Supramolecular helical self-assembly of small peptides
Rajat Subhra
Giri
 and
Bhubaneswar
Mandal
and
Bhubaneswar
Mandal
 *
*
Department of Chemistry, Laboratory of Peptide and Amyloid Research, Indian Institute of Technology Guwahati, Assam-781039, India. E-mail: bmandal@iitg.ac.in
First published on 26th November 2021
Abstract
The self-assembly of peptides forming various micro to nanostructures has several applications in nanobiotechnology. β-Sheet based peptides are well known as they are abundant in nature, e.g., silk and amyloid fibrils. Therefore, the self-assembly of small β-sheet peptides and their potent applications are well explored to date. On the other hand, various helical secondary structures are also abundant in nature. Like β-sheet peptides, small peptide-based helical assemblies are not explored enough, but they also attract attention for fabricating various artificial nanomaterials in recent times. This highlight focuses on single-crystal X-ray diffraction (SC-XRD) based analysis of the supramolecular arrangement, conformation, and higher-order assembly of small helical peptides. We outline the role of building blocks and their structural beauty to adopt helical assembly, including single-, double- and triple-stranded helices. We also describe their micro or nano-level structures obtained from the solution and their potent applications as drug delivery vehicles, as porous materials for N2 adsorption, and for nanomaterial fabrication.
Introduction
Molecular self-assembly is a unique pathway where molecules self-assemble by various non-covalent interactions without external force.1 Most interestingly, such a molecular self-assembly process is also observed in nature, e.g., right-handed α-helix of proteins,2 right-handed DNA double helix,3 and triple helix of collagen,4 spider silks,5 and tobacco mosaic virus (TMV).6 The self-assembly of small peptides through the ‘bottom-up approach’ to form various interesting micro to nano-level structures7,8 (e.g., nano-rods, nano-tubes, nanofibrils, nanoribbons, nanotapes, nanospheres, and nanobelts) has exciting applications in nanobiotechnology, material science, medicinal chemistry9,10 drug-delivery,11–13 gene expression,14,15 ion channels,16 tissue engineering,17–19 biosensors,20,21 smart materials, and devices.22–26 Additionally, due to their structural diversity,27,28 biocompatibility,29,30 bioactivity, biodegradability,31 self-association ability,32 molecular recognition,33 flexible mechanical stability and electronic properties,34–36 peptides are attractive entities in the delivery of xenobiotics, optoelectronics, piezoelectrics and pyroelectrics, optical waveguides, sensing, and photocatalysts.25,26,34–38 The self-assembly of peptides happens through various supramolecular non-covalent interactions to build several secondary structures. The most abundant secondary structures found in proteins are the β-sheet and α-helix. β-Sheet structures are abundant in spider silks and amyloid fibrils.39,40 The accumulation of amyloid fibrils in the brain causes various neurodegenerative diseases, e.g., Alzheimer's disease, Parkinson's disease, Huntington's disease, and prion disease.41–44 The amyloid structure deciphered from XRD, NMR, and cryoEM experiments39,40,45–47 is a cross-β-sheet with a tubular hollow core. However, recently, amyloid fibril with a cross-α structure48 (a 22-membered phenol-soluble modulin α3 peptide) has also been reported.Additionally, globular proteins and collagen protein (abundant in mammals) are enriched with helices. The self-assembled helical arrangements, e.g., the triple helix of collagen49,50 (three left-handed helical polypeptide chains tightly bound to form a right-handed superhelix), play an essential role in biological systems, e.g., cell adhesion and growth.51,52 Collagen is a fibrous protein found in connective tissue, e.g., ligaments, bones, tendons, and skin. Inspired by this beauty of self-assembled structures from biological systems, scientists have tried to de novo design and develop short helical peptides using several building blocks (mimicking natural or unnatural amino acids) over the last few decades. This peptide modification via mimicking the backbone or side chains to build supramolecular helical association provided a new direction in functional material design and their applications. Although the construction of helical assembly from small peptides is more challenging than the self-assembled β-sheet structure, the possibility of generating several valuable properties in helical peptides motivates us to explore this research field.
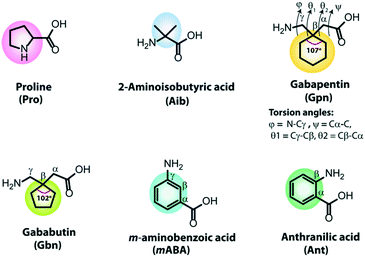
Structural feature of some building blocks
In developing de novo designed artificial helical peptides, the building blocks of peptides play an essential role. Natural or unnatural amino acids containing a pre-organized kink moiety may act as a turn inducer in the peptide backbone. Further, these peptide backbones may self-organize aided by various non-covalent interactions to construct a supramolecular helical assembly. Therefore, the design and construction of chemically modified building blocks are challenging tasks. Naturally occurring amino acids, proline and glycine, act as turn inducers due to their structural rigidity and flexibility. Over the last few decades, some unnatural building blocks have been developed as turn inducers, e.g., α-aminoisobutyric acid (Aib) and its higher homologs (α,α-diethylglycine (Deg), α,α-di-n-propylglycine (Dpg) and α,α-di-n-butylglycine (Dbg)), cycloalkyl side-chains of dialkylglycine (Acnc, n = 3–12, number of atoms in the ring),53–57trans-2-aminocyclopentanecarboxylic acid (ACPC), trans-2-aminocyclohexanecarboxylic acid (ACHC),58–61 chiral α,α-dialkylated amino acids (α-methyl-α-ethylglycine, and α-methyl phenylalanine),62,63 α/β/γ cyclic or acyclic amino acids,64–66 and so on. But, here, we discussed some building blocks involved in short peptides to form helical structures. Due to its structural diversity and high crystallinity, the most frequently used turn and helix inducer is α-aminoisobutyric acid (Aib). It is a modified alanine amino acid as the Cα proton of Ala is replaced with a methyl group, i.e., two geminal methyl groups are present at the Cα of Aib (Fig. 1). The observed backbone torsion angles ϕ and ψ of the Aib residue are −60° (±20°) and −30° (±20°), suggesting the adaptation of a right-handed α-helix or 310 helical structure. However, its ϕ and ψ angles may also fall in the left-handed α-helix or 310 helix region in the Ramachandran plot, and thus it may adapt left-handed conformations.54,67,68Another important non-proteogenic building block is γ-aminobutyric acid (GABA), which acts as a neurotransmitter. Due to the diversity of conformation orientation around the C–C bond of GABA, it produces several modified γ-residues, e.g., gabapentin (Gpn) and gababutin (Gbn). Gpn is an achiral cyclic β,β′-disubstituted γ-amino acid(1-(aminomethyl)cyclohexaneacetic acid) used as an antiepileptic drug (Fig. 1). The Gpn residue adopts a gauche–gauche conformation due to the presence of two symmetrical geminal substitutions at the Cβ atom, which restricts the torsion angles θ1 (Cγ–Cβ) and θ2 (Cβ–Cα).55 It is observed that Gpn-based peptides exhibited a C7 to C14 intramolecular H-bonded structure.69 Moreover, Gbn contains a five-membered cyclic ring at the Cβ position, and it is more rigid than Gpn (six-membered cyclic ring at Cβ) as the interior angle of the Gbn ring (102.247°) is more rigid than the Gpn ring (107.157°).70 Interestingly, Gbn-based peptides exhibit a similar structural conformation and folding patterns to Gbn-based peptides.
Another substituted γ-aminobutyric acid is meta-amino benzoic acid which has an all-trans extended configuration (Fig. 1). It is a rigid, unnatural γ-amino acid, contains a six-membered aromatic ring and has a two intervening dihedral angles θ1 (Cβ–Cγ) and θ2 (Cα–Cβ).66,71 Similarly, anthranilic acid (Ant) or ortho-amino benzoic acid (Fig. 1) is a constrained β-amino acid with a dihedral angle θ (Cα–Cβ), which also acts as a turn inducer.
Construction of a single helix
The naturally occurring single helical structure was noted in proteins and RNA that plays an essential role in biological systems. Double helical DNA structures influenced by hydrophobic and electrostatic interactions form a coiled-coil structure vital for gene transcription and molecular recognition. Additionally, long-chain polypeptide helical structures help in designing functional materials. Similarly, creating a single helical structure from small peptides is useful in nanobiotechnology and materials research. Recently, Gazit et al. reviewed the functional nanomaterials made up of short helical peptides.72,73 But, in this highlight, we describe the formation of supramolecular helical peptides by various non-covalent interactions based on the XRD data. We categorized them into di, tri, and tetrapeptide helices and discussed their developments in the last two decades.Single amino acid-based single helix
Xing, Zhao and co-workers reported the transmission of helicity from the molecularly resolved level to the macroscopically resolved level through the inversion of the supramolecular chirality of N-terminal Fmoc protected single amino acids.74 In the crystalline state, Fmoc protected L-Asp and D-Asp exhibited an M-helix and P-helix, respectively, along the H-bonded array (Fig. 2a). But interestingly, both L and D isomers showed inverted handedness helical structures (P-helix for L-Asp and M-helix for D-Asp) with the co-assembly of melamine (Mm) in solution. Here, Mm plays a crucial role in inverting the chirality of the helix through H-bonding with carboxylic acid groups of aspartic acid.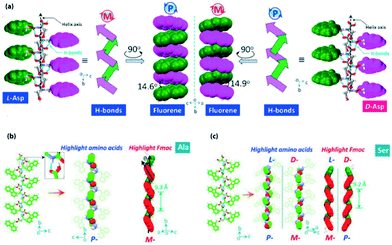 | ||
| Fig. 2 (a) Supramolecular tilt chirality with a P-helix and M-helix for L- and D-Asp, respectively (reproduced from ref. 74 with permission from American Chemical Society). (b and c) Packing of Fmoc-Ala and Fmoc-Ser with tilted helical chirality of aryl groups (reproduced from ref. 75 with permission from Elsevier). | ||
Hao, Xing, and co-workers described the helical secondary structure and supramolecular tilted chirality of N-terminal aromatic amino acids.75 They developed a process to determine helical sequences in N-terminal aromatic amino acids. Fmoc-amino acids produced a super-helical structure by three types of asymmetric H-bond modalities. The modalities are amide → carboxylic acid and carboxylic acid → carboxylic acid modality (responsible for the P-handed helix) and carboxylic acid → amide modality (responsible for the M-handed helix). The tilted supramolecular helical structures of Fmoc-Ala and Fmoc-Ser are represented, for example (Fig. 2b and c).
Dipeptide single helix
Görbitz reported four hydrophobic dipeptides (Leu–Leu, Leu–Phe, Phe–Leu, and Phe–Phe), which formed nanotubes (Fig. 3a–d). These peptides crystallized to form helices with four to six peptide molecules per turn through hydrogen bonding interactions in a head-to-tail fashion.76 Interestingly, two dimers of Leu–Leu and Leu–Phe formed rectangular central hydrophilic channels with van der Waals' dimensions of 2.5 × 6.0 Å. In contrast, the dimers of Phe–Leu formed a pseudo-tetragonal hydrophilic channel with dimensions of 4.0 × 6.0 Å. Phe–Phe formed a bigger hydrophilic channel with a diameter of 10 Å. As the channel size of Phe–Phe is large, therefore water molecules and ions can be transported.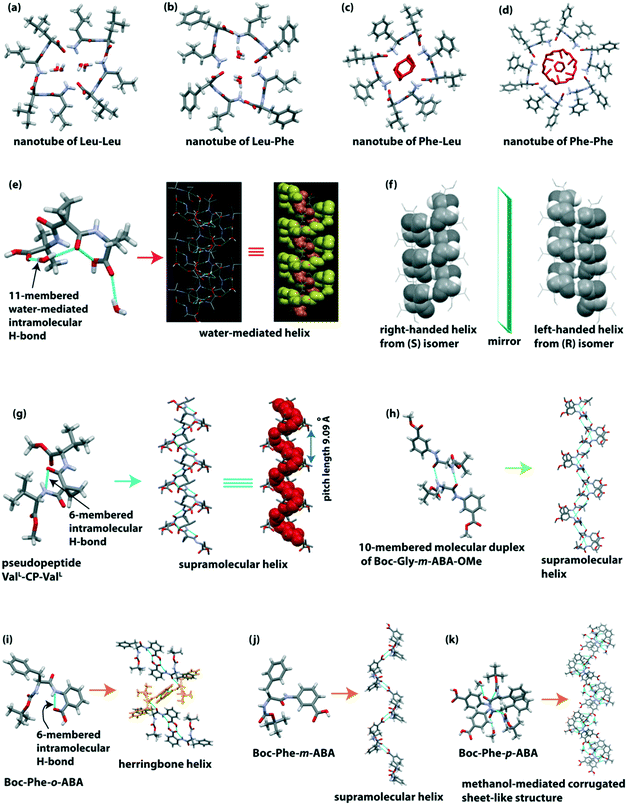 | ||
| Fig. 3 Hydrophilic channels of helical hydrophobic dipeptides (a) Leu–Leu, (b) Leu–Phe, (c) Phe–Leu and (d) Phe–Phe. (e) Water-mediated 1D helix of the pseudopeptide bis(N-α-amido-α-aminoisobutyric acid)-1,1-cyclopropane dicarboxylate (reproduced from ref. 77 with permission from Elsevier). (f) Right and left-handed helixes of the enantiomeric pseudopeptide bis(N-α-amido-L-valine methyl ester) malonate (reproduced from ref. 78 with permission from John Wiley and Sons). (g) Supramolecular single helix of the pseudopeptide bis-(N-α-amido-L-valine methyl ester)-1,1-cyclopropane dicarboxylate (ValL-CP-ValL). (h) Supramolecular single helix of the pseudopeptide Boc-Gly-m-ABA-OMe. (i) Herringbone helix of the (α,β) hybrid peptide Boc-Phe-o-aminobenzoic acid. (j) Single helix of the (α,γ) hybrid peptide Boc-Phe-m-aminobenzoic acid. (k) Corrugated sheet-like structure formed through the bridging methanol of the (α,δ) hybrid peptide Boc-Phe-p-aminobenzoic acid. | ||
Banerjee et al. developed a water-mediated one dimensional (1D) helical assembly from the water-soluble pseudopeptide bis(N-α-amido-α-aminoisobutyric acid)-1,1-cyclopropane dicarboxylate.77 The pseudopeptide adopted a folded turn conformation stabilized by a cyclic 9-membered intramolecular H-bond and a water-mediated cyclic 11-membered H-bond (Fig. 3e). Here, two water molecules play an essential role in forming the outer and inner supramolecular helix. The pitch of the hydrated outer helix is 9.52 Å, and the interior dimension of the helical channel is 6.36 × 5.44 Å. The internal channel of the helix is hydrophilic as polar COOH and CONH groups are present, while the exterior is hydrophobic as hydrophobic side chains of Aib and cyclopropyl groups are present (Fig. 3e). Interestingly, a 1D helical array of water molecules is formed, which is stabilized by this supramolecular helix, i.e., this pseudopeptide-based helix acts as a host.
They further constructed right and left-handed helical nanofibers from the self-association of pseudopeptides.78 The reported pseudopeptides are bis(N-α-amido-L-valine methyl ester) malonate (S), and its enantiomer bis(N-α-amido-D-valine methyl ester) malonate (R), and the racemic mixture of R and S isomers (R, S). Both S and R enantiomers adopted a six-membered intramolecular H-bonded (N–H⋯O (amide), 2.20 Å) turn-like conformation. Next, these S and R isomers self-assembled through intermolecular H-bonding to construct a supramolecular right-handed structure and its mirror-image left-handed helical structure (Fig. 3f), respectively. Interestingly, the racemic pseudopeptide (R, S) did not show any six-membered intramolecular H-bond and supramolecular helical assembly. Rather, the R and S form interconnected to form supramolecular β-sheet structures through intermolecular H-bonds. From the morphological view, S and R isomers produced a right-handed (width 50 nm) and a left-handed (width 49 nm) helical nanofiber, whereas the racemic (R, S) compound produced straight nanofibers with average width of 62 nm. The modified pseudopeptides exhibited a β-sheet structure without forming any intramolecular H-bond in the crystalline state and straight nanofibers in solution. Therefore, the presence of a six-membered intramolecular H-bond is the crucial factor in creating the helical structure. Thus not only the molecular structure and chirality dictate the tuning of the morphology and chirality of the nanostructure but molecular scaffolds also play an essential role in directing the shape and chiral nature of the nanostructures. Another exciting feature is that the helical nanofibers act as a template to fabricate dipeptide-capped (β-Ala-L-Tyr dipeptide) gold nanoparticles (GNPs) on the outer surface of the helical nanofibers.
The same group further explored the pseudopeptide-based helical or sheet structures by changing the supramolecular synthons.79 In this case, the pseudopeptide bis-(N-α-amido-L-valine methyl ester)-1,1-cyclopropane dicarboxylate (ValL-CP-ValL) adopted a six-membered intramolecular H-bonded (N–H⋯O (amide), 1.83 Å) turn-like structure which further self-assembled to construct a supramolecular helix with a helical pitch length of 9.09 Å (Fig. 3g). Interestingly, when 1,1-cyclopropane dicarboxamide was replaced with a 1,1-cyclobutane dicarboxamide residue in the central core of the pseudopeptide, it did not exhibit any intramolecular H-bond, but it adopted a β-sheet structure. Notably, the C–C–C angle (θ) of the central core moiety is responsible for forming a helix or β-sheet. When the C–C–C angle (θ) is ≥114°, it helps in the formation of a six-membered intermolecular H-bond followed by a supramolecular helical structure. But when the θ angle is <114°, only an intermolecular H-bonded β-sheet structure is observed. Such observations indeed improve our understanding of supramolecular organizations of dipeptides.
Pramanik and co-workers demonstrated the diversity of the supramolecular self-association of the terminally modified pseudopeptide Gly-m-aminobenzoic acid (Gly-m-ABA).80 The introduction of m-ABA helps in improving π-stacking interactions during the self-assembly process. Boc-Gly-m-ABA-OMe crystalized with two molecules in the asymmetric unit. These two enantiomeric molecules interconnected by intermolecular H-bonding in an anti-parallel manner to build a 10-membered molecular duplex (Fig. 3h). These duplexes are consecutively inter-connected via intermolecular H-bonds to form a supramolecular helix (Fig. 3h). Next, only the N-terminal protected pseudopeptide Boc-Gly-m-ABA-OH exhibited an infinitely running staircase through intermolecular H-bonding. Interestingly, the zwitterionic form of this pseudopeptide (H3+N-Gly-m-ABA–COO−) interconnected through intermolecular H-bonding in a head-to-tail fashion to form a β-sheet structure. This structure is also stabilized by π–π interactions (phenyl rings of m-ABA) and H-bonds with water molecules. So, this study reveals that protection and deprotection influence the peptide self-assembly process as the number of non-covalent interactions changes.
Haldar et al. reported a dipeptide-based helical structure of Phe and aromatic β/γ/δ amino acid-containing peptide.81 SC-XRD revealed that the hybrid dipeptide Boc-Phe-o-aminobenzoic acid adopted a rigid conformation through two intramolecular hydrogen bonds (five-membered intramolecular H-bond between N of Phe and NH of o-amino benzoic acid; six-membered intramolecular H-bond between NH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of o-amino benzoic acid). This peptide further self-associated to build a supramolecular herringbone helical architecture (Fig. 3i) via intermolecular H-bonding interactions. Next, the peptide Boc-Phe-m-aminobenzoic acid did not contain any intramolecular H-bond but the subunits interconnected through intermolecular H-bonding to adopt a single helix (Fig. 3j) in higher-order packing. Interestingly, the peptide Boc-Phe-p-aminobenzoic acid self-assembled to construct a corrugated sheet-like structure (Fig. 3k) by intermolecular H-bonding interaction and the intervening bridging methanol. The o-aminobenzoic acid-containing peptide exhibited a rose-like morphology, and the meta and para-aminobenzoic acid-containing peptides exhibited a twisted fiber-like morphology in methanol–water (1
O of o-amino benzoic acid). This peptide further self-associated to build a supramolecular herringbone helical architecture (Fig. 3i) via intermolecular H-bonding interactions. Next, the peptide Boc-Phe-m-aminobenzoic acid did not contain any intramolecular H-bond but the subunits interconnected through intermolecular H-bonding to adopt a single helix (Fig. 3j) in higher-order packing. Interestingly, the peptide Boc-Phe-p-aminobenzoic acid self-assembled to construct a corrugated sheet-like structure (Fig. 3k) by intermolecular H-bonding interaction and the intervening bridging methanol. The o-aminobenzoic acid-containing peptide exhibited a rose-like morphology, and the meta and para-aminobenzoic acid-containing peptides exhibited a twisted fiber-like morphology in methanol–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution. Interestingly, Boc-Phe-o-aminobenzoic acid adsorbs more N2 gas (22 cm3 g−1) than Boc-Phe-m-aminobenzoic acid (11 cm3 g−1), which indicates that the higher-order self-assembled herringbone helix of Boc-Phe-o-aminobenzoic acid has a higher void space (3.02 nm) than that of the single helix of Boc-Phe-m-aminobenzoic acid (2.84 nm). This result indicates that a porous material is formed from helical peptides.
1) solution. Interestingly, Boc-Phe-o-aminobenzoic acid adsorbs more N2 gas (22 cm3 g−1) than Boc-Phe-m-aminobenzoic acid (11 cm3 g−1), which indicates that the higher-order self-assembled herringbone helix of Boc-Phe-o-aminobenzoic acid has a higher void space (3.02 nm) than that of the single helix of Boc-Phe-m-aminobenzoic acid (2.84 nm). This result indicates that a porous material is formed from helical peptides.
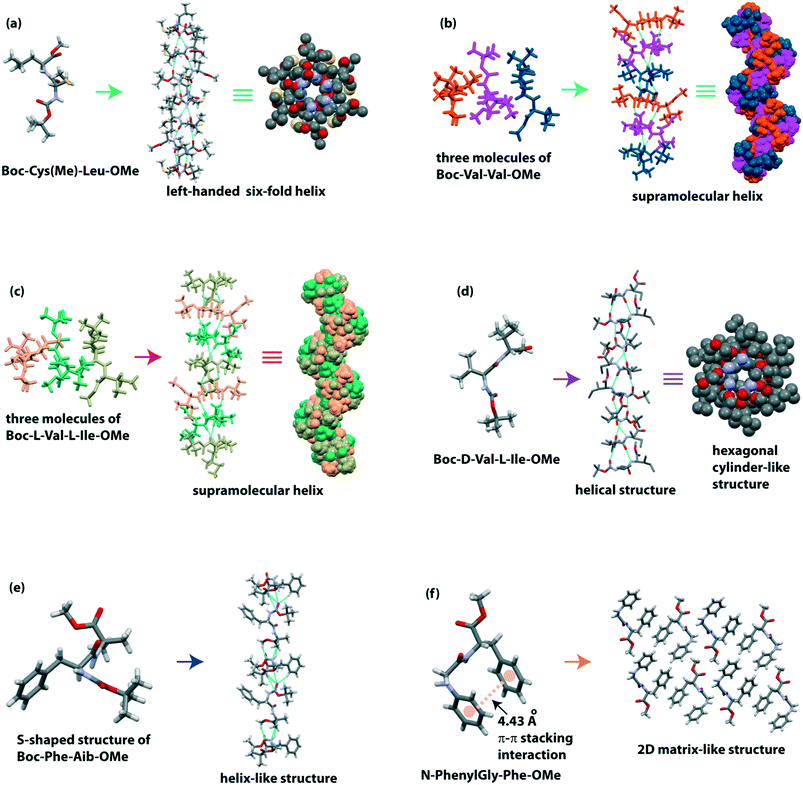
Moretto and co-workers illustrated the 3D structure of the terminally protected dipeptide Boc-Cys(Me)-Leu-OMe.82 This peptide H-bonded to build a left-handed supramolecular six-fold helix with 2.5 Å diameter (Fig. 4a) in the crystalline state. This molecule hierarchically self-assembled to form a parallelepiped shape with an empty inner cavity containing a rod-like structure (around 10 cm long) in either ethyl acetate, acetonitrile, or acetone medium. Interestingly, these nano-rods could encapsulate the water-soluble gold nanoparticles. This dipeptide also formed organogel, which further co-assembled with fullerene (C60) and multiwalled carbon nanotubes (MWCNTs) to create a robust, water-insoluble, carbon-based solid material by a vacuum-drying process. This material acts as a catalyst for reducing azo compound to amine and benzoic acid to benzyl alcohol.
Zelenovskiy and colleagues monitored the structural diversity of L-Phe-L-Phe and D-Phe-D-Phe experimentally and theoretically.83 They observed that the DD isomer self-associated to build a left-handed helix (P65 space group). In contrast, the LL isomer self-associated, forming a right-handed helix (P61 space group), which is two-fold longer than the left-handed helix of the DD isomer. Morphologically, the length of LL microtubes was found to be twice that of DD microtubes, but the average diameter of LL microtubes was 20% lower than that of DD microtubes.
We reported the crystal structure of two hydrophobic dipeptides, Boc-Val-Val-OMe and Boc-Ile-Ala-OMe, identical to the C-terminus of Alzheimer's Aβ39–40 and Aβ41–42, respectively.84 Although Görbitz and colleagues previously reported the crystal structure of Boc-Val-Val-OMe,85 we also found a similar secondary structure, i.e., it adopted a twisted parallel β-sheet structure followed by a H-bonded helical structure (Fig. 4b). Interestingly, Boc-Ile-Ala-OMe exhibited a parallel β-sheet structure, which further constructed a cross-β-sheet structure in higher-order assembly. Morphologically, both peptides self-associated to form two ended spear-like and hexagonal hollow tube-like structures, respectively.
Recently, our group demonstrated the nanostructures from alternating L/L and D/L amino acids containing dipeptides.86 The peptide Boc-L-Val-L-Ile-OMe (L/L), identical to the C-terminus of Alzheimer's Aβ40–41, exhibited three molecules in the asymmetric unit, and they intermolecularly H-bonded to create a twisted β-sheet structure which further builds a helical assembly (Fig. 4c) in higher-order packing. On the other hand, the peptide Boc-D-Val-L-Ile-OMe (D/L) showed an intermolecular H-bonded helical structure followed by a hexagonal cylinder-like architecture with ∼2.5 Å diameter around a six-fold screw axis (Fig. 4d). This packing difference is also reflected in the morphology and thermal stability of these two peptides. The L/L peptide exhibited a net nano-rod, and the D/L peptide showed a hexagonal hollow nano tube-like structure. Interestingly, the closed-packed nano-rods are more thermally stable than the nano-tubes on dry heating. The introduction of a D-amino acid in the peptide increases the proteolytic stability of the nano-tubes in biological systems.
Haldar and colleagues observed the diverse role of the phenyl group of dipeptides in the self-assembly process.87 The peptide Boc-Gly-Aib-OMe adopted a kink-like confirmation and further formed a sheet-like structure. But, when Gly is replaced with more crowded Phe, the peptide Boc-Phe-Aib-OMe adopted an ‘S’-shape structure which created a helix-like structure (Fig. 4e). Here, the π–π stacking interaction of phenyl rings in a face-to-edge (T-shape) fashion also stabilized the supramolecular helix. Moreover, peptide N-PhenylGly-Phe-OMe formed a kink-like conformation through backbone intramolecular H-bonding (five-membered) interaction and face-to-face π–π stacking interaction of aromatic rings of Phe and N-phenylGly. Next, the peptide formed a column-like structure, further building a 2D matrix-like system (Fig. 4f). Interestingly, this dipeptide developed metallogels with metals, e.g., Cu(II), Zn(II), and Pb(II) selectively (Table 1).
| Entry | Peptide sequence | Flexible/kink moieties | Secondary structure | Supramolecular self-assembly | Ref. |
|---|---|---|---|---|---|
| 1 | Leu–Leu, Leu–Phe, Phe–Leu, and Phe–Phe | Helix | 76 | ||
| 2 | Bis(N-α-amido-α-aminoisobutyric acid)-1,1-cyclopropane dicarboxylate | Aib and cyclopropane | Intramolecular 9-memberd H-bonded turn and water-mediated cyclic 11-membered H-bond turn | Single helix | 77 |
| 3 | Bis(N-α-amido-L-valine methyl ester)malonate (S), and its enantiomer bis(N-α-amido-D-valine methyl ester)malonate (R) | Malonate | Six-membered intramolecular H-bonded turn | Right-handed and its mirror-image left-handed helical structure, respectively | 78 |
| 4 | Bis-(N-α-amido-L-valine methyl ester)-1,1-cyclopropane dicarboxylate (ValL-CP-ValL) | 1,1-Cyclopropane dicarboxylate | Six-membered intramolecular H-bonded turn | Single helix | 79 |
| 5 | Gly-m-ABA | Gly, m-ABA | 10-Membered molecular duplex | Single helix | 80 |
| 6 | Boc-Gly-m-ABA-OH | Gly, m-ABA | 10-Membered molecular duplex | Running staircase | 80 |
| 7 | H3+N-Gly-m-ABA–COO− | Gly, m-ABA | Water mediated β-sheet structure | 80 | |
| 8 | Boc-Phe-o-ABA-OH | o-ABA | Five-membered intramolecular H-bonded turn | Herringbone helix | 81 |
| 9 | Boc-Phe-m-ABA-OH | m-ABA | No intramolecular turn | Single helix | 81 |
| 10 | Boc-Phe-p-ABA-OH | Corrugated sheet-like structure with intervening bridging methanol | 81 | ||
| 11 | Boc-Cys(Me)-Leu-OMe | Left-handed supramolecular six-fold helix | 82 | ||
| 12 | L-Phe-L-Phe | Right-handed helix | 83 | ||
| 13 | D-Phe-D-Phe | Left-handed helix | 83 | ||
| 14 | Boc-Val-Val-OMe | Twisted parallel β-sheet | Helical structure | 84 and 85 | |
| 15 | Boc-Ile-Ala-OMe | Parallel β-sheet | Cross-β-sheet structure | 84 | |
| 16 | Boc-L-Val-L-Ile-OMe | Twisted β-sheet structure | Helical structure | 86 | |
| 17 | Boc-D-Val-L-Ile-OMe | β-Sheet | Helical structure | 86 | |
| 18 | Boc-Gly-Aib-OMe | Aib | Kink-like structure | Sheet-like structure | 87 |
| 19 | Boc-Phe-Aib-OMe | Aib | S-Shaped structure | Single helix | 87 |
| 20 | N-PhenylGly-Phe-OMe | Kink-like structure | 2D matrix-like structure | 87 |
Tripeptide single helix
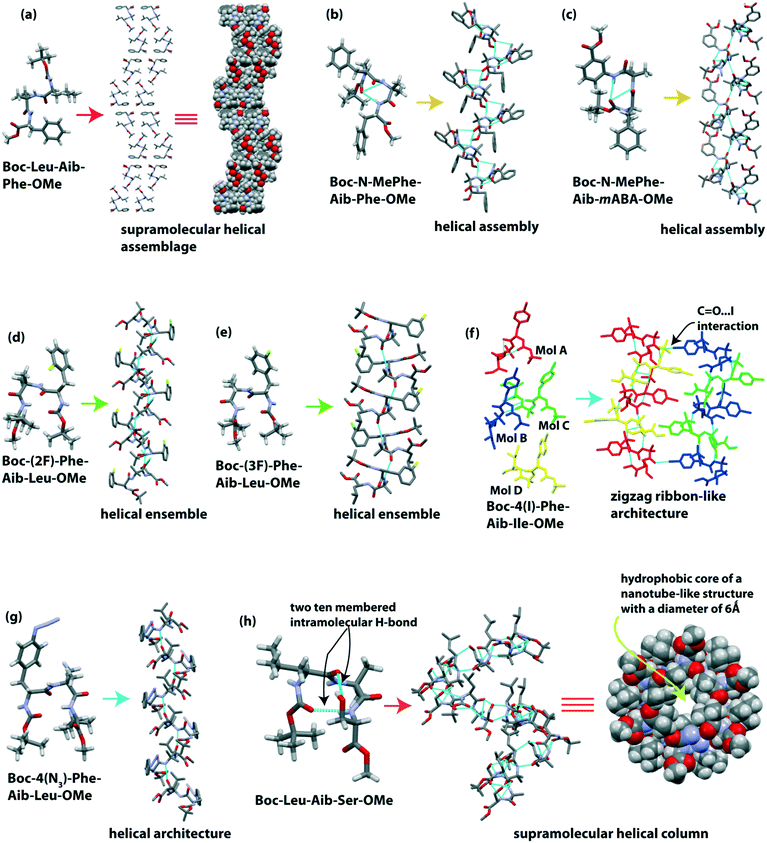
Banerjee et al. reported the formation of supramolecular helical assemblage from the self-assembly of the tripeptide Boc-Leu(1)-Aib(2)-Phe(3)-OMe.88 The backbone torsion angles (i.e., ϕ and ψ values) of this tripeptide fall in the helical region in the Ramachandran plot. Next, a helical assembly (Fig. 5a) is formed through van der Waal interactions. This peptide molecule self-associated in ethyl acetate solution to form ribbon-like fibrillar assemblies.Dutt Konar reported that N-methylated amino acid and Aib containing tripeptides exhibited a helical structure in the solid-state.89 The two tripeptides Boc-N-MePhe(1)-Aib(2)-Phe(3)-OMe and Boc-N-MePhe(1)-Aib(2)-mABA(3)-OMe adopted a type III and III′ β-turn conformation, respectively, in the crystalline state (Fig. 5b and c).
Each β-turn subunit stacked in an anti-parallel (head-to-tail) fashion through intermolecular H-bonding to create a supramolecular helix (Fig. 5b and c). N-Methylated phenylalanine acts as an inhibitor of β-sheet aggregation as the H of NH is replaced with a Me group, i.e., reduction of H-bond interactions.
Further, Dutt Konar and colleagues focused on side-chain modified amino acids in the peptide sequence for the self-assembly process.90 The isomeric mono-fluorinated (positional isomer) phenylalanine based tripeptides Boc-(2F)-Phe-Aib-Leu-OMe and Boc-(3F)-Phe-Aib-Ile-OMe adopted type II′ and II β-turn (Fig. 5d and e) conformations, respectively. Next, each subunit intermolecularly H-bonded in an anti-parallel fashion to create a helical ensemble (Fig. 5d and e). But the peptide Boc-(4F)-Phe-Aib-Ile-OMe exhibited a semi-cylindrical structure followed by a corrugated β-sheet-like structure.
The same group further monitored a set of N-terminally situated side-chain modified Phe containing tripeptides.91 In the solid-state, Boc-4(I)-Phe-Aib-Ile-OMe showed four molecules in the asymmetric unit, and from this, two molecules adopted a type III β-turn structure, and another two adopted an open strand conformation (Fig. 5f). Each type III β-turn and open strand interconnected through four types of intermolecular H-bonds to build two different strands of a zigzag ribbon (Fig. 5f). Meanwhile Boc-4(N3)-Phe-Aib-Leu-OMe adopted a type II β-turn conformation, creating a single helical structure (Fig. 5g). These results suggest that, like the backbone, the side-chains of amino acids in peptides also play an essential role in the self-association process.
Haldar et al. reported a supramolecular helix of the tripeptide Boc-Leu-Aib-Ser-OMe.92 The tripeptide adopted a rigid type III′ β-turn like structure (Fig. 5h) through two 10-membered cyclic intramolecular H-bonds. One intramolecular (N–H⋯O) hydrogen bond formed between Boc C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and Ser NH, and another intramolecular (O–H⋯O) hydrogen bond formed between the backbone C
O and Ser NH, and another intramolecular (O–H⋯O) hydrogen bond formed between the backbone C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Leu and the side-chain OH of Ser. The subunits self-assembled in a helical manner to build a hydrophobic core of a nanotube-like structure with a diameter of 6 Å (Fig. 5h). They observed that the hydrophobic tail of coumarin and naphthalene diimide derivatives could incorporate inside the hydrophobic core of this nano-tube to form a supramolecular polymer.
O of Leu and the side-chain OH of Ser. The subunits self-assembled in a helical manner to build a hydrophobic core of a nanotube-like structure with a diameter of 6 Å (Fig. 5h). They observed that the hydrophobic tail of coumarin and naphthalene diimide derivatives could incorporate inside the hydrophobic core of this nano-tube to form a supramolecular polymer.
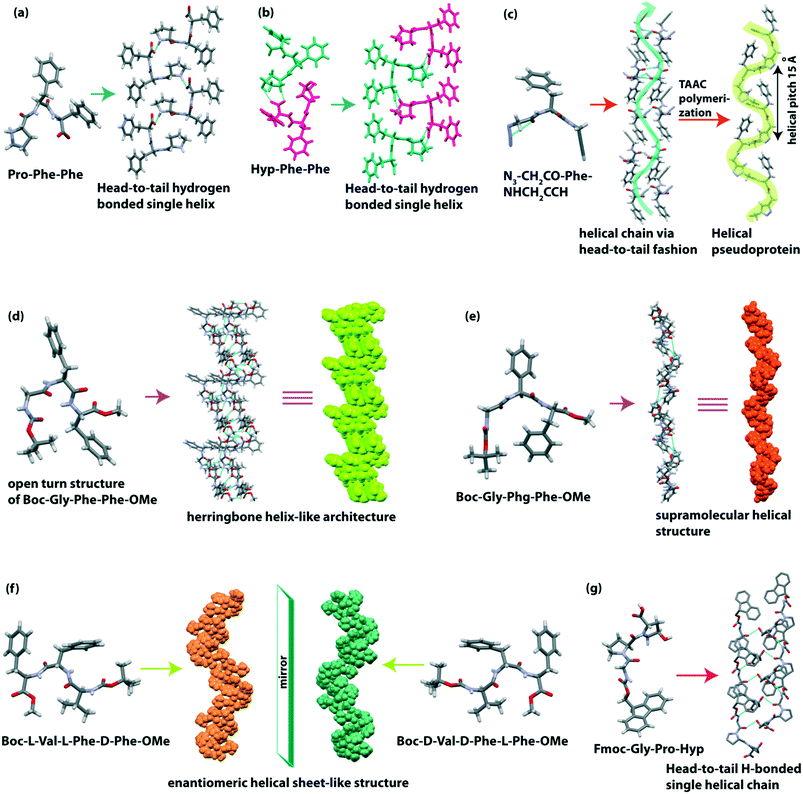
Gazit and co-workers observed that the most aggregation-prone natural amino acid containing tripeptide Pro-Phe-Phe self-assembled to form a supramolecular rigid helical-like sheet (Fig. 6a) through a head-to-tail assembly.93 When Pro is replaced with Hyp, a more tightly bound helical sheet (Fig. 6b) structure is formed in the peptide Hyp-Phe-Phe as Hyp has an OH group which takes part in H-bonding interaction. From the morphological view, this Pro-Phe-Phe peptide self-aggregated to form a helical fiber network and bound with amyloid binding dye ThT.
Sureshan et al. reported a cross-α amyloid-like helical-sheet structure of the triazole-linked covalent pseudoprotein (Gly–Phe–Gly repeats) from the dipeptide monomer N3–CH2CO–Phe–NHCH2CCH through the TAAC polymerization of single-crystal-to-single-crystal (SCSC).94 The pseudoprotein showed a left-handed helical sheet (Fig. 6c), and the two adjacent helices are stabilized through non-covalent interaction. Similarly, the dipeptide (N3–CH2CO–Phe–NHCH2CCH) also formed a helical arrangement in a head-to-tail fashion by C–H⋯N and C–H⋯π interactions (Fig. 6c). Interestingly, the helicity of this pseudoprotein was obtained in the crystalline state, but in the solution state, both mixtures of helix and β-sheet were present.
We recently described the effect of the side-chain of amino acids on the peptide backbone and their supramolecular structure.95 The designed tripeptide Boc-Gly(1)-Phe(2)-Phe(3)-OMe adopted a rare conformation known as the ‘open turn’ structure (Fig. 6d) which is similar to the type II β-turn structure, but it does not contain an intramolecular (4 → 1) H-bond between i (Boc-CO) and i + 3 (NH of Phe 3). The backbone torsion angles (φ1 = −56.1, ψ1 = 145.8 and φ2 = 59.8, ψ2 = 29.7) also supported the formation of the deviated type II β-turn structure, but due to the absence of an intramolecular H-bond, it formed the ‘open turn’ conformation. Next, each ‘open turn’ subunit self-assembled to build a supramolecular herringbone helix-like architecture (Fig. 6d) through C–H⋯O interactions. On the other hand, the analogous peptide Boc-Gly-Phg-Phe-OMe adopted a parallel β-sheet secondary structure, which further self-assembled to create a helical structure (Fig. 6e) through C–H⋯O and C–H⋯π interactions. The obtained secondary structure and supramolecular self-assembled structure of these two peptides are different due to their backbone orientation governed by the side-chain interaction of central Phe or Phg. Significantly, the presence or absence of one extra methylene (–CH2–) group of central Phe or Phg changes the conformation of these peptides. They formed two different flower-like architectures in 30% acetonitrile–water medium.
Further, our group reported the supramolecular self-association of all possible stereoisomers of Boc-Val-Phe-Phe-OMe, identical to the hydrophobic core sequence of the amyloid β (Aβ18–20) peptide.96 In the crystalline state, the peptide Boc-L-Val-L-Phe-D-Phe-OMe and its enantiomer Boc-D-Val-D-Phe-L-Phe-OMe adopted parallel-β sheet structures, and they further formed an enantiomeric helical sheet-like architecture (Fig. 6f) through non-covalent interactions. Although the self-assembled homo and heterochiral tripeptides were found to bind with amyloid binding dye ThT, their morphology is different. Here, chirality plays a vital role in molecular self-association and diversity of morphology.
Adler-Abramovich et al. demonstrated that a mimicked collagen repeating unit Fmoc-Gly-Pro-Hyp exhibited a left-handed polyproline II superhelical structure (Fig. 6g) similar to the native single strand of the collagen helix.97 Head-to-tail and side-by-side H-bonding interactions help to build such a type of higher-order structure. An aromatic “zipper-like” interaction is also observed between the Fmoc groups of two adjacent helical sheets. Interestingly, this tripeptide co-assembled with a dipeptide hydrogelator, Fmoc-Phe-Phe, to form a hybrid hydrogel comprising twisted helical fibrils (Table 2).
| Entry | Peptide sequence | Flexible/kink moieties | Secondary structure | Supramolecular self-assembly | Ref. |
|---|---|---|---|---|---|
| 1 | Boc-Leu-Aib-Phe-OMe | Aib | Helical assembly | 88 | |
| 2 | Boc-N-MePhe-Aib-Phe-OMe | Aib, N-MePhe | Type III β-turn | Single helix | 89 |
| 3 | Boc-N-MePhe-Aib-mABA-OMe | Aib, mABA | Type III′ β-turn | Single helix | 89 |
| 4 | Boc-(2F)-Phe-Aib-Leu-OMe | Aib | Type II′ β-turn | Helical ensemble | 90 |
| 5 | Boc-(3F)-Phe-Aib-Ile-OMe | Aib | Type II β-turn | Helical ensemble | 90 |
| 6 | Boc-(4F)-Phe-Aib-Ile-OMe | Aib | Type II'β-turn | Corrugated β-sheet | 90 |
| 7 | Boc-4(I)-Phe-Aib-Ile-OMe | Aib | Type III β-turn | Zigzag ribbon | 91 |
| 8 | Boc-4(N3)-Phe-Aib-Leu-OMe | Aib | Type II β-turn | Single helical structure | 91 |
| 9 | Boc-Leu-Aib-Ser-OMe | Aib | Type III′ β-turn | Single helix | 92 |
| 10 | Pro-Phe-Phe | Pro | Helical-like sheet | 93 | |
| 11 | Hyp-Phe-Phe | Hyp | Helical-like sheet | 93 | |
| 12 | N3–CH2CO–Phe–NHCH2CCH | Single helix | 94 | ||
| 13 | Boc-Gly(1)-Phe(2)-Phe(3)-OMe | Gly | Open turn | Herringbone helix | 95 |
| 14 | Boc-Gly-Phg-Phe-OMe | Gly | Parallel β-sheet | Helical structure | 95 |
| 15 | Boc-L-Val-L-Phe-D-Phe-OMe and its enantiomer Boc-D-Val-D-Phe-L-Phe-OMe | Parallel-β sheet | Enantiomeric helical sheet | 96 | |
| 16 | Fmoc-Gly-Pro-Hyp | Gly, Pro, Hyp | Left-handed polyproline II helix | Left-handed polyproline II helix | 97 |
Tetrapeptide single helix
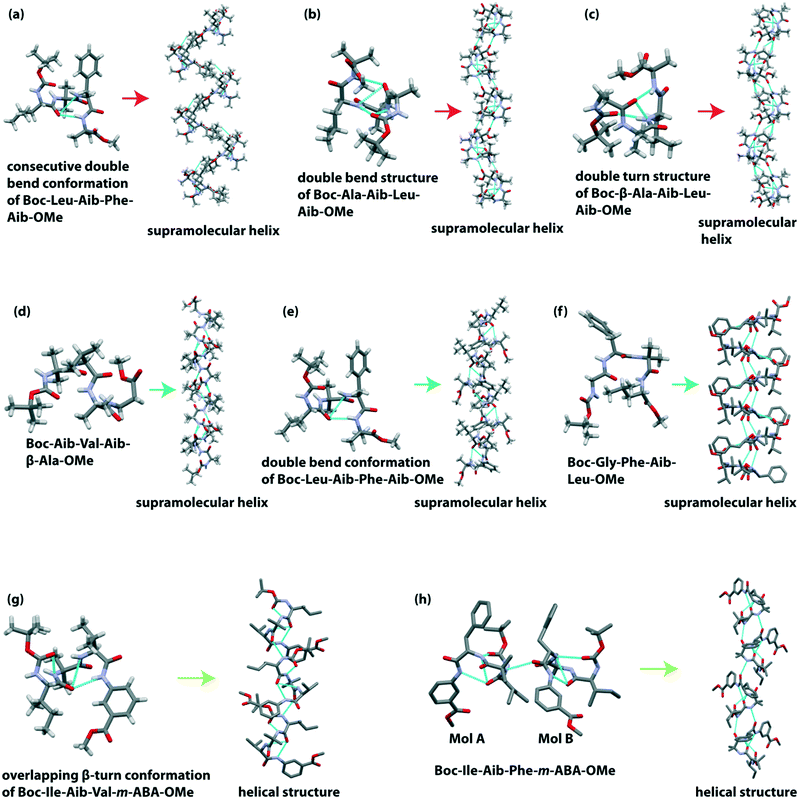
Banerjee et al. reported a hydrogen-bonded supramolecular helical structure of the tetrapeptide Boc-Leu(1)-Aib(2)-Phe(3)-Aib(4)-OMe.98 This peptide adopted a consecutive double bend conformation through intramolecular H-bonding between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Leu(1) and NH of Phe(3) (7-membered ring) and between the C
O of Leu(1) and NH of Phe(3) (7-membered ring) and between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Leu(1) and NH of Aib(4) (10-membered ring) in the solid-state. Additionally, the backbone torsion angle suggested that these two bends are type I β-turn. Further, each subunit is regularly interconnected through intermolecular H-bonding to construct a supramolecular helical architecture (Fig. 7a). An amyloid-like fibril morphology is obtained from SEM analysis.
O of Leu(1) and NH of Aib(4) (10-membered ring) in the solid-state. Additionally, the backbone torsion angle suggested that these two bends are type I β-turn. Further, each subunit is regularly interconnected through intermolecular H-bonding to construct a supramolecular helical architecture (Fig. 7a). An amyloid-like fibril morphology is obtained from SEM analysis.
They further demonstrated a supramolecular helix of the tetrapeptide Boc-Ala(1)-Aib(2)-Leu(3)-Aib(4)-OMe.99 This peptide also adopted a double bend structure via the formation of a type III β-turn and an unusual β-turn involved between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Ala(1) and NH of Aib(4). Next, these two overlapping β-turns self-organized to build an infinite 1D helical column (Fig. 7b) through intermolecular H-bonding interaction between the NH of Ala(1) and Aib(2) residues and C
O of Ala(1) and NH of Aib(4). Next, these two overlapping β-turns self-organized to build an infinite 1D helical column (Fig. 7b) through intermolecular H-bonding interaction between the NH of Ala(1) and Aib(2) residues and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Leu(3) and Aib(4) residues. Backbone torsion angles also supported the acceptance of the helical region in the Ramachandran plot. This peptide also exhibited an amyloid-like fibril assembly.
O of Leu(3) and Aib(4) residues. Backbone torsion angles also supported the acceptance of the helical region in the Ramachandran plot. This peptide also exhibited an amyloid-like fibril assembly.
Next, based on the SC-XRD experiment, they observed that the unnatural amino acid (Aib and β-Ala) containing terminally protected tetrapeptide Boc-β-Ala(1)-Aib(2)-Leu(3)-Aib(4)-OMe adopted a new type of double turn structure.100 The introduction of the flexible β-amino acid (β-Ala) residue at the N-terminal resulted in an intramolecular H-bonded 11-membered ring between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Boc and NH of Leu(3). Similarly, another distorted type I β-turn is formed through the intramolecular H-bond (10-membered ring) between the C
O of Boc and NH of Leu(3). Similarly, another distorted type I β-turn is formed through the intramolecular H-bond (10-membered ring) between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of β-Ala(1) and NH of Aib(4). Interestingly, each subunit further interlinked through an intermolecular H-bond to form a supramolecular helix (Fig. 7c). This peptide self-assembled to form an amyloid-like fibrillar morphology.
O of β-Ala(1) and NH of Aib(4). Interestingly, each subunit further interlinked through an intermolecular H-bond to form a supramolecular helix (Fig. 7c). This peptide self-assembled to form an amyloid-like fibrillar morphology.
The same group further reported the unnatural amino acid (Aib and β-Ala) containing supramolecular single helix of the tetrapeptide Boc-Aib(1)-Val(2)-Aib(3)-β-Ala(4)-OMe with an average diameter of 10 Å.101 In the crystalline state, this peptide exhibited two overlapping β-turns through intramolecular H-bonds. Next, each peptide subunit self-aligned to build a helical assembly (Fig. 7d) through intermolecular H-bonds. Interestingly, this helical assembly revealed a rod-like architecture (diameter ∼10 Å). The TEM image also supported the formation of nano-rods (diameter ∼10–40 nm) in solution.
Later, they studied an aggregating peptide, Leu-Val-Phe-Phe, with the hydrophobic core sequence (17–20) of amyloid-β (Aβ), responsible for Alzheimer's disease.102 This terminally protected peptide (Boc-Leu-Val-Phe-Phe-OMe) and its Ala substituted analogs Boc-Leu-Val-Phe-Ala-OMe and Boc-Leu-Ala-Phe-Ala-OMe self-assembled through intermolecular H-bonding to form a β-sheet structure. Interestingly, on incorporating the non-proteogenic amino acid Aib in the sequence (Boc-Leu-Aib-Phe-Aib-OMe), the structure changes from a β-sheet (native peptide) to a 310 helix. This alternating Aib containing peptide adopted a double bend conformation (two consecutive β-turn) via two intramolecular H-bonds in the crystalline state. Notably, the measured backbone torsion angles (ϕ1 = −70.2° and ψ1 = −16.9° for Leu(1), ϕ2 = −57.8° and ψ2 = −21.3° for Aib(2), ϕ3 = −56.4° and ψ3 = −35.9° for Phe(3)) indicated that the peptide belongs to the 310 helical region in the Ramachandran plot. These 310 helical subunits are further inter-stacked by multiple intermolecular H-bonds to form a supramolecular helix (Fig. 7e).
Pramanik et al. described a supramolecular helix from the tetrapeptide Boc-Gly(1)-Phe(2)-Aib(3)-Leu(4)-OMe where Aib is located at the 3rd position in the sequence.103 This peptide folded to adopt a distorted type II β-turn conformation. The hierarchical self-assembly of each β-turn unit (in anti-parallel mode) constructed a supramolecular helix (Fig. 7f) via intermolecular H-bonding. A bunch of non-twisted fibrillar morphology was observed under FESEM.
Pramanik et al. observed that Aib at the 2nd position in a tetrapeptide sequence with the general formula Boc-Xx(1)-Aib(2)-Yy(3)-Zz(4)-OMe (Xx, Yy and Zz are natural amino acids) adopted an overlapping double turn structure which self-assembled in a head to tail fashion to form a supramolecular helical assembly via intermolecular H-bonds.104 The peptide Boc-Ile-Aib-Leu-Ala-OMe exhibited two type III β-turns, but Boc-Phe-Aib-Leu-Ala-OMe and Boc-Ile-Aib-Leu-Phe-OMe exhibited one type III and another type I β-turn to construct an overlapping double β-turn structure. On the other hand, the peptide Boc-Leu-Aib-Phe-Ala-OMe showed two molecules (A and B) in the asymmetric unit. Molecules A and B formed double bend structures by two β-turns of type III and type III′, respectively. Interestingly, the self-assembled helical peptides showed a flat non-twisted tape-like morphology.
Pramanik and co-workers described the helical structure formation from two tetrapeptides that contain two natural and two unnatural amino acids in their sequence.105 Boc-Ile-Aib-Val-m-ABA-OMe crystalized to adopt an overlapping β-turn conformation by forming type III β-turn and type I β-turn. These pre-organized double bend subunits are stacked by intermolecular H-bonding to form a supramolecular helical structure (Fig. 7g). On the other hand, Boc-Ile-Aib-Phe-m-ABA-OMe crystallized to give two molecules (A and B) in the asymmetric unit. Like the previous peptide, molecule A of this peptide also exhibited double β-turn structures (type III and type I β-turn), whereas molecule B showed a diastereoisomeric type III′ β-turn structure. These two molecules were stacked to build a helical assembly (Fig. 7h). Both peptides self-aggregated to form amyloid-like fibrils.
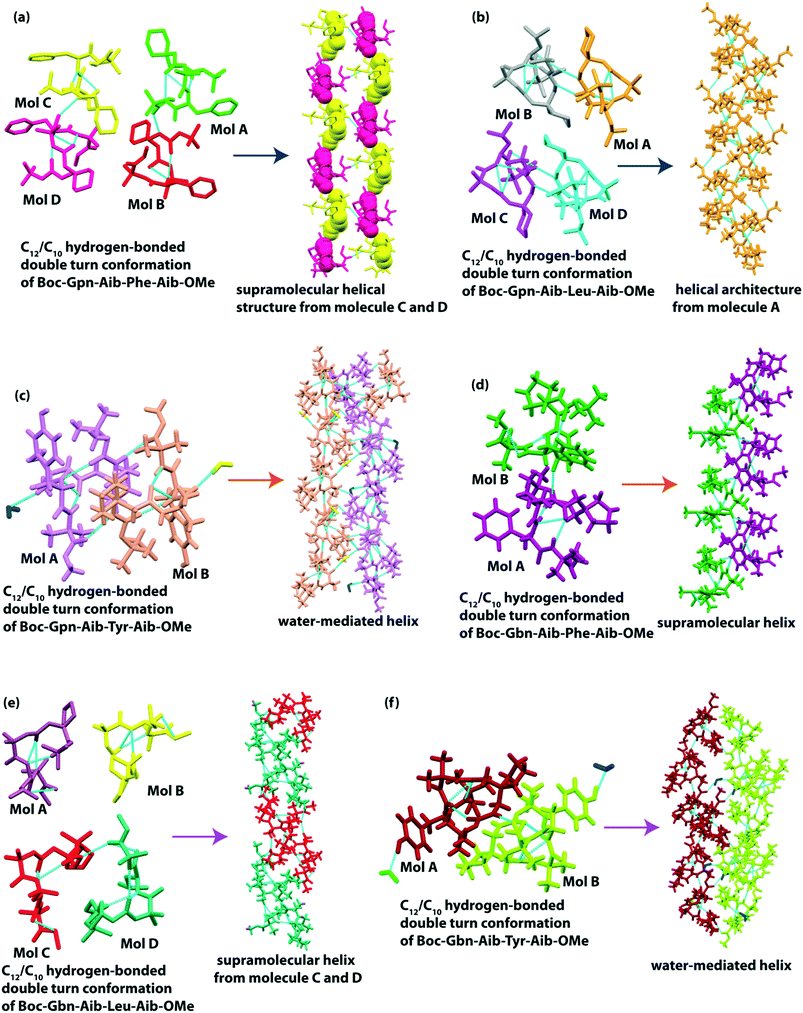
Das and co-workers reported supramolecular helical structures from structurally constrained γ-amino acid, gabapentin (Gpn), containing hybrid peptides, Boc-Gpn(1)-Aib(2)-Xaa(3)-Aib(4)-OMe (where Xaa = Phe or Leu or Tyr (ref. 106)). All three peptides adopted C12 and C10 intramolecularly H-bonded double-turn conformations in the solid-state. The unusual 12-membered C12 turn is formed between (Boc) C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N (Xaa = Phe or Leu or Tyr) H-bonding interactions, and the C10 turn is formed between (Gpn(1)) C
O⋯H–N (Xaa = Phe or Leu or Tyr) H-bonding interactions, and the C10 turn is formed between (Gpn(1)) C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N (Aib(4)) H-bonding interactions. The Phe containing peptide exhibited four molecules (A–D) in the asymmetric unit and molecules A and B and molecules C and D hydrogen bonded intermolecularly to form two helical assemblies (Fig. 8a), respectively. Meanwhile, the Leu containing peptide also showed four molecules in the asymmetric unit, but each molecule individually formed a supramolecular helix (Fig. 8b). Interestingly, the Tyr-containing peptide crystallized in two molecules (A and B) with two water molecules in the asymmetric unit. The helical assembly is stabilized by the interaction of OH of Tyr and intervening bridged water molecules (Fig. 8c). Here, the side chain of the third amino acid residue plays a vital role in building up diverse supramolecular architectures.
O⋯H–N (Aib(4)) H-bonding interactions. The Phe containing peptide exhibited four molecules (A–D) in the asymmetric unit and molecules A and B and molecules C and D hydrogen bonded intermolecularly to form two helical assemblies (Fig. 8a), respectively. Meanwhile, the Leu containing peptide also showed four molecules in the asymmetric unit, but each molecule individually formed a supramolecular helix (Fig. 8b). Interestingly, the Tyr-containing peptide crystallized in two molecules (A and B) with two water molecules in the asymmetric unit. The helical assembly is stabilized by the interaction of OH of Tyr and intervening bridged water molecules (Fig. 8c). Here, the side chain of the third amino acid residue plays a vital role in building up diverse supramolecular architectures.
Later, the same group observed the supramolecular helical association from another structurally constrained γ-amino acid, gababutin (Gbn), based hybrid tetrapeptide.70 Like previous peptide sequences, they kept the same sequence, with the only difference being the use of Gbn in place of Gpn, i.e., Boc-Gbn(1)-Aib(2)-Xaa(3)-Aib(4)-OMe (where Xaa = Phe or Leu or Tyr). Here, all three peptides also adopted a C12/C10 helical double turn conformation, and in higher-order assembly, they built a helical assembly (Fig. 8d and e). The peptide Boc-Gbn(1)-Aib(2)-Tyr(3)-Aib(4)-OMe formed a water-mediated single helical (Fig. 8f) assembly through (water) O⋯H–O Tyr(3) interactions. The phenolic OH of Tyr plays an essential role in developing such a helical structure among peptide and water molecules (Table 3).
| Entry | Peptide sequence | Flexible/kink moieties | Secondary structure | Supramolecular self-assembly | Ref. |
|---|---|---|---|---|---|
| 1 | Boc-Leu-Aib-Phe-Aib-OMe | Aib | Double turn structure (type I β-turn) | Right-handed helical structure | 98 |
| 2 | Boc-Ala-Aib-Leu-Aib-OMe | Aib | Double turn structure (one is type III β-turn) | 1D helical column | 99 |
| 3 | Boc-β-Ala-Aib-Leu-Aib-OMe | Aib and β-Ala | Double turn structure (one is type I β-turn) | Single helix | 100 |
| 4 | Boc-Aib-Val-Aib-β-Ala-OMe | Aib | Two overlapping β-turns | Helical assembly | 101 |
| 5 | Boc-Leu-Val-Phe-Ala-OMe and Boc-Leu-ala-Phe-Ala-OMe | β-Sheet structure | 102 | ||
| 6 | Boc-Leu-Aib-Phe-Aib-OMe | Aib | Double bend conformation | Single helix | 102 |
| 7 | Boc-Gly-Phe-Aib-Leu-OMe | Aib | Type II β-turn | Single helix | 103 |
| 8 | Boc-Ile-Aib-Leu-Ala-OMe | Aib | Overlapping double turn (type III β-turn) | Helical assembly | 104 |
| 9 | Boc-Phe-Aib-Leu-Ala-OMe and Boc-Ile-Aib-Leu-Phe-OMe | Aib | Overlapping double turn (type III and type I β-turn) | Helical assembly | 104 |
| 10 | Boc-Leu-Aib-Phe-Ala-OMe molecule A and B | Aib | (Double bend structures) two type III for molecule A and type III′ β-turn for B | Helical assembly | 104 |
| 11 | Boc-Ile-Aib-Val-m-ABA-OMe | Aib, m-ABA | Overlapping β-turn (type III and type I β-turn) | Single helix | 105 |
| 12 | Boc-Ile-Aib-Phe-m-ABA-OMe, two molecules (A and B) | Aib, m-ABA | Double β-turn (type III and type I β-turn) for molecule A and type III′ β-turn for molecule B | Single helix | 105 |
| 13 | Boc-Gpn-Aib-Phe-Aib-OMe | Gpn, Aib | C12/C10 double turn | Single helix | 106 |
| 14 | Boc-Gpn-Aib-Leu-Aib-OMe | Gpn, Aib | C12/C10 double turn | Single helix | 106 |
| 15 | Boc-Gpn-Aib-Tyr-Aib-OMe | Gpn, Aib | C12/C10 double turn | Water mediated helix | 106 |
| 16 | Boc-Gbn-Aib-Phe-Aib-OMe | Gbn, Aib | C12/C10 double turn | Single helix | 70 |
| 17 | Boc-Gbn-Aib-Leu-Aib-OMe | Gbn, Aib | C12/C10 double turn | Single helix | 70 |
| 18 | Boc-Gbn-Aib-Tyr-Aib-OMe | Gbn, Aib | C12/C10 double turn | Water mediated helix | 70 |
Construction of a double helix
The construction of a double-helical structure is more challenging than a single helical structure from small peptides due to the orientation and stability (governed by supramolecular interactions) of two helical strands in the double helix. In nature, only DNA contains a double-helical structure, stabilized by an intermolecular H-bond between the nucleic acid base pair. But ongoing research revealed that small di/tri/tetra peptides formed supramolecular double-helical assemblies that further created hollow channels or cylindrical structures. Such a porous double-helical assembly can act as a host to accommodate N2, CO2, and CH4 gas. Herein, we describe the double-helical structures of di, tri, and tetrapeptides sequentially and their applications.Dipeptide double helix
Görbitz described the left-handed double-helical structures obtained from the crystal structure of the Val–Ala class (Val–Ala, Val–Val, Val–Ile, Ala–Val, Ala–Ile, Ile–Ala, and Ile–Val).107–109 Five types of intermolecular H-bonds stabilize the double helices. All these self-assembled peptides formed a one dimensional (1D) channel. Later, Soldatov and colleague calculated the diameter of the channel (Ala–Val (5.4Å) > Val–Ala (5.0Å) > Ala–Ile (4.3Å) > Val–Val (4.0Å) > Ile–Ala (3.6Å) > Ile–Val (3.4Å) > Val–Ile (3.0Å)).110 The channels are right-handed helical shaped, and the order of channel helicity is Val–Ala > Ile–Ala > Ile–Val > Ala–Val ≈ Ala–Ile ≈ Val–Val > Val–Ile. We didn’t discuss the Val–Ala class peptides too elaborately in this highlight, as Görbitz represented them nicely in his concept review.109Later, Comotti and Görbitz and co-workers reported hydrophobic microporous organic materials from unnatural amino acid (L-aminobutyric acid (Abu) and L-norvaline (Nva)) containing dipeptides.111 They synthesized ten dipeptides, e.g., Abu–Abu, Abu–Nva, Abu–Val, Abu–Leu, Abu–Ile, Nva–Abu, Nva–Nva, Nva–Val, Nva–Leu, and Nva–Ile and the structure was determined by SC-XRD experiments. All peptides exhibited a heft-handed double-helical H-bonding pattern in a head-to-tail fashion with a three-fold screw symmetry. A right-handed helical crystal channel is formed from each peptide with a pore size of 2.3–5.1 Å (Fig. 9a–j). Interestingly, these channels could accommodate guest molecules, e.g., CO2 and CH4 gas.
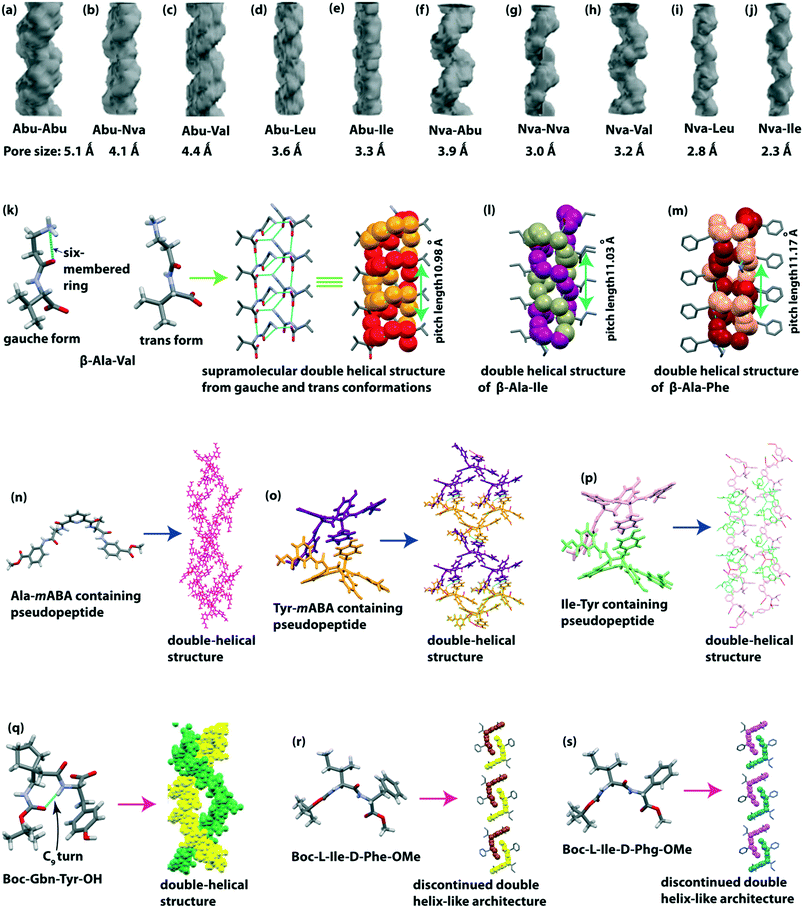 | ||
| Fig. 9 Supramolecular double-helical structure of dipeptides (a) Abu–Abu, (b) Abu–Nva, (c) Abu–Val, (d) Abu–Leu, (e) Abu–Ile, (f) Nva–Abu, (g) Nva–Nva, (h) Nva–Val, (i) Nva–Leu, (j) Nva–Ile (reproduced from ref. 111 with permission from John Wiley and Sons), (k) β-Ala-Val, (l) β-Ala-Ile, and (m) β-Ala-Phe. Double-helical structure of pseudopeptides (n) pyridinedicarboxylic acid and Ala-mABA-OMe, (o) pyridinedicarboxylic acid and Tyr-mABA-OMe, and (p) pyridinedicarboxylic acid and Ile-Tyr-OMe. (q) Double helical structure of the C9-turn containing peptide Boc-Gbn-Tyr-OH (reproduced from ref. 115 with permission from Royal Society of Chemistry). Discontinued double helix-like structure of (r) Boc-L-Ile-D-Phe-OMe and (s) Boc-L-Ile-D-Phg-OMe. | ||
Banerjee and co-workers illustrated that the N-terminally located flexible unnatural amino acid β-Ala containing dipeptides (β-Ala-Xaa, Xaa = Val/Ile/Phe) exhibited a supramolecular double-helical structure (helical pitch length ∼1 nm) through intra and intermolecular H-bonding interactions in higher-order assembly.112 These dipeptides exist in two conformations (gauche, θ ∼ 60 and trans, θ ∼ 180) around the backbone –C(β)–C(α)– bond of the flexible β-Ala moiety, in the asymmetric unit. Interestingly, the gauche conformation adopted an intramolecularly H-bonded six-membered cycle (NH3+⋯C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (amide)), whereas the trans conformation remained in the extended form. The presence of both gauche and trans conformations of β-Ala around –C(β)–C(α)– helps in the formation of the double-helical architecture (Fig. 9k–m). When the β-Ala residue is located at the C-terminal, the peptide Phe-β-Ala exhibited a supramolecular single helical architecture, i.e., the position of the β-Ala residue in the peptide sequence is also crucial for creating a double or single helical assembly. The obtained supramolecular double-helical arrangements showed significant thermal stability.
O (amide)), whereas the trans conformation remained in the extended form. The presence of both gauche and trans conformations of β-Ala around –C(β)–C(α)– helps in the formation of the double-helical architecture (Fig. 9k–m). When the β-Ala residue is located at the C-terminal, the peptide Phe-β-Ala exhibited a supramolecular single helical architecture, i.e., the position of the β-Ala residue in the peptide sequence is also crucial for creating a double or single helical assembly. The obtained supramolecular double-helical arrangements showed significant thermal stability.
Keeping in mind the application of porous biomaterials in various orthopedic purposes, Dutt Konar designed and synthesized two enantiomeric pseudopeptides composed of one pyridinedicarboxylic acid and a dipeptidyl fragment H-Xaa-mABA-OMe (Xaa: L-Ala or L-Tyr).113,114 In crystal analysis, each subunit is interconnected via H-bonding to form a dimeric entity, where the central pyridine moieties interdigitate slightly from each other. Next, each dimeric entity assembled to create a double helical structure, stabilized by intermolecular H-bonding and π–π interactions (Fig. 9n and o). These double-helical columns of the L-Tyr containing pseudopeptide further formed 3D channels with a diameter of 12.5 × 15.053 Å. Another pyridinedicarboxylic acid and Ile–Tyr containing pseudopeptide also formed a double-helical structure in a similar manner (Fig. 9p). Interestingly, the double helix of the Tyr–mABA containing pseudopeptide adsorbed N2 gas (65 cc g−1) 15 times more than the Ala–mABA containing pseudopeptide (4.5 cc g−1) due to the presence of extra H-bonding interaction of the side chain of phenolic OH of Tyr. From the morphological view, the Ala and Ile based pseudopeptides formed a rod-like structure, but the Tyr–mABA containing pseudopeptide developed a nanoporous morphology. So, this study reveals that side-chain to side-chain interactions play an important role in creating porous biomaterials.
Das et al. illustrated a supramolecular double-helical superstructure of the conformationally rigid moiety (gababutin, Gbn) containing dipeptide Boc-Gbn-Tyr-OH.115 The SC-XRD analysis revealed that it adopted a nine-membered C9 turn structure through intramolecular H-bonding between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Boc and NH of Tyr. Next, two turn units are connected in an antiparallel manner between the side-chain OH group of Tyr and NH of Gbn to form a dimer which further propagated to form a helical strand, stabilized by (Gbn) C
O of Boc and NH of Tyr. Next, two turn units are connected in an antiparallel manner between the side-chain OH group of Tyr and NH of Gbn to form a dimer which further propagated to form a helical strand, stabilized by (Gbn) C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–O(HOOC–Tyr) and (Tyr–COOH)H–O⋯H–N (Gbn) H-bonding interactions. Next, the helical strands intertwisted among themselves to form a double-helical structure (Fig. 9q). Here, Tyr H–O⋯H–N(Gbn) H-bonding interaction plays an essential role in developing the twist of helical strands. Interestingly, this double helix behaves as a potential energy storage material with a specific capacitance of 3500 μF g−1 at 0.6 A g−1.
O⋯H–O(HOOC–Tyr) and (Tyr–COOH)H–O⋯H–N (Gbn) H-bonding interactions. Next, the helical strands intertwisted among themselves to form a double-helical structure (Fig. 9q). Here, Tyr H–O⋯H–N(Gbn) H-bonding interaction plays an essential role in developing the twist of helical strands. Interestingly, this double helix behaves as a potential energy storage material with a specific capacitance of 3500 μF g−1 at 0.6 A g−1.
Recently we reported the supramolecular helical and double helical architecture of alternating L/L and L/D amino acid-containing peptides. Boc-L-Ile-L-Phe-OMe (L/L) and Boc-L-Ile-L-Phg-OMe (L/L) exhibited a single helical architecture and a cylinder-like structure around a water molecule, respectively.116 On the other hand, alternating L/D amino acid-containing dipeptides, Boc-L-Ile-D-Phe-OMe and Boc-L-Ile-D-Phg-OMe, self-organized to build a discontinued double helix-like architecture through C–H⋯O and C–H⋯π interactions (Fig. 9r and s). The observed different supramolecular structures are also explained by the presence of configurationally different chirality amino acids and the effect of side chains of amino acids (especially Phe and Phg) on the peptide backbone. Due to the packing difference, Boc-L-Ile-L-Phe-OMe and Boc-L-Ile-L-Phg-OMe peptides self-assembled to exhibit helical ribbon-like and nanotube-like (inner diameter ∼261.9 nm and outer diameter ∼986.2 nm) structures, whereas D/L peptides showed nano rod-like structures in 50% acetonitrile–water solution.
Tripeptide double helix
Haldar and co-workers designed both N- and C-protected three tripeptides, Boc-Tyr-Aib-Xaa-OMe (Xaa = Leu/Ile/Ala), which contain the helix inducer unnatural amino acid Aib at the central position of the sequence.117 The SC-XRD analysis revealed that the Leu and Ile containing peptides adopted a type II β and type II′ β turn, respectively, with an intramolecular H-bond between i (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Boc) and i + 3 (NH of leu/Ile) to form a 10-membered cyclic ring (Fig. 10a and b). Although the backbone torsion angles of the Ala containing peptide are similar to the type II β turn, it cannot form an intramolecular H-bond between i and i + 3, but it forms a water-mediated turn-like structure (Fig. 10c). Each peptide interconnected through an intermolecular H-bond to form a supramolecular helical strand, further building a double-helical superstructure through non-covalent interactions. Interestingly, these double helixes could take up N2 gas (18.31 cc g−1) in their double helical pore, being applicable as porous biomaterials.
O of Boc) and i + 3 (NH of leu/Ile) to form a 10-membered cyclic ring (Fig. 10a and b). Although the backbone torsion angles of the Ala containing peptide are similar to the type II β turn, it cannot form an intramolecular H-bond between i and i + 3, but it forms a water-mediated turn-like structure (Fig. 10c). Each peptide interconnected through an intermolecular H-bond to form a supramolecular helical strand, further building a double-helical superstructure through non-covalent interactions. Interestingly, these double helixes could take up N2 gas (18.31 cc g−1) in their double helical pore, being applicable as porous biomaterials.
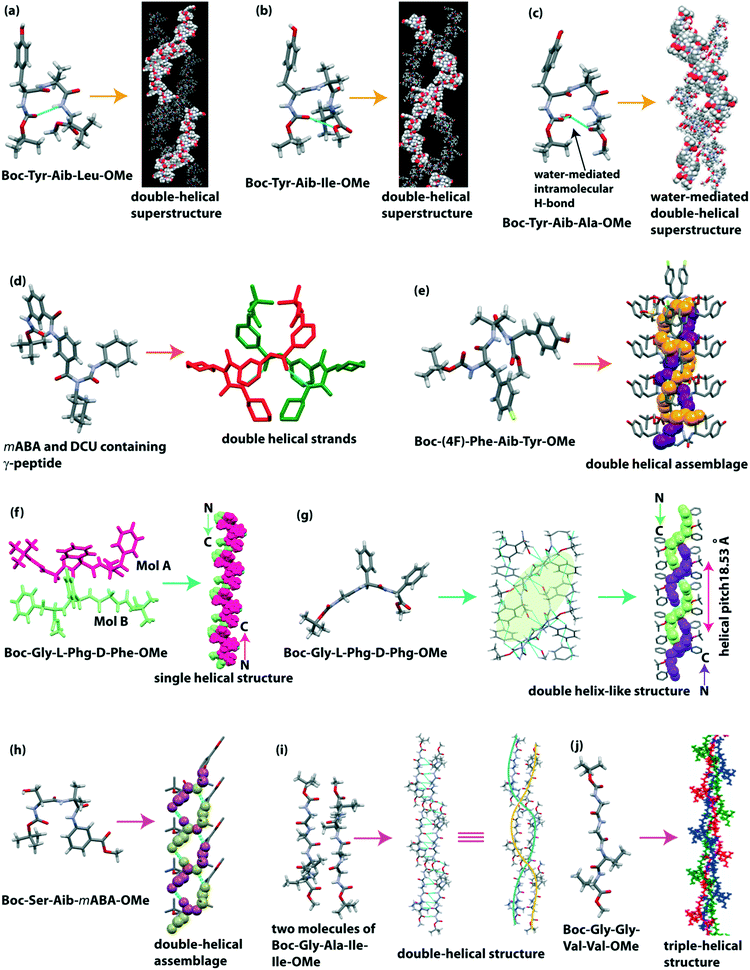 | ||
| Fig. 10 Double helical superstructure of tripeptides (reproduced from ref. 117 with permission from Royal Society of Chemistry) (a) Boc-Tyr-Aib-Leu-OMe and (b) Boc-Tyr-Aib-Ile-OMe. (c) Water-mediated double-helical superstructure of Boc-Tyr-Aib-Ala-OMe. (d) Helical duplex of m-aminobenzoic acid and N,N′-dicyclohexylurea containing γ-peptide. (e) Double helical assemblage of Boc-(4F)-Phe-Aib-Tyr-OMe. Single helical and double-helical structure of (f) Boc-Gly-L-Phg-D-Phe-OMe and (g) Boc-Gly-L-Phg-D-Phg-OMe. (h) Double-helical assemblage of Boc-β-hydroxy-Ala-Aib-mABA-OMe. Double and triple helical structure of tetrapeptides (i) Boc-Gly-Ala-Ile-Ile-OMe and (j) Boc-Gly-Gly-Val-Val-OMe (reproduced from ref. 123 with permission from American Chemical Society). | ||
They observed that the m-aminobenzoic acid and N,N′-dicyclohexylurea containing γ-peptide dimerized in chloroform to build a parallel double helix, which is stabilized through intermolecular H-bonding and π–π stacking interactions.118,119 It also adopted an anti-parallel sheet-like structure, which further self-assembled to create a corrugated sheet-like structure in methanol. Interestingly, due to the change in solvent polarity (methanol to chloroform), the higher-order association of the helical strand also changed from a sheet to a double-helical structure (Fig. 10d). The morphology also varied (plate-like matrices in methanol to polydisperse microspheres in chloroform) by changing the solvent polarity. Interestingly, this double helix interacted with a potent bacteriostatic antibiotic, sulfamethoxazole (Table 4).
| Entry | Peptide sequence | Flexible/kink moieties | Secondary structure | Supramolecular self-assembly | Ref. |
|---|---|---|---|---|---|
| 1 | Val–Ala class (Val–Ala, Val–Val, Val–Ile, Ala–Val, Ala–Ile, Ile–Ala, and Ile–Val) | Double helical channel | 107–110 | ||
| 2 | Abu–Abu, Abu–Nva, Abu–Val, Abu–Leu, Abu–Ile, Nva–Abu, Nva–Nva, Nva–Val, Nva–Leu, and Nva–Ile | Double helical channel | 111 | ||
| 3 | β-Ala-Val | β-Ala | Double helix | 112 | |
| 4 | β-Ala-Ile | β-Ala | Double helix | 112 | |
| 5 | β-Ala-Phe | β-Ala | Double helix | 112 | |
| 6 | Phe-β-Ala | β-Ala | Single helix | 112 | |
| 7 | Pyridinedicarboxylic acid and Ala-mABA-OMe containing pseudopeptide | Double helix | 113 | ||
| 8 | Pyridinedicarboxylic acid Tyr-mABA-OMe containing pseudopeptide | Double helix | 114 | ||
| 9 | Pyridinedicarboxylic acid and Ile–Tyr containing pseudopeptide | Double helix | 114 | ||
| 10 | Boc-Gbn-Tyr-OH | Gbn | C9-turn | Double helix | 115 |
| 11 | Boc-L-Ile-L-Phe-OMe | Single helical structure | 116 | ||
| 12 | Boc-L-Ile-L-Phg-OMe | Cylinder-like structure around a water molecule | 116 | ||
| 13 | Boc-L-Ile-D-Phe-OMe and Boc-L-Ile-D-Phg-OMe | Discontinued double helix-like structure | 116 | ||
| 14 | Boc-Try-Aib-Leu-OMe | Aib | Type II β-turn | Double helix | 117 |
| 15 | Boc-Try-Aib-Ile-OMe | Aib | Type II′ β-turn | Double helix | 117 |
| 16 | Boc-Try-Aib-Ala-OMe | Aib | Hydrated turn | Double helix | 117 |
| 17 | m-Aminobenzoic acid and N,N′-dicyclohexylurea containing γ-peptide | mABA, DCU | Double helix | 118 | |
| 18 | Boc-4-F-Phe-Aib-Tyr-OMe | Aib | Double helix | 120 | |
| 19 | Boc-Gly-L-Phg-D-Phe-OMe | Single helix | 121 | ||
| 19 | Boc-Gly-L-Phg-D-Phg-OMe | Double helix | 121 | ||
| 20 | Boc-β-hydroxy-Ala-Aib-mABA-OMe | Aib and mABA | Type II′ β-turn | Double helix | 122 |
| 21 | Boc-Gly-Ala-Ile-Ile-OMe | Antiparallel β-sheet | Double-helix | 123 | |
| 22 | Boc-Gly-Gly-Val-Val-OMe | Gly–Gly | Antiparallel β-sheet | Triple-helix | 123 |
Dutt Konar and co-workers reported a side-chain modified Phe containing tripeptide (Boc-(4F)-Phe-Aib-Tyr-OMe) double helical assemblage, which is stabilized by two intermolecular H-bonds.120 The peptide molecule adopted an open strand conformation with three backbone kinks. Next, each subunit was inter-linked in anti-parallel mode to construct a double-helical assemblage (Fig. 10e) where the NH of Aib of one strand was connected with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Boc of another stand. Similarly, the NH of 4F-Phe of one strand was interlinked with the C
O of Boc of another stand. Similarly, the NH of 4F-Phe of one strand was interlinked with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Aib of another strand. Interestingly, the double-helical columns built a 3D channel framework with an internal dimension 14.472 × 13.377 Å. These double-helical entities self-associated in solution to create a flower-like morphology.
O of Aib of another strand. Interestingly, the double-helical columns built a 3D channel framework with an internal dimension 14.472 × 13.377 Å. These double-helical entities self-associated in solution to create a flower-like morphology.
We described single and double helix-like superstructure formation from terminally protected alternating D/L amino acid containing tripeptides.121 In the crystalline state, Boc-Gly-L-Phg-D-Phe-OMe exhibited two molecules (A and B) in the asymmetric unit, and they formed an anti-parallel β-sheet structure via intermolecular H-bonding. A single helical structure is formed through C–H⋯π and C–H⋯O interactions (Fig. 10f). Interestingly when the C-terminal D-Phe is replaced with D-Phg (one methylene less in the side chain than that of Phe), the supramolecular superstructure of the peptide Boc-Gly-L-Phg-D-Phg-OMe is changed. It showed an anti-parallel β-sheet secondary structure, which further self-assembled to build a supramolecular double-helix-like architecture through C–H⋯π and C–H⋯O interactions with a helical pitch of 18.53 Å (Fig. 10g). These peptides self-aggregated to make a bunch of flower-petal-like and a flower-like architecture, respectively, in solution.
Tetrapeptide double helix
Dutt Konar noticed that the crystal structure of the peptide Boc-β-hydroxy-Ala-Aib-mABA-OMe (β-hydroxy-Ala = Ser) exhibited a double helical structure in higher-order assembly.122 Each type II′ β-turn subunit is interconnected through intermolecular H-bonding between the amide NH of Aib of one molecule and CO of Aib of another molecule to build a single strand of double-helical assemblage (Fig. 10h). Next, another double helix strand is formed by intermolecular interaction between the OH group of the side chain of the Ser residue of one molecule and the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Ser of the following molecule. Interestingly, here the OH group of Ser plays a vital role in the formation of the double helix.
O of Ser of the following molecule. Interestingly, here the OH group of Ser plays a vital role in the formation of the double helix.
Nanda and co-workers reported the crystal structure of hydrophobic tetra peptides Boc-Gly-Ala-Ile-Ile-OMe and Boc-Gly-Gly-Val-Val-OMe, similar to the amyloid β fragments Aβ29–32 and Aβ37–40.123 Boc-Gly-Ala-Ile-Ile-OMe exhibited two molecules (A and B) in the asymmetric unit, and they interconnected in an antiparallel manner to form a dimer. Interestingly, B molecules interlinked themselves in a head-to-tail fashion through C–H⋯O and a rare C⋯C interaction (between sp2 hybridized carbonyl carbon of C-terminal Ile and sp3 hybridized tertiary carbon of the Boc group) to form a helical column. Next, each B molecule is H-bonded with molecule A to form a double-helix-like structure stabilized by C–H⋯O and C⋯C interactions (Fig. 10i). On the other hand, Boc-Gly-Gly-Val-Val-OMe showed one molecule in the asymmetric unit, and they connected to form an anti-parallel β-sheet. Further, they built a triple-helix-like structure through van der Waal interactions (Fig. 10j). The presence of two consecutive Gly–Gly units helps to form a bent structure and also stabilizes the triple helix. Interestingly, both peptides exhibited amyloidogenic nature.
Construction of a triple helix
The construction of a supramolecular triple-helical structure from small peptides is a challenging task. However, the long-chain collagen triple helix is ubiquitous in the extracellular matrix in animals and plays a vital role in cell adhesion and growth. This protein is mainly enriched with three amino acids e.g., Gly, Pro, and Hyp (hydroxyproline), with ‘Gly–Pro–Hyp’ tripeptide repeats in each strand. Scientists have been developing an artificial peptide-based triple helix by using natural or unnatural amino acids as building blocks, as collagen-like peptides are also used in pathogenic substances to eliminate contamination and improve thermal stability.124–126Banerjee et al. first reported the formation of a triple helix from the water-soluble tetrapeptide Tyr(1)-Aib(2)-Tyr(3)-Val(4).127 Here, the Tyr residue helps to improve the H-bonding ability at the side chain of the phenolic OH group. The peptide crystallized in one molecule with two water molecules in the asymmetric unit and adopted an ‘S’ shaped conformation. These ‘S’ shaped molecules are connected with two intermolecular H-bonds to construct a right-handed helical strand. Additionally, one phenolic OH of Tyr is H-bonded with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Val(4), and another phenolic OH of Tyr is H-bonded with the NH of Val(4) to form a single helical strand. Next, a supramolecular triple helix was generated by stacking three individual right-handed helical strands (Fig. 11a). The strands were elongated through the intermolecular H-bonding interactions of the intervening bridging water molecule.
O of Val(4), and another phenolic OH of Tyr is H-bonded with the NH of Val(4) to form a single helical strand. Next, a supramolecular triple helix was generated by stacking three individual right-handed helical strands (Fig. 11a). The strands were elongated through the intermolecular H-bonding interactions of the intervening bridging water molecule.
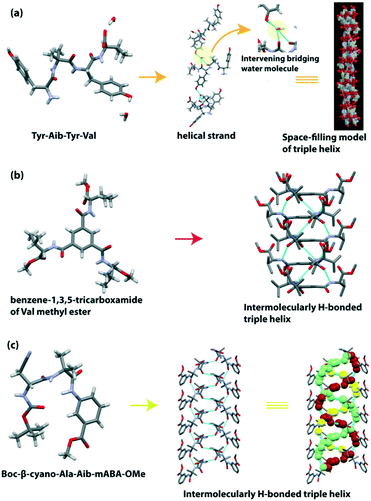 | ||
| Fig. 11 (a) Water-mediated triple helix of the tetrapeptide Tyr-Aib-Tyr-Val (reproduced from ref. 127 with permission from Royal Society of Chemistry). (b) Triple helical structure of the pseudopeptide benzene-1,3,5-tricarboxamide of Val methyl ester. (c) Triple-helical assemblage of the tripeptide Boc-β-cyano-Ala-Aib-mABA-OMe. | ||
Later, they demonstrated the formation of the triple helical structure of chiral benzene-1,3,5-tricarboxamide of Val methyl ester.128 This disc-shaped compound interconnected through intermolecular H-bonding and π–π stacking interactions among the central aromatic moieties (inter-ring centroid distance = 3.546 Å) to create a right-handed supramolecular triple helical structure (Fig. 11b). Morphology analysis suggested that this peptide is regularly intertwined to create triple-helical nanofibers with 130–150 nm diameter. Its enantiomer also twisted to form reverse-handed triple-helical nanofibers with a diameter of 165–185 nm.
Dutt Konar observed that the terminally protected tripeptide Boc-β-cyano-Ala-Aib-mABA-OMe adopted a type II′ β-turn structure, and further self-associated to form a supramolecular triple-helical assemblage (Fig. 11c).122 The two β-turn subunits are interconnected through intermolecular H-bonding between the NH group of Aib of one molecule and the CO group of Aib of another molecule to form helical strands. Then these strands are further stabilized through intermolecular H-bonding between the NH group of β-cyano-Ala of one molecule and the CO group of β-cyano-Ala of another molecule to form a triple-helical assemblage. Notably, the triple helix is stabilized by the electron-withdrawing effect of the cyano group of β-cyanoalanine, but interestingly, it does not take part in intermolecular H-bonding interaction. Such an effect of hydroxyproline in the collagen triple helix was also observed (Table 5).129
Conclusions and future perspective
In conclusion, this research highlight mainly covers small peptide-based supramolecular helical self-association in the solid-state. The structural feature of some essential building blocks that help form the helical assemblies of short peptides is discussed. In the entire highlight, the research focuses on the molecular structure, conformation, and higher-order helical (single/double/triple helix) self-association of single amino acid derivatives, di-, tri-, and tetra-peptides in the crystalline state. Supramolecular non-covalent interactions, especially intra and intermolecular H-bonding (N–H⋯O) interactions, C–H⋯O, C–H⋯N, rare C⋯C, C–H⋯π, π–π stacking interactions, and intra/intermolecular (O–H⋯O) hydrogen bonding involving the side-chain OH of Ser and Tyr residue, which also plays a vital role in forming the supramolecular helical structure, are described. Supramolecular helical assembly involving water molecules interconnecting two peptides through intermolecular (O–H⋯O) hydrogen bonding interactions is discussed. Although predicting the structural features from the sequence is not straightforward, the overall structural analysis suggests a few exciting trends. It seems that the peptides in native form do not like to form a crystal, probably because nature does not like native peptides to get crystallized in the body. However, the protected forms and peptides containing non-canonical amino acid(s) form crystals relatively easily (of course, the contribution of the solvent system must not be neglected).Similarly, the formation of supramolecular helical assembly does not follow any particular rule. However, the presence of a turn inducer moiety (e.g., Pro, Ant, m-ABA, Aib, etc.) in the peptide sequence helps to form a helical assembly. The side-chain of amino acids in the peptide sequence and weak supramolecular non-covalent interactions also play a crucial role in developing the helical assembly.
The reported helical peptides are attractive for their innovative applications in biotechnology and material science. An enantiomeric single-helical pseudopeptide acts as a template for fabricating dipeptide-capped gold nanoparticles, i.e., the pseudopeptide is a promising entity for the construction of 1D arrays of gold nanoparticles.78 A carbon-based solid material obtained from the co-assembly of organogel of a dipeptide-fullerene-multiwalled carbon nano-tube acts as a catalyst to reduce azo compounds to amine and benzoic acid to benzyl alcohol.82 A dipeptide with a column-like structure forms metallogel with Cu(II), Zn(II), and Pb(II) selectively, indicating its future application for developing peptidic functional materials.87 Some helical tripeptides form nano-tubes with a hydrophobic core that acts as a hydrophobic host to include a hydrophobic guest to generate a supramolecular polymer.92 Supramolecular single and double helical peptides with significant thermal stability can adsorb N2, CO2, and CH4 gas, indicating the formation of porous biomaterials which may be applicable for gas storage systems.81,108,111,117 A porous material pyridine dicarboxylic acid-containing pseudopeptide with a double-helical structure can adsorb I2 gas in the crystalline state.113 Gbn containing double-helical dipeptides act as a prospective energy storage material with a specific capacitance115 of 3500 μF g−1 at 0.6 A g−1. A double-helical γ-peptide also interacts with potent bacteriostatic antibiotic sulfamethoxazole, suggesting that the peptide behaves like a drug delivery cargo.118 Moreover, such helical di/tri/tetra-peptides hierarchically self-associated in solution to form various nanostructures, e.g., nano-rods, nano-tubes, and nanofibers, which can bind with amyloid binding dyes ThT and Congo red.
Many applications of supramolecular helical (single/double/triple helix) structures from short peptides (di/tri/tetrapeptides) in various bioactive fields have been observed. Although the construction of helical organization from small peptides at will is challenging due to their conformational flexibility, they are attractive in current research for developing smart biomaterials. The mechanical properties of single crystals of small molecules are well explored, but peptide single crystals are not explored yet. Mechanically flexible crystals are used in pharmaceuticals, solar cells, transistors, optoelectronics, flexible smart devices, and so on. Till now, the piezo-, pyro-, and ferroelectric properties of small helical peptides are not explored enough, although they have enough electron conductivity. These peptide molecules are eligible entities for the formation of smart flexible materials in the near future. Recently, collagen mimicking tripeptides Pro-Phe-Phe and Hyp-Phe-Phe exhibited significant piezoelectricity.130 These tripeptides act as a power generator to produce max current >50 nA and potential >1.2 V. On the other hand, the current research indicates that short helical peptides can form metal–organic frameworks (MOFs). For example, the terminal 3-pyridyl group containing peptide Gly-Pro-Pro coordinated with Ag(I) in aqueous alcohol to produce two helical nano channels (diameter >2 nm) through the formation of polyproline II type helices.131 These channels recognize anions, chiral organic molecules, and bio-oligomers. Terminal pyridyl moieties containing short α,γ-hybrid peptide 12-helices coordinated with the silver ion to form a porous metal–helix framework which can adsorb CO2.132 The creation of peptide-based MOFs may be helpful for structural elucidation of bio-oligomers and storage of gases in the near future. Finally, most of the exciting applications of peptide-based nanomaterials are due to their specific morphology. However, delineating the relationship of the crystal structure with its morphology is not straightforward due to environmental influence, such as solvent composition. Therefore, predicting and generating a particular morphology of these materials from their amino acid constituents at will are still challenging. Inspired by “AlphaFold”,133 artificial intelligence-based technologies may soon be available to overcome such challenges.
Author contributions
RSG prepared the initial draft. BM supervised the project, edited the manuscript, and managed the funding.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We are thankful to the Department of Biotechnology, Govt. of India, for financial support (sanction no. BT/PR29978/MED/30/2037/2018).Notes and references
- G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418–2421 CrossRef PubMed.
- L. Pauling, R. B. Corey and H. R. Branson, Proc. Natl. Acad. Sci. U. S. A., 1951, 378, 205–211 CrossRef PubMed.
- J. D. Watson and F. H. C. Crick, Nature, 1953, 171, 737–738 CrossRef PubMed.
- D. R. Eyre, Science, 1980, 207, 1315–1322 CrossRef PubMed.
- J. A. Kluge, O. Rabotyagova, G. G. Leisk and David L. Kaplan, Trends Biotechnol., 2008, 26, 244–251 CrossRef PubMed.
- R. E. Franklin, Nature, 1956, 177, 928–930 CrossRef.
- D. Mandal, A. N. Shirazi and K. Parang, Org. Biomol. Chem., 2014, 12, 3544–3561 RSC.
- E. Gazit, Chem. Soc. Rev., 2007, 36, 1263–1269 RSC.
- Y. Li, Y. Wang, G. Huang and J. Gao, Chem. Rev., 2018, 118, 5359–5391 CrossRef PubMed.
- N. Habibi, N. Kamaly, A. Memic and H. Shafiee, Nano Today, 2016, 11, 41–60 CrossRef PubMed.
- S. K. Maji, D. Schubert, C. Rivier, S. Lee, J. E. Rivier and R. Riek, PLoS Biol., 2008, 6, e17 CrossRef PubMed.
- F. Zhao, M. L. Ma and B. Xu, Chem. Soc. Rev., 2009, 38, 883–891 RSC.
- D. G. Fatouros, D. A. Lamprou, A. J. Urquhart, S. N. Yannopoulos, I. S. Vizirianakis, S. Zhang and S. Koutsopoulos, ACS Appl. Mater. Interfaces, 2014, 6, 8184–8189 CrossRef PubMed.
- P. Stano, P. Carrara, Y. Kuruma, T. P. de Souza and P. L. Luisi, J. Mater. Chem., 2011, 21, 18887–18902 RSC.
- M. Li, D. C. Green, J. L. R. Anderson, B. P. Binks and S. Mann, Chem. Sci., 2011, 2, 1739–1745 RSC.
- J. Sanchez-Quesada, M. P. Isler and M. R. Ghadiri, J. Am. Chem. Soc., 2002, 124, 10004–10005 CrossRef CAS PubMed.
- V. Jayawarna, M. Ali, T. A. Jowitt, A. E. Miller, A. Saiani, J. E. Gough and R. V. Ulijn, Adv. Mater., 2006, 18, 611–614 CrossRef CAS.
- S. Zhang, Adv. Cancer Res., 2008, 99, 335–362 CrossRef CAS.
- Y. Yang, U. Khoe, X. Wang, A. Horii, H. Yokoi and S. Zhang, Nano Today, 2009, 4, 193–210 CrossRef CAS.
- M. Yemini, M. Reches, J. Rishpon and E. Gazit, Nano Lett., 2005, 5, 183–186 CrossRef CAS.
- Y. Ding, D. Li, K. Zhao, W. Du, J. Zheng and M. Yang, Biosens. Bioelectron., 2013, 48, 281–286 CrossRef CAS PubMed.
- Z. S. Yao, M. Mito, T. Kamachi, Y. Shiota, K. Yoshizawa, N. Azuma, Y. Miyazaki, K. Takahashi, K. Zhang, T. Nakanishi, S. Kang, S. Kanegawa and O. Sato, Nat. Chem., 2014, 6, 1079–1083 CrossRef CAS.
- S. Günes, H. Neugebauer and N. S. Sariciftci, Chem. Rev., 2007, 107, 1324–1338 CrossRef.
- Q. Tang, L. Li, Y. Song, Y. Liu, H. Li, W. Xu, Y. Liu, W. Hu and D. Zhu, Adv. Mater., 2007, 19, 2624–2628 CrossRef CAS.
- L. Catalano, D. P. Karothu, S. Schramm, E. Ahmed, R. Rezgui, T. J. Barber, A. Famulari and P. Naumov, Angew. Chem., Int. Ed., 2018, 57, 17254–17258 CrossRef CAS PubMed.
- J. Harada, T. Shimojo, H. Oyamaguchi, H. Hasegawa, Y. Takahashi, K. Satomi, Y. Suzuki, J. Kawamata and T. Inabe, Nat. Chem., 2016, 8, 946–952 CrossRef CAS PubMed.
- D. T. Bong, T. D. Clark, J. R. Granja and M. R. Ghadiri, Angew. Chem., Int. Ed., 2001, 40, 988–1011 CrossRef CAS PubMed.
- C. H. Görbitz, M. Nilsen, K. Szeto and L. W. Tangen, Chem. Commun., 2005, 4288–4290 RSC.
- J. D. Hartgerink, E. Beniash and S. I. Stupp, Science, 2001, 294, 1684–1688 CrossRef CAS.
- M. Reches and E. Gazit, Science, 2003, 300, 625–627 CrossRef CAS.
- J. Chen and X. Zou, Bioact. Mater., 2019, 4, 120–131 CrossRef.
- N. Hendler, N. Sidelman, M. Reches, E. Gazit, Y. Rosenberg and S. Richter, Adv. Mater., 2007, 19, 1485–1488 CrossRef CAS.
- X. Gao and H. Matsui, Adv. Mater., 2005, 17, 2037–2050 CrossRef CAS PubMed.
- V. Nguyen, R. Zhu, K. Jenkins, R. Yang and Y. Jin, Nat. Commun., 2016, 7, 13566 CrossRef CAS PubMed.
- L. Adler-Abramovich and E. Gazit, Chem. Soc. Rev., 2014, 43, 6881–6893 RSC.
- L. Adler-Abramovich, Z. A. Arnon, X. M. Sui, I. Azuri, H. Cohen, O. Hod, L. Kronik, L. J. W. Shimon, H. D. Wagner and E. Gazit, Adv. Mater., 2018, 30, 1704551 CrossRef.
- J. Harada, Y. Kawamura, Y. Takahashi, Y. Uemura, T. Hasegawa, H. Taniguchi and K. Maruyama, J. Am. Chem. Soc., 2019, 141, 9349–9357 CrossRef PubMed.
- Y. Zhang, M. A. Hopkins, D. J. Liptrot, H. Khanbareh, P. Groen, X. Zhou, D. Zhang, Y. Bao, K. Zhou, C. R. Bowen and D. R. Carbery, Angew. Chem., Int. Ed., 2020, 59, 1–6 CrossRef.
- O. S. Makin and L. C. Serpell, FEBS J., 2005, 272, 5950–5961 CrossRef CAS.
- A. M. Squires, G. L. Devlin, S. L. Gras, A. K. Tickler, C. E. MacPhee and C. M. Dobson, J. Am. Chem. Soc., 2006, 128, 11738–11739 CrossRef CAS.
- D. J. Selkoe, Nature, 2003, 426, 900–904 CrossRef CAS PubMed.
- A. S. Detoma, S. Salamekh, A. Ramamoorthy and M. H. Lim, Chem. Soc. Rev., 2012, 41, 608–621 RSC.
- A. Paul, K. C. Nadimpally, T. Mondal, K. Thalluri and B. Mandal, Chem. Commun., 2015, 51, 2245–2248 RSC.
- A. Paul, S. Kumar, S. Kalita, S. Kalita, D. Sarkar, A. Bhunia, A. Bandyopadhyay, A. C. Mondal and B. Mandal, Chem. Sci., 2021, 12, 2853–2862 RSC.
- M. Schmidt, C. Sachse, W. Richter, C. Xu, M. Fandrich and N. Grigorieff, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 19813–19818 CrossRef CAS.
- R. Zhang, X. Hu, H. Khant, S. J. Ludtke, W. Chiu, M. F. Schmid, C. Frieden and J.-M. Lee, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 4653–4658 CrossRef CAS.
- Y. Miller, B. Ma, C.-J. Tsai and R. Nussinov, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14128–14133 CrossRef CAS PubMed.
- E. Tayeb-Fligelman, O. Tabachnikov, A. Moshe, O. Goldschmidt-Tran, M. R. Sawaya, N. Coquelle, J.-P. Colletier and M. Landau, Science, 2017, 355, 831–833 CrossRef PubMed.
- G. N. Ramachandran and G. Kartha, Nature, 1955, 176, 593 CrossRef CAS.
- T. Luo and K. L. Kiick, Eur. Polym. J., 2013, 49, 2998–3009 CrossRef CAS.
- A. Konietzny, J. Bär and M. Mikhaylova, Front. Cell. Neurosci., 2017, 11, 147 CrossRef.
- M. D. Shoulders and R. T. Raines, Annu. Rev. Biochem., 2009, 78, 929–958 CrossRef CAS.
- R. Kaul and P. Balaram, Bioorg. Med. Chem., 1999, 7, 105–117 CrossRef CAS.
- J. Venkatraman, S. C. Shankaramma and P. Balaram, Chem. Rev., 2001, 101, 3131–3152 CrossRef CAS.
- P. G. Vasudev, S. Chatterjee, N. Shamala and P. Balaram, Chem. Rev., 2011, 111, 657–687 CrossRef CAS.
- C. Toniolo, M. Crisma, F. Formaggio and C. Peggion, Biopolymers, 2001, 60, 396–419 CrossRef CAS.
- S. Aravinda, N. Shamala and P. Balaram, Chem. Biodiversity, 2008, 5, 1238–1262 CrossRef CAS.
- D. H. Appella, L. A. Christianson, I. L. Karle, D. R. Powell and S. H. Gellman, J. Am. Chem. Soc., 1996, 118, 13071–13072 CrossRef CAS.
- D. H. Appella, L. A. Christianson, D. A. Klein, D. R. Powell, L. Huang, J. J. Barchi and S. H. Gellman, Nature, 1997, 387, 381–384 CrossRef CAS PubMed.
- D. H. Appella, L. A. Christianson, I. L. Karle, D. R. Powell and S. H. Gellman, J. Am. Chem. Soc., 1999, 121, 6206–6212 CrossRef CAS.
- D. H. Appella, L. A. Christianson, D. A. Klein, M. A. Richards, D. R. Powell and S. H. Gellman, J. Am. Chem. Soc., 1999, 121, 7574–7581 CrossRef CAS.
- C. Toniolo, M. Crisma, F. Formaggio, A. Polese, M. Doi, T. Ishida, E. Mossel, Q. Broxterman and J. Kamphuis, Biopolymers, 1996, 40, 523–527 CrossRef CAS.
- Y. Lapeña, P. Lopez, C. Cativiela, B. Kaptein, Q. B. Broxterman, J. Kamphuis, E. Mossel, C. Peggion, F. Formaggio, M. Crisma and C. Toniolo, J. Chem. Soc., Perkin Trans. 2, 2000, 2, 631–636 RSC.
- A. Roy, P. Prabhakaran, P. K. Baruah and G. J. Sanjayan, Chem. Commun., 2011, 47, 11593–11611 RSC.
- R. V. Nair, K. N. Vijayadas, A. Roy and G. J. Sanjayan, Eur. J. Org. Chem., 2014, 7763–7780 CrossRef CAS.
- S. Kar, M. G. B. Drew and A. Pramanik, J. Mol. Struct., 2010, 963, 160–167 CrossRef CAS.
- I. L. Karle and P. Balaram, Biochemistry, 1990, 29, 6747–6756 CrossRef CAS.
- C. Toniolo and E. Benedetti, Macromolecules, 1991, 24, 4004–4009 CrossRef CAS.
- P. G. Vasudev, S. Chatterjee, N. Shamala and P. Balaram, Acc. Chem. Res., 2009, 42, 1628–1639 CrossRef CAS PubMed.
- M. Konda, R. G. Jadhav, S. Maiti, S. M. Mobin, B. Kauffmann and A. K. Das, Org. Biomol. Chem., 2018, 16, 1728–1735 RSC.
- G. Srinivasulu, M. H. V. Ramana Rao, S. Kiran Kumar and A. C. Kunwar, ARKIVOC, 2004, 8, 69–86 Search PubMed.
- S. Mondal and E. Gazit, ChemNanoMat, 2016, 2, 323–332 Search PubMed.
- S. Bera and E. Gazit, Protein Pept. Lett., 2019, 26, 88–97 CrossRef CAS.
- J. Liang, A. Hao, P. Xing and Y. Zhao, ACS Nano, 2021, 15, 5322–5332 CrossRef CAS PubMed.
- Z. Wang, A. Hao and P. Xing, Chin. Chem. Lett., 2021, 32, 1390–1396 CrossRef CAS.
- C. H. Görbitz, Chem. – Eur. J., 2001, 7, 5153–5159 CrossRef.
- S. Guha, M. G. B. Drew and A. Banerjee, Tetrahedron Lett., 2006, 47, 7951–7955 CrossRef CAS.
- S. Guha, M. G. B. Drew and A. Banerjee, Small, 2008, 4, 1993–2005 CrossRef CAS.
- S. Guha, M. G. B. Drew and A. Banerjee, Cryst. Growth Des., 2010, 10, 4716–4721 CrossRef CAS.
- A. Dutta, S. Kar, R. Fröhlich, P. Koley and A. Pramanik, ARKIVOC, 2009, 2, 31–43 Search PubMed.
- S. Maity, P. Jana, S. K. Maity, P. Kumar and D. Haldar, Cryst. Growth Des., 2012, 12, 422–428 CrossRef CAS.
- D. Mazzier, F. Carraro, M. Crisma, M. Rancan, C. Toniolo and A. Moretto, Soft Matter, 2016, 12, 238–245 RSC.
- P. S. Zelenovskiy, A. S. Nuraeva, S. Kopyl, S. G. Arkhipov, S. G. Vasilev, V. S. Bystrov, D. A. Gruzdev, M. Waliczek, V. Svitlyk, V. Y. Shur, L. Mafra and A. L. Kholkin, Cryst. Growth Des., 2019, 19, 6414–6421 CrossRef CAS.
- R. S. Giri and B. Mandal, CrystEngComm, 2018, 20, 4441–4448 RSC.
- Ø. Jacobsen, H. G. Gebreslasie, J. Klaveness, P. Rongved and C. H. Görbitz, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2011, 67, 278–282 CrossRef.
- R. S. Giri, S. Pal, S. Roy, G. Dolai, S. R. Manne, S. Paul and B. Mandal, Pept. Sci., 2021, 113, e24176 CAS.
- S. Kumar, S. K. Nandi, S. Suman and D. Haldar, CrystEngComm, 2020, 22, 7975–7982 RSC.
- D. Haldar, S. K. Maji, W. S. Sheldrick and A. Banerjee, Tetrahedron Lett., 2002, 43, 2653–2656 CrossRef CAS.
- A. Dutt Konar, CrystEngComm, 2013, 15, 10569–10578 RSC.
- A. Sharma, S. Goswami, R. Rajagopalan and A. Dutt Konar, Supramol. Chem., 2015, 27, 669–678 CrossRef CAS.
- A. S. Gangele, S. Goswami, A. K. Bar, P. Tiwari, S. Konar and A. Dutt Konar, Cryst. Growth Des., 2016, 16, 2130–2139 CrossRef.
- A. Paikar, A. Pramanik, T. Das and D. Haldar, Polym. Chem., 2017, 8, 396–403 RSC.
- S. Bera, S. Mondal, B. Xue, L. J. W. Shimon, Y. Cao and E. Gazit, Nat. Mater., 2019, 18, 503–509 CrossRef CAS PubMed.
- K. Hema and K. M. Sureshan, Angew. Chem., Int. Ed., 2020, 59, 8854–8859 CrossRef CAS PubMed.
- R. S. Giri and B. Mandal, CrystEngComm, 2019, 21, 236–243 RSC.
- G. Dolai, R. S. Giri, S. Roy and B. Mandal, Pept. Sci., 2021,(113), e24229 CAS.
- M. Ghosh, S. Bera, S. Schiffmann, L. J. W. Shimon and L. Adler-Abramovich, ACS Nano, 2020, 14, 9990–10000 CrossRef CAS PubMed.
- D. Haldar, S. K. Maji, M. G. B. Drew, A. Banerjee and A. Banerjee, Tetrahedron Lett., 2002, 43, 5465–5468 CrossRef CAS.
- S. K. Maji, A. Banerjee, M. G. B. Drew, D. Haldar and A. Banerjee, Tetrahedron Lett., 2002, 43, 6759–6762 CrossRef CAS.
- A. Banerjee, S. K. Maji, M. G. B. Drew, D. Haldar and A. Banerjee, Tetrahedron Lett., 2003, 44, 699–702 CrossRef CAS.
- D. Haldar, A. Banerjee, M. G. B. Drew, A. K. Das and A. Banerjee, Chem. Commun., 2003, 1406–1407 RSC.
- D. Haldar, M. G. B. Drew and A. Banerjee, Tetrahedron, 2006, 62, 6370–6378 CrossRef CAS.
- A. Dutta, A. Dutt, M. G. B. Drew and A. Pramanik, Supramol. Chem., 2008, 20, 625–633 CrossRef CAS.
- S. Kar, A. Dutta, M. G. B. Drew, P. Koley and A. Pramanik, Protein Pept. Lett., 2009, 16, 1063–1073 CrossRef CAS.
- A. Dutta, M. G. B. Drew and A. Pramanik, Helv. Chim. Acta, 2010, 93, 1025–1037 CrossRef CAS.
- M. Konda, T. Ghosh, S. M. Mobin and A. K. Das, New J. Chem., 2019, 43, 4830–4834 RSC.
- C. H. Görbitz, Curr. Opin. Solid State Mater. Sci., 2002, 6, 109–116 CrossRef.
- C. H. Görbitz, New J. Chem., 2003, 27, 1789–1793 RSC.
- C. H. Görbitz, Chem. – Eur. J., 2007, 13, 1022–1031 CrossRef.
- D. V. Soldatov, I. L. Moudrakovski, E. V. Grachev and J. A. Ripmeester, J. Am. Chem. Soc., 2006, 128, 6737–6744 CrossRef CAS.
- V. N. Yadav, A. Comotti, P. Sozzani, S. Bracco, T. Bonge-Hansen, M. Hennum and C. H. Görbitz, Angew. Chem., Int. Ed., 2015, 54, 15684–15688 CrossRef.
- S. Guha, M. G. B. Drew and A. Banerjee, Org. Lett., 2007, 9, 1347–1350 CrossRef CAS.
- A. Dutt Konar, CrystEngComm, 2013, 15, 2466–2473 RSC.
- P. Tiwari, S. Biswas, R. Verma, A. Sharma and A. Dutt Konar, ChemistrySelect, 2018, 3, 262–272 CrossRef CAS.
- M. Konda, T. Ghosh, S. M. Mobin and A. K. Das, New J. Chem., 2019, 43, 4830–4834 RSC.
- S. Roy, R. S. Giri, G. Dolai and B. Mandal, J. Mol. Struct., 2020, 1221, 128877 CrossRef CAS.
- P. Jana, S. Maity, S. K. Maity and D. Haldar, Chem. Commun., 2011, 47, 2092–2094 RSC.
- S. K. Maity, S. Maity, P. Jana and D. Haldar, Chem. Commun., 2012, 48, 711–713 RSC.
- R. Sarkar, M. Debnath, K. Maji and D. Haldar, RSC Adv., 2015, 5, 76257–76262 RSC.
- A. Sharma, P. Tiwari and A. Dutt Konar, J. Mol. Struct., 2018, 1161, 44–54 CrossRef CAS.
- R. S. Giri and B. Mandal, CrystEngComm, 2019, 21, 5618–5625 RSC.
- A. Dutt Konar, CrystEngComm, 2012, 14, 6689–6694 RSC.
- S. Misra, P. Singh, R. N. Mahata, P. Brandao, S. Roy, A. K. Mahapatra and J. Nanda, J. Phys. Chem. B, 2021, 125, 4274–4285 CrossRef CAS PubMed.
- J. A. Fallas, J. Dong, Y. J. Tao and J. D. Hartgerink, J. Biol. Chem., 2012, 287, 8039–8047 CrossRef CAS.
- S. P. Boudko and J. Engel, J. Mol. Biol., 2004, 335, 1289–1297 CrossRef CAS.
- A. V. Persikov, J. A. M. Ramshaw and B. Brodsky, J. Biol. Chem., 2005, 280, 19343–19349 CrossRef CAS.
- A. K. Das, D. Haldar, R. P. Hegde, N. Shamala and A. Banerjee, Chem. Commun., 2005, 1836–1838 RSC.
- P. P. Bose, M. G. B. Drew, A. K. Das and A. Banerjee, Chem. Commun., 2006, 3196–3198 RSC.
- I. R. Babu and K. N. Ganesh, J. Am. Chem. Soc., 2001, 123, 2079–2080 CrossRef CAS.
- S. Bera, S. Guerin, H. Yuan, J. O'Donnell, N. P. Reynolds, O. Maraba, W. Ji, L. J. W. Shimon, P.-A. Cazade, S. A. M. Tofail, D. Thompson, R. Yang and E. Gazit, Nat. Commun., 2021, 12, 2634 CrossRef CAS PubMed.
- T. Sawada, A. Matsumoto and M. Fujita, Angew. Chem., 2014, 126, 7356–7360 CrossRef.
- R. Misra, A. Saseendran, S. Dey and H. N. Gopi, Angew. Chem., 2019, 58, 2251–2255 CrossRef CAS.
- J. Jumper, et al. , Nature, 2021, 596, 583–589 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |