022-10). Balb/c mice, aged 7–9 weeks, were housed in individual ventilated cages and allowed to access food and water ad libitum. LNPs were lyophilized in the presence of 20% w/v of sucrose. Lyophilized LNPs were reconstituted in TBS. Then, 50 μL of Fluc mRNA-LNP (40 μg mL−1 Fluc mRNA) was injected intramuscularly into the quadriceps (n = 3) (2 μg mRNA dose). Prior to bioluminescence imaging at 0, 4 and 24 h post injection, mice were injected subcutaneously with 200 μL D-luciferin. After 12 min, bioluminescence images were acquired using a IVIS Lumina II imaging system. The Living Image 4.4 software package was used for quantitative analysis.Results and discussion
mRNA LNP formulations and characterization
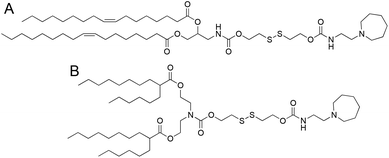
We selected two structurally analogous ionizable lipids, i.e., S-Ac7-Dog and S-Ac7-DHDa, from an ongoing combinatorial library screening campaign of ionizable lipids for the mRNA LNP formulation.16 The molecular structure of both ionizable lipids is depicted in Fig. 2. S-Ac7-Dog and S-Ac7-DHDa contain an azepanyl moiety as an ionizable amine headgroup and a reducible disulfide bond in the linker region between the head group and the lipid tails. S-Ac7-Dog contains two oleoyl lipid tails, whereas S-Ac7-DHDa contains two branched hexyldecanoic acid lipid tails. The connection between the lipid region and the disulfide linker region also differs between both lipids.LNP formulations contained, besides ionizable lipids, cholesterol, distearoylphosphatidylcholine (DSPC) as a phospholipid and 1,2-dimyristoyl-rac-glycero-3-methylpolyethylene glycol, with 2 kDa molecular weight of the PEG chain (DMG-PEG). Cholesterol and DSPC serve to enhance the LNP stability and facilitate endosomal membrane rupturing. DMG-PEG serves for colloidal stabilization. All lipids were dissolved in ethanol at a molar ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5. The latter is a composition often reported in the literature3,8,17,18 (Table S1†). mRNA was dissolved in an aqueous acetate buffer at pH 4.
1.5. The latter is a composition often reported in the literature3,8,17,18 (Table S1†). mRNA was dissolved in an aqueous acetate buffer at pH 4.
mRNA LNP formulations were produced by mixing ethanolic and aqueous solutions in a microfluidic turbulent mixing device.
mRNA encoding for enhanced green fluorescent protein (eGFP) was used as a reporter protein for analysing the in vitro testing of the mRNA transfection by flow cytometry. mRNA encoding for firefly luciferase (Fluc), as a reporter protein, was used for measuring the in vivo mRNA transfection efficiency by bioluminescence imaging. Ethanol was removed after microfluidic mixing by dialysis against nuclease-free (RNase) water. We deliberately did not use buffer salts during dialysis to mitigate the risk of pH changes upon freezing and lyophilization by evaporation and crystallization of buffer components.7 Dynamic light scattering analysis revealed a diameter of 109 ± 4 nm for S-Ac7-Dog LNP and 148 ± 5 nm for S-Ac7-DHDa LNPs loaded with eGFP mRNA. The S-Ac7-Dog LNP had a pKa of 6.85 and the S-Ac7-DHDa LNP had a pKa of 6.47.16
Both LNPs showed a slightly positive electrophoretic mobility analysis, at a physiological pH of 7.4, indicating slightly positive zeta-potential values for S-Ac7-Dog LNPs and slightly negative values for S-Ac7-DHDa LNPs. A RiboGreen RNA assay indicated that that mRNA was fully (i.e., 100%) encapsulated in both LNP formulations. LNP formulations containing mRNA encoding for Fluc showed similar characteristics.
mRNA LNP lyophilization and biophysical characterization upon reconstitution
The produced LNP formulations were divided into 5 parts. One part was stored at 4 °C, and the other parts were subjected to lyophilization. Half of the latter were supplemented with 20% w/v sucrose as a lyoprotectant prior to lyophilization. The other half was lyophilized in the absence of lyoprotectants.We selected sucrose as a widely used lyoprotectant.19–21 This choice was supported by a screening campaign, during which we tested two monosaccharides (i.e., glucose and fructose), one sugar alcohol (mannitol) and three disaccharides (i.e., sucrose, lactose and trehalose). LNPs supplemented with 20% w/v of mannitol were not colloidally stable after freezing and thawing, probably due to the crystallization of mannitol from solution upon freezing.19,22 Lyophilization with fructose and glucose resulted in a collapsed cake, probably due to the lower glass transition temperature of monosaccharides compared to disaccharides.23 Notably, lactose is a reducing sugar and might affect the stability of ionizable lipids that contain a reduction-sensitive disulfide bond. Hence, sucrose and trehalose were found suitable for lyophilization. Ultimately, we selected sucrose as it is already present in the Pfizer-BioNTech and Moderna COVID-19 mRNA LNP vaccine formulations as a cryoprotectant to maintain the LNP integrity during freezing.5
LNP formulations were subjected to batch freeze-drying and spin freeze-drying, respectively. For batch freeze-drying, all vials were placed on the shelf of the freeze-dryer and lyophilized in a single run. The total drying time was 51 hours. For spin freeze-drying, each vial was placed separately inside a single-vial spin freeze-dryer, spin-frozen and dried under vacuum for 5 hours. During the spin-freezing step, a thin layer spread over the entire vial wall was achieved by rotating the vial rapidly along its longitudinal axis. This thin frozen product layer enabled an 8 times faster drying time compared to conventional freeze-drying. The product temperature was controlled by using an infrared camera in combination with an infrared heater in a closed feedback loop. The cake structure (Fig. 3) in the freeze-dried vials containing 20% w/v sucrose lyoprotectant had an intact appearance without any sign of collapse or cracks. A minor shrinkage of the cake occurred due to the release of drying tension. Spin freeze-drying resulted in a thin cake structure layered across the inner vial wall. This thin cake structure offers the advantage of fast reconstitution in an aqueous medium due to a higher contact surface in comparison to the thick cake obtained by batch lyophilization.
Next, all vials were reconstituted in Tris buffered saline (TBS) by gentle vortexing, targeting an mRNA concentration of 40 μg mL−1.
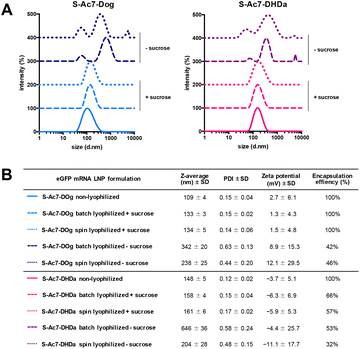
Biophysical characterization of the reconstituted mRNA LNP formulations by DLS, electrophoretic mobility analysis and RiboGreen assay is reported in Fig. 4. Formulations (batch and spin), lyophilized in the presence of sucrose as a lyoprotectant, exhibited a 10–25 nm increase in the hydrodynamic diameter, but maintained a narrow size distribution (PDI < 0.2) and identical zeta-potential values. Lyophilization of LNPs in the absence of cryoprotectants showed a strong increase in particle diameter and high PDI values, indicating the formation of macroscopic aggregates. The type of ionizable lipid did not influence the LNP size and zeta-potential values. S-Ac7-Dog LNP maintained a 100% mRNA encapsulation efficiency upon reconstitution. S-Ac7-DHDa LNPs, by contrast, released about 40% of the initially encapsulated mRNA into the solution, independent of the type of lyophilization process.
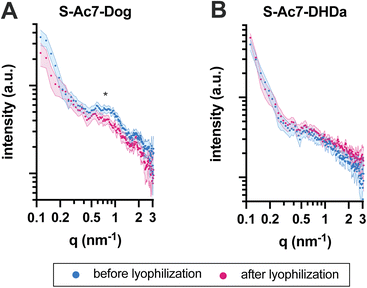
The self-assembled nanostructure of the LNPs before and after lyophilization was investigated by small-angle X-ray scattering (SAXS) (Fig. 5). All LNPs exhibited an upturn in scattering intensity at low q values (<0.3 nm−1), attributed to the scattering from lipid emulsions with a defined volume at dimensions larger than the resolution of the experimental SAXS setup. The curve for S-Ac7-Dog LNPs before lyophilization also exhibited a correlation peak at around q ≈ 0.7 nm−1 (Fig. 5A), indicating the presence of structural organization within the LNPs with a spacing of around 9.0 nm. This is attributed to the long-range arrangement of mRNA phosphate backbone chains in relation to one another, potentially residing in water pockets within the LNP core, correlating well with reports in the literature.8 Notably, such correlation peak was less pronounced in the SAXS curve of S-Ac7-Dog LNPs after lyophilization, indicating structural rearrangements most likely caused by the removal of water from the LNPs’ core and collapse of the internal water pockets. The correlation peak was not observed for the S-Ac7-DHDa LNPs (Fig. 5B), suggesting a lack of internal nanostructure within the core of these particles.
In vitro transfection efficiency of lyophilized mRNA LNP formulations
In vitro eGFP expression by cells treated with mRNA LNP formulations before and after lyophilization was analyzed by flow cytometry on three different cell lines: MC38 (mouse colon cancer), CT26 (mouse colon cancer) and HEK293T (human embryonic kidney). DSPE-Cy5 was included as a fluorescent dye in the LNP formulations for measuring LNP uptake by cells.Lyophilization in the absence of cryoprotectants induced, relative to freshly prepared LNPs, a drastic drop in eGFP expression and LNP uptake in all cell lines (Fig. 6). By contrast, lyophilization of mRNA LNPs in the presence of cryoprotectants fully maintained the magnitude of eGFP expression and cellular uptake of LNPs. Batch lyophilization and spin lyophilization were equally performing in this context. Interestingly, the fraction of mRNA that is released from S-Ac7-DHDa LNPs upon lyophilization and reconstitution did not majorly impact the transfection efficiency. eGFP expression, however, did differ between cell lines and LNP formulations. The underlying reason for this is the subject of ongoing research and is beyond the scope of this paper.
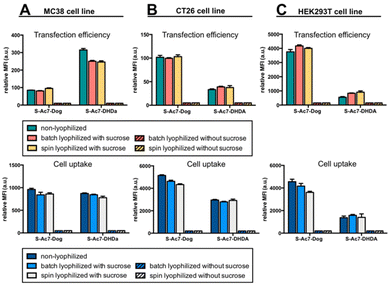 | ||
| Fig. 6 In vitro transfection efficiency and cell uptake of eGFP mRNA LNP formulations on (A) MC38, (B) CT26 and (C) HEK293T cell lines. | ||
In vivo transfection efficiency of lyophilized mRNA LNP formulations
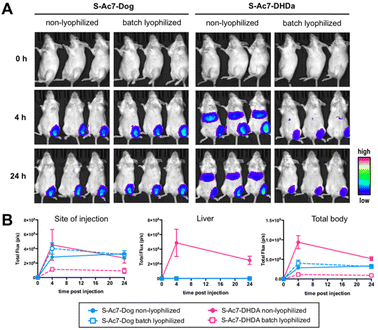
Batch lyophilization was selected to limit the number of laboratory animals. BALB/c mice were administered with an intramuscular injection into the quadricep of mRNA LNP containing 2 μg of mRNA. Fluc expression was measured 4 and 24 h post injection by bioluminescence imaging (Fig. 7A). The total bioluminescence signal (total flux (photons per second)) and the signal of the injection site and liver were quantified (Fig. 7B). Freshly prepared S-Ac7-Dog mRNA LNPs induced a strong bioluminescence signal at the site of injection, whereas S-Ac7-DHDA LNPs induced a strong signal at the site of injection and in distal tissues, likely in the liver and the spleen. The underlying reason for the qualitative differences in the biodistribution of the Fluc expression between both mRNA LNP formulations might be due to the differences in cationic charge density and is subject of our ongoing studies. Lyophilization did not alter the Fluc expression of S-Ac7-Dog mRNA LNPs. By contrast, the Fluc expression of S-Ac7-DHDA mRNA LNPs at the site of injection was strongly reduced upon lyophilization. Moreover, an even stronger reduction of the Fluc bioluminescence was observed in distal tissues.Conclusions
We explored conventional batch lyophilization and an innovative continuous lyophilization process for the lyophilization of mRNA LNP formulations. Lyophilization of mRNA LNPs in the absence of lyoprotectants induced the full release of the mRNA payload from the LNP upon reconstitution in an aqueous medium, deteriorated the colloidal stability and fully abrogated the capacity of mRNA LNPs to transfect cells. Sucrose (20% w/v) as a lyoprotectant allowed the reconstitution of the LNP in an aqueous medium with minor influence on the LNP size and zeta potential. LNPs comprising S-Ac7-Dog as an ionizable lipid retained their mRNA payload stably encapsulated upon lyophilization and reconstitution, whereas LNPs comprising S-Ac7-DHDa released part of their mRNA payload. This difference might, as suggested by SAXS analysis, be attributed to the structural differences in the LNP core between S-Ac7-Dog and S-Ac7-DHDa LNP, which render the latter LNP prone to mRNA release upon dehydration during the lyophilization process. In vitro translation of eGFP-encoding mRNA was not affected by lyophilization for any of the LNP formulation. The lyophilization of S-Ac7-Dog LNPs comprising mRNA encoding for Fluc did not alter the amplitude and biodistribution of the Fluc bioluminescence upon intramuscular injection. S-Ac7-DHDa mRNA LNPs showed a loss in Fluc expression efficiency upon lyophilization and an altered biodistribution of the bioluminescence signal, which again might be attributed to the structural changes in the LNP core due to lyophilization and redispersion.Our findings demonstrate that lyophilization of mRNA LNPs, comprising a reduction-sensitive ionizable lipid, is feasible, also using an innovative rapid and continuous lyophilization process. The structure of ionizable lipid strongly influenced the quality of the lyophilized product. Whether the difference in the biodistribution of the Fluc expression, between S-Ac7-Dog and S-Ac7-DHDa LNP, which only differ in the structure of their alkyl tails, and the inability of S-Ac7-DHDa LNPs to be successfully lyophilized are connected remains elusive and will be subject of future investigations.
Author contributions
A. L., J. L., E. D., Z. Z., M. G. and Y. C. performed experimental work. T. R. M. D. and B. G. D. supervised the work. All authors contributed to writing and editing of the manuscript.Conflicts of interest
Y. C., S. D. and B. G. D. are inventor of a patent on the ionizable lipids used in this work. S. D. is employee of a company commercializing these lipids. T. R. M. D. is co-founder of a company commercializing the spin lyophilization technology used in this work.Acknowledgements
B. G. D. G. acknowledges funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant no. 817938). The Hercules foundation is recognized for funding the Xenocs Xeuss 3.0 X-ray scattering equipment (FWO Hercules Grant AUGE/17/29).References
- T. Ye, Z. Zhong, A. García-Sastre, M. Schotsaert and B. G. De Geest, Current Status of COVID–19 (Pre)Clinical Vaccine Development, Angew. Chem., 2020, 132(43), 19045–19057 CrossRef
.
- S. Jain, A. Venkataraman, M. E. Wechsler and N. A. Peppas, Messenger RNA-Based Vaccines: Past, Present, and Future Directions in the Context of the COVID-19 Pandemic, Adv. Drug Delivery Rev., 2021, 179, 114000 CrossRef CAS PubMed
.
- M. D. Buschmann, M. J. Carrasco, S. Alishetty, M. Paige, M. G. Alameh and D. Weissman, Nanomaterial Delivery Systems for MRNA Vaccines, Vaccines, 2021, 9(1), 65 CrossRef CAS PubMed
.
- P. P. Piccaluga, A. Di Guardo, A. Lagni, V. Lotti, E. Diani, M. Navari and D. Gibellini, COVID-19 Vaccine: Between Myth and Truth, Vaccines, 2022, 10(3), 349 CrossRef CAS PubMed
.
- M. N. Uddin and M. A. Roni, Challenges of Storage and Stability of Mrna-Based Covid-19 Vaccines, Vaccines, 2021, 9(9), 1–9 Search PubMed
.
- D. J. A. Crommelin, T. J. Anchordoquy, D. B. Volkin, W. Jiskoot and E. Mastrobattista, Addressing the Cold Reality of MRNA Vaccine Stability, J. Pharm. Sci., 2021, 110(3), 997–1001 CrossRef CAS PubMed
.
- L. Schoenmaker, D. Witzigmann, J. A. Kulkarni, R. Verbeke, G. Kersten, W. Jiskoot and D. J. A. Crommelin, MRNA-Lipid Nanoparticle COVID-19 Vaccines: Structure and Stability, Int. J. Pharm., 2021, 601, 120586 CrossRef CAS PubMed
.
- M. Y. Arteta, T. Kjellman, S. Bartesaghi, S. Wallin, X. Wu, A. J. Kvist, A. Dabkowska, N. Székely, A. Radulescu, J. Bergenholtz and L. Lindfors, Successful Reprogramming of Cellular Protein Production through MRNA Delivered by Functionalized Lipid Nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(15), E3351–E3360 CAS
.
- X. Tang and M. J. Pikal, Design of Freeze-Drying Processes for Pharmaceuticals: Practical Advice, Pharm. Res., 2004, 21(2), 191–200 CrossRef CAS PubMed
.
- F. Emami, A. Vatanara, E. J. Park and D. H. Na, Drying Technologies for the Stability and Bioavailability of Biopharmaceuticals, Pharmaceutics, 2018, 10(3), 1–22 CrossRef PubMed
.
- P. J. Van Bockstal, S. T. F. C. Mortier, L. De Meyer, J. Corver, C. Vervaet, I. Nopens and T. De Beer, Mechanistic Modelling of Infrared Mediated Energy Transfer during the Primary Drying Step of a Continuous Freeze-Drying Process, Eur. J. Pharm. Biopharm., 2017, 114, 11–21 CrossRef CAS PubMed
.
- R. Pisano, A. Arsiccio, L. C. Capozzi and B. L. Trout, Achieving Continuous Manufacturing in Lyophilization: Technologies and Approaches, Eur. J. Pharm. Biopharm., 2019, 142(June), 265–279 CrossRef CAS PubMed
.
- L. Leys, B. Vanbillemont, P. J. Van Bockstal, J. Lammens, G. Nuytten, J. Corver, C. Vervaet and T. De Beer, A Primary Drying Model-Based Comparison of Conventional Batch Freeze-Drying to Continuous Spin-Freeze-Drying for Unit Doses, Eur. J. Pharm. Biopharm., 2020, 157(May), 97–107 CrossRef CAS PubMed
.
- J. Lammens, N. M. Goudarzi, L. Leys, G. Nuytten, P. J. Van Bockstal, C. Vervaet, M. N. Boone and T. De Beer, Spin Freezing and Its Impact on Pore Size, Tortuosity and Solid State, Pharmaceutics, 2021, 13(12), 1–16 CrossRef PubMed
.
- L. De Meyer, P. J. Van Bockstal, J. Corver, C. Vervaet, J. P. Remon and T. De Beer, Evaluation of Spin Freezing versus Conventional Freezing as Part of a Continuous Pharmaceutical Freeze-Drying Concept for Unit Doses, Int. J. Pharm., 2015, 496(1), 75–85 CrossRef CAS PubMed
.
- Manuscript in preparation.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Lipid Nanoparticles for MRNA Delivery, Nat. Rev. Mater., 2021, 6(12), 1078–1094 CrossRef CAS PubMed
.
- K. H. Moss, P. Popova, S. R. Hadrup, K. Astakhova and M. Taskova, Lipid Nanoparticles for Delivery of Therapeutic RNA Oligonucleotides, Mol. Pharm., 2019, 16(6), 2265–2277 CrossRef CAS PubMed
.
- M. D. Howard, X. Lu, M. Jay and T. D. Dziubla, Optimization of the Lyophilization Process for Long-Term Stability of Solidlipid Nanoparticles, Drug Dev. Ind. Pharm., 2012, 38(10), 1270–1279 CrossRef CAS PubMed
.
- P. Zhao, X. Hou, J. Yan, S. Du, Y. Xue, W. Li, G. Xiang and Y. Dong, Long-Term Storage of Lipid-like Nanoparticles for MRNA Delivery, Bioact. Mater., 2020, 5(2), 358–363 CrossRef PubMed
.
- R. Lball, P. Bajaj and K. A. Whitehead, Achieving Long-Term Stability of Lipid Nanoparticles: Examining the Effect of PH, Temperature, and Lyophilization, Int. J. Nanomed., 2017, 12, 305–315 Search PubMed
.
- J. Horn and W. Friess, Detection of Collapse and Crystallization of Saccharide, Protein, and Mannitol Formulations by Optical Fibers in Lyophilization, Front. Chem., 2018, 6(JAN), 4 CrossRef PubMed
.
-
Methods in Molecular Biology 1257, ed. W. F. Wolkers and H. Oldenhof, Cryopreservation and Freeze-Drying Protocols, Springer-Verlag, New York, 2015 Search PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2bm02031a |
| ‡ Authors with equal contribution. |
| This journal is © The Royal Society of Chemistry 2023 |