Open Access Article
Open Access ArticleComprehensive exploration of the anticancer activities of procaine and its binding with calf thymus DNA: a multi spectroscopic and molecular modelling study†
Mohd. Sajid Ali *a,
Mohammad Abul Farahb,
Hamad A. Al-Lohedana and
Khalid Mashay Al-Anazib
*a,
Mohammad Abul Farahb,
Hamad A. Al-Lohedana and
Khalid Mashay Al-Anazib
aDepartment of Chemistry, College of Science, King Saud University, P.O. Box-2455, Riyadh-11451, Saudi Arabia. E-mail: smsajidali@gmail.com; Fax: +966-14679972; Tel: +966-598878428
bDepartment of Zoology, College of Science, King Saud University, P.O. Box-2455, Riyadh-11451, Saudi Arabia
First published on 1st March 2018
Abstract
Procaine is an anesthetic drug commonly administrated topically or intravenously for use in local anesthesia. Promisingly, some anticancer activities of procaine have also been reported. Therefore, the mechanism of interaction between anesthetic drug procaine with ct-DNA was determined collectively by means of various spectroscopic and molecular docking methods. Minor groove 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding of procaine to the ct-DNA was evidenced from absorption spectroscopy, fluorescence quenching, DNA melting, competitive binding measurements with EB and DAPI dyes, viscosity and CD spectroscopy together with molecular docking simulations and DFT calculations. Molecular docking on five different B-DNA structures (taken from the Protein Data Bank) shows that procaine binds in the AT rich region of all five B-DNA structures. Thermodynamic parameters, evaluated using van't Hoff's isotherm, shown that the interaction was feasible and the binding forces involved were hydrophobic as well as hydrogen bonding which were, further, confirmed by molecular docking. The frontier molecular orbitals (HOMO and LUMO) of procaine and DNA bases have been calculated by DFT method and the chemical potential (μ), chemical hardness (η) and fraction number of electrons (ΔN) from procaine to DNA bases were evaluated, which have shown that procaine acts as an electron donor to the DNA bases. Simultaneously, anticancer activities of procaine alone and in combination with doxorubicin were observed on the MCF-7 breast cancer cell line. The results showed that the combined treatment with both procaine and doxorubicin enhanced the cytotoxic and apoptotic inducing potential of doxorubicin.
1 binding of procaine to the ct-DNA was evidenced from absorption spectroscopy, fluorescence quenching, DNA melting, competitive binding measurements with EB and DAPI dyes, viscosity and CD spectroscopy together with molecular docking simulations and DFT calculations. Molecular docking on five different B-DNA structures (taken from the Protein Data Bank) shows that procaine binds in the AT rich region of all five B-DNA structures. Thermodynamic parameters, evaluated using van't Hoff's isotherm, shown that the interaction was feasible and the binding forces involved were hydrophobic as well as hydrogen bonding which were, further, confirmed by molecular docking. The frontier molecular orbitals (HOMO and LUMO) of procaine and DNA bases have been calculated by DFT method and the chemical potential (μ), chemical hardness (η) and fraction number of electrons (ΔN) from procaine to DNA bases were evaluated, which have shown that procaine acts as an electron donor to the DNA bases. Simultaneously, anticancer activities of procaine alone and in combination with doxorubicin were observed on the MCF-7 breast cancer cell line. The results showed that the combined treatment with both procaine and doxorubicin enhanced the cytotoxic and apoptotic inducing potential of doxorubicin.
1. Introduction
Procaine, an anesthetic drug, has been used in dentistry for a long time. Several other uses of procaine have also been reported, such as for the cure of depression, inflammation, obesity, etc.1–5 Apart from these uses, procaine has an enormous range of applications, particularly, in cancer related research. It has been found to inhibit the triggering as well as the induction of thermotolerance in mouse fibroblast LM cells.6 Furthermore, it was found to improve the therapeutic index of chemotherapy agent cisplatin around two-fold.7 Procaine in a small dose of 2 mM has shown cytotoxicity to leukemic cells with further enhancement in the cytotoxicity at 42 °C.8 The most noticeable role of procaine related to the cancer chemotherapy is the demethylation of DNA.9,10 DNA methylation has enormous applications in gene regulation. A hallmark of cancer is the deregulation of the DNA methylation machinery and aberrant DNA methylation patterns. “DNA methyltransferases (DNMTs) catalyzes the transfer of a methyl group from S-adenosyl-L-methionine to the carbon-5 position of cytosine residues that results in an epigenetic change”.11 “These enzymes regulate gene expression, for example, hypermethylation of the promoter lead to transcriptional silencing of tumor suppressor genes. Therefore, DNMT inhibitors or demethylating agents are promising new drugs for the treatment of diseases such as cancer and brain disorders”.11,12 DNA demethylation is a process in which inhibitor suppresses the activity of DNA methyltransferases.13Owing to the promising roles of the procaine in molecular biology it is important to understand the interaction of this pharmacologically important drug with nucleic acid. DNA is a significant genomic constituent of life that transports most of the hereditary data and enables the biological synthesis of proteins and enzymes through the replication and transcription.14 The investigation of binding of DNA with small molecules is also of importance among the researchers to investigate its modes of binding. Small molecules, such as drugs, can binds to the DNA either with covalent mode of binding or non-covalently. Covalent binding mode is ascribed to the formation of covalent adducts between DNA and ligand which generally takes place via alkylation or inter- and intrastrand crosslinking and is irreversible in nature which can restrict the activity, such as, transcription and replication of DNA and a consequent cell death.15,16 Non-covalent binding includes three different modes which are (1) intercalative binding, which comprises of the insertion of the ligand inside the DNA helix which may cause in the discrepancy in the DNA structure and functions; (2), electrostatic binding or external binding between the negatively charged DNA phosphate backbone and positive end of the molecules; and (3) groove binding that occurs either in major groove or minor groove depending on the size of the ligand as the bigger molecules tend to bind at major groove and smaller molecules generally occupies the minor groove.17 Intercalative and groove binding occur near or inside the DNA double helix, while the electrostatic binding takes place outside the groove.18,19
As procaine is an established anesthetic drug with many other uses including anticancer activities, therefore, in this study we have seen the interaction of procaine with ct-DNA by using theoretical as well as experimental methods in order to understand the mechanism of the drug binding. The anticancer capability of procaine was also seen by elucidating its cytotoxic and apoptotic effects on human breast cancer (MCF-7) cells. Finally, an effort was made to determine the influence of procaine combined with doxorubicin, an anthracycline antibiotic which is one of the most effective anticancer agents used for the treatment of breast cancer.
2. Experimental
Sodium salt of ct-DNA (D1501, type I, fibers, 41.9 mol% G–C and 58.1 mol% A–T) and procaine hydrochloride (99%) (Scheme S1†) were purchased from Sigma, USA. Rests of the chemicals were of high purity grades or molecular biology grades and the details of all the materials used are given in ESI.† Stock solutions of ct-DNA were prepared by reported methods and the ratio of absorbance at 260 nm and 280 nm was in the range of 1.8–1.9. For experimental binding studies, we have used UV-visible, fluorescence, circular dichroism spectroscopies and viscometry.20 For molecular modelling the geometries of procaine and DNA bases were optimized at DFT/BP RI by ORCA.21 Autodock 4.2.3 program was used to perform docking calculations of B-DNAs with procaine.22 Anticancer activities of procaine alone and in combination of doxorubicin on MCF-7 breast cancer cell lines were performed using well established methods. Detailed information of all experiments and computational studies is given in ESI.†3. Results and discussions
3.1. Experimental investigation of interaction of procaine with ct-DNA
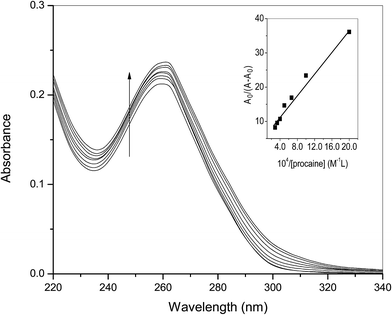
The association constant (Ka) between ct-DNA and procaine can be calculated by using Benesi–Hildebrand equation16,24 which is given as:
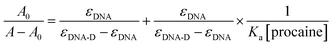 | (1) |
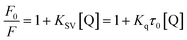 | (2) |
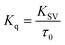 | (3) |
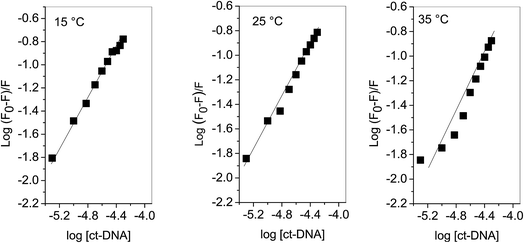
KSV can be obtained from the linear regression of eqn (2) for which the plots of F0/F versus [Q] at three temperatures are given in Fig. 2. Calculated values of KSV, which are given in Table 1, show a decreasing trend on increasing the temperature conclusive of involvement of static quenching mechanism in procaine–ct-DNA interaction.
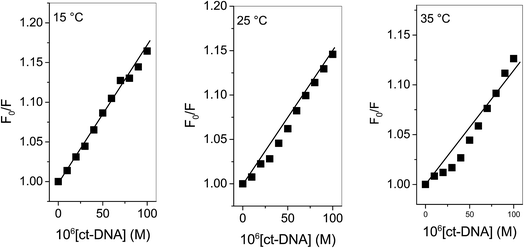 | ||
| Fig. 2 Stern–Volmer plots of ct-DNA interaction with procaine at various temperatures. [procaine] = 30 × 10−6 M. | ||
| T (K) | Stern–Volmer quenching constants | Binding parameters | Thermodynamic parameters | ||||
|---|---|---|---|---|---|---|---|
| KSV (M−1) | Kq (M−1 s−1) | n | Kb (M−1) | ΔG (kJ M−1) | ΔH (kJ M−1) | ΔS (J M−1 K−1) | |
| 288 | 3.3 × 103 | 5.8 × 1011 | 1.03 | 5.1 × 103 | −20.5 | −6.4 | 48.9 |
| 298 | 2.8 × 103 | 4.9 × 1011 | 1.04 | 4.8 × 103 | −21.0 | ||
| 308 | 2.2 × 103 | 3.9 × 1011 | 1.04 | 4.3 × 103 | −21.5 | ||
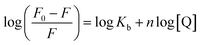 | (4) |
Plots of log(F0 − F)/F versus log[Q] (Fig. 3) were used to evaluate the binding constant and number of binding sites which are given in Table 1. The Kb obtained at 25 °C is in very close agreement from the one obtained using absorption spectroscopy. The value of n also validates that there is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding between procaine and ct-DNA.
1 binding between procaine and ct-DNA.
The involvement of various forces (hydrophobic, van der Waals, electrostatic and hydrogen bonding) between a ligand and macromolecule can be understood by assessing the thermodynamic parameters. Change in enthalpy (ΔH) and entropy (ΔS) of interaction process along with free energy change (ΔG) are promising factors to define the binding modes involved in the interaction.
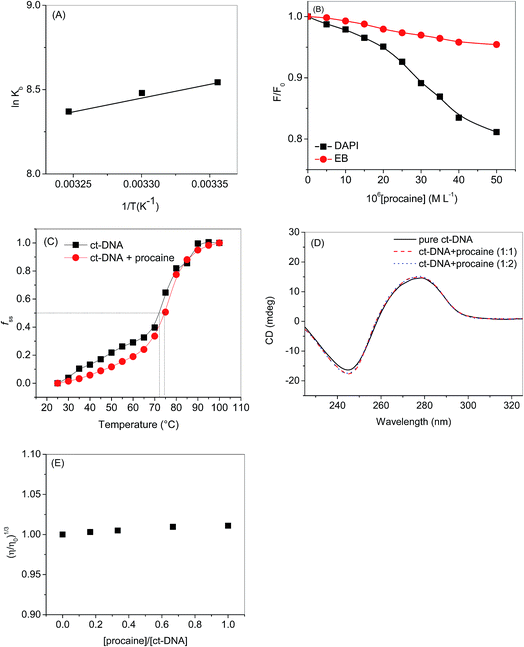
If the ΔH does not change too much on the temperature variation than both ΔH and ΔS can be evaluated from the van't Hoff equation (eqn (5)):
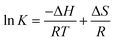 | (5) |
| ΔG = ΔH − TΔS | (6) |
van't Hoff plot of interaction of ct-DNA with procaine are given in Fig. 4(A). The values of ΔG were found to be negative at all temperatures envisage the feasibility of interaction. From enthalpy and entropy point of views the binding process was exothermic in nature i.e., negative ΔH with positive ΔS. It is well understood that hydrophobic interaction is characterized by the positive values of both ΔH and ΔS. Involvement of van der Waals forces and hydrogen bonding results in the negative values of both ΔH and ΔS whereas in case of electrostatic interaction the value of ΔH is either very less or zero.29 In present case the negative value of ΔH and positive value of ΔS suggests that the main forces involved in the binding of procaine with ct-DNA are hydrophobic forces and hydrogen bonding that may play major roles in the interaction.30 The presence of other interaction forces cannot be ruled out, though, it can be said that the interaction forces are dominated by hydrophobic forces and hydrogen bonding.31
3.2. Computational analyses of interaction of procaine with DNA
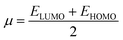 | (7) |
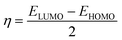 | (8) |
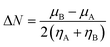 | (9) |
Fig. 6 is showing the molecular orbital plots of procaine along with the four DNA nucleotides. The charge density of HOMO of procaine is mainly accumulated on the nitrogen atom attached to the aliphatic carbon atoms whereas in case of LUMO charged densities moved to the benzene ring and the nitrogen attached to that. In case of nucleotides i.e., adenine, guanine, cytosine and thymine the charge densities are accumulated in on the aromatic rings in both HOMO and LUMO.
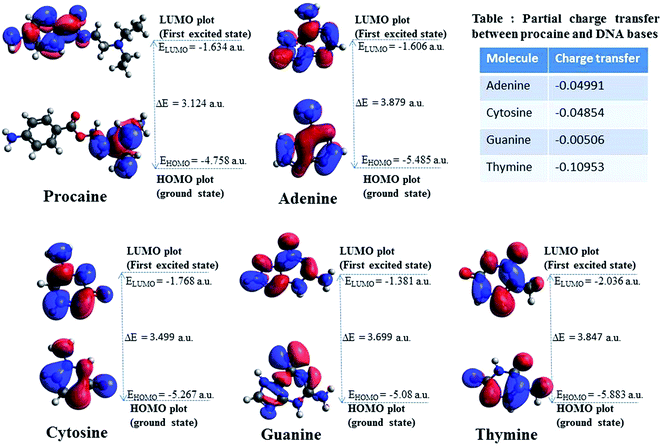 | ||
| Fig. 6 HOMO–LUMO composition of the frontier molecular orbitals of procaine and DNA bases. (Inset table) The calculated charge transfer values between procaine and DNA bases. | ||
The values of the chemical potential (inset table of Fig. 6) indicate the transfer of electrons from a high chemical potential (less electronegative) system to a low chemical potential (high electronegative) system.55 It can be stated that procaine acts as electron donor to the DNA bases.
3.3. Anticancer and anti-proliferative activities of procaine
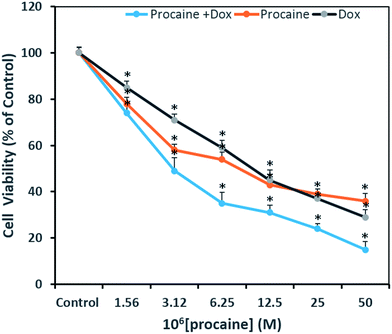
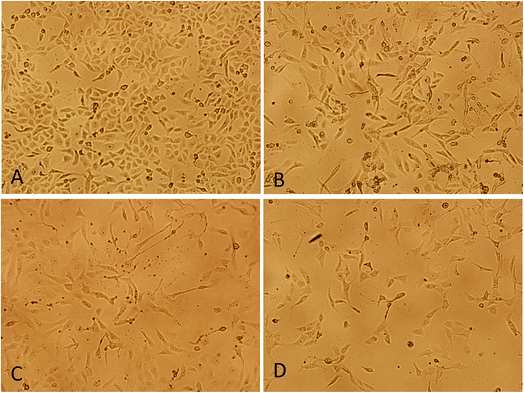
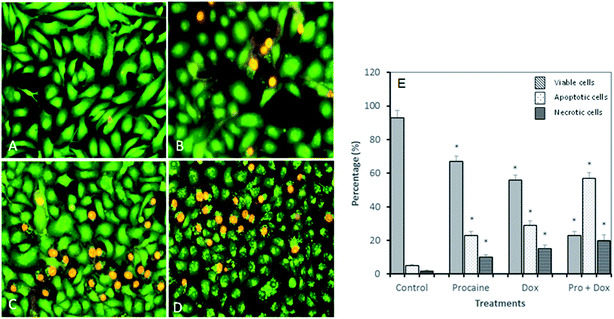
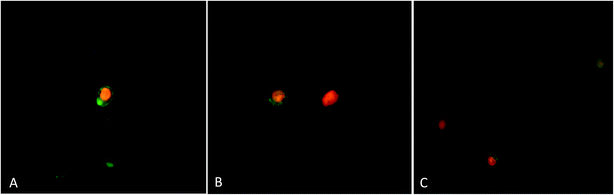
A concentration dependent decrease in the cell viability was evident. Procaine and doxorubicin inhibited cell proliferation by 71% and 64%, respectively at highest concentration of 50 μM. While, combined treatment reduced cell growth by 85% and higher concentration. These data suggest that procaine enhanced the antiproliferative property of doxorubicin.
Our results proved that procaine inhibits the proliferation of MCF-7 cells alone or in the presence of doxorubicin, thereby increasing its cytotoxicity. Previously, procaine has been reported to exert DNA demethylation and inhibits the growth of MCF-7 cells.9 Recently, procaine has been shown to induce mitochondrial dysfunction, elevated ROS level and apoptosis in human neuroblastoma cell line.56 On the other hand, doxorubicin was reported to exhibit its cytotoxic effect by intercalating between DNA base pairs and by inhibition of TOPO II enzyme, protein synthesis and overproduction of ROS.57 Unfortunately, doxorubicin is associated with several serious adverse effects such as myelosuppression, mucositis, alopecia, chronic cardiotoxicity that is limiting factor of the chemotherapeutic use of doxorubicin.58 Therefore, a wide variety of approaches has been investigated to minimize the side effects of doxorubicin and to improve its therapeutic use. Some reports emphasize that combining more than one anticancer drug can minimize the side effects of anticancer agent used alone. In this context, Li et al., demonstrated that lidocaine sensitizes the cytotoxicity of cisplatin against MCF-7 cells.59 However, in the present study, it is not clear by what mechanism procaine enhances the effects of doxorubicin, but these results support the possible use of procaine and its derivatives in cancer therapy with some other anticancer agents.
4. Conclusions
Assessment of ct-DNA binding properties (experimental and theoretical) of procaine along with its anticancer activities has been carried out in this study. Analyses of experimental investigations inferred that there was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
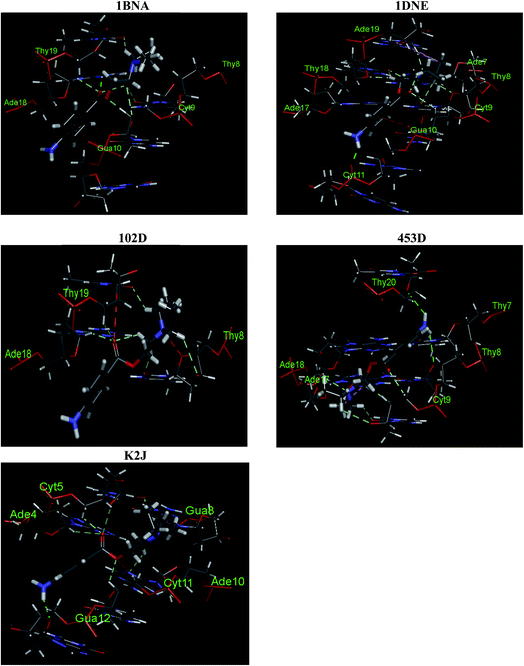
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding between ct-DNA and procaine and the preferred binding mode was minor groove binding which involves hydrogen bonding, and hydrophobic forces. Molecular docking on five different structures of B-DNA and procaine interaction also favors the experimental observations and in all five B-DNA structures, minor groove, which is AT rich region, was the principle binding site, however, from molecular docking the involvement of electrostatic forces in the interaction was also observed. Molecular orbital calculations using DFT methods were used to calculate the chemical potentials of procaine and DNA bases which stated that procaine acts as electron donor to the DNA bases. Procaine was also found to have cytotoxic effects towards MCF-7 breast cancer cell lines. The results also showed that the combined treatment with both procaine and doxorubicin enhanced the cytotoxic and apoptosis inducing potential of doxorubicin. The results support the promising use of procaine and its derivatives in cancer therapy with some other anticancer agents.
1 binding between ct-DNA and procaine and the preferred binding mode was minor groove binding which involves hydrogen bonding, and hydrophobic forces. Molecular docking on five different structures of B-DNA and procaine interaction also favors the experimental observations and in all five B-DNA structures, minor groove, which is AT rich region, was the principle binding site, however, from molecular docking the involvement of electrostatic forces in the interaction was also observed. Molecular orbital calculations using DFT methods were used to calculate the chemical potentials of procaine and DNA bases which stated that procaine acts as electron donor to the DNA bases. Procaine was also found to have cytotoxic effects towards MCF-7 breast cancer cell lines. The results also showed that the combined treatment with both procaine and doxorubicin enhanced the cytotoxic and apoptosis inducing potential of doxorubicin. The results support the promising use of procaine and its derivatives in cancer therapy with some other anticancer agents.
Conflicts of interest
There are no conflicts to declare.Abbreviations
| ct-DNA | Calf thymus DNA |
| CD | Circular dichroism |
| EB | Ethidium bromide |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| DFT | Density functional theory |
| HOMO | Highest occupied molecular orbital |
| LUMO | Lowest unoccupied molecular orbital |
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding the work through the research group project no. RGP-148.References
- E. J. Olsen, L. Bank and L. F. Jarvik, J. Gerontol., 1978, 33, 514–520 CrossRef CAS PubMed.
- R. Jakobs, M. U. Adamek, A. C. Von Bubnoff and J. F. Riemann, Scand. J. Gastroenterol., 2000, 35, 1319–1323 CrossRef CAS PubMed.
- L. J. Casarett and C. D. Klaassen, Casarett and Doull's Toxicology: The Basic Science of Poisons, McGraw-Hill, 2001 Search PubMed.
- W. W. Zung, D. Gianturco and E. Pfieffer, Psychopharmacol. Bull., 1976, 12, 50–51 CAS.
- M. D. A. R. P. Bulcao, M. Roehrs, C. Paniz, F. L. Cervi, F. V. Thiesen, M. B. Leal and S. C. Garcia, Rev. Cienc. Farm. Basica Apl., 2011, 32, 297–303 Search PubMed.
- A. W. T. Konings, Cancer Res., 1985, 45, 2016 CAS.
- M. Esposito, R. A. Fulco, P. Collecchi, A. Zicca, A. Cadoni, F. Merlo, R. Rosso and A. Sobrero, JNCI, J. Natl. Cancer Inst., 1990, 82, 677–684 CrossRef CAS PubMed.
- Y. Rong, X. Xia and B. Lin, Chin. J. Cancer Res., 1993, 5, 70–73 CrossRef.
- A. Villar-Garea, M. F. Fraga, J. Espada and M. Esteller, Cancer Res., 2003, 63, 4984–4989 CAS.
- M. Tada, F. Imazeki, K. Fukai, A. Sakamoto, M. Arai, R. Mikata, T. Tokuhisa and O. Yokosuka, Hepatol. Int., 2007, 1, 355–364 CrossRef PubMed.
- M. Rius and F. Lyko, Oncogene, 2012, 31, 4257–4265 CrossRef CAS PubMed.
- J. M. Foulks, K. M. Parnell, R. N. Nix, S. Chau, K. Swierczek, M. Saunders, K. Wright, T. F. Hendrickson, K. K. Ho, M. V. McCullar and S. B. Kanner, J. Biomol. Screening, 2012, 17, 2–17 CrossRef CAS PubMed.
- N. Martinet, B. Y. Michel, P. Bertrand and R. Benhida, MedChemComm, 2012, 3, 263–273 RSC.
- Y. Ni, M. Wei and S. Kokot, Int. J. Biol. Macromol., 2011, 49, 622–628 CrossRef CAS PubMed.
- C. Silvestri and J. S. Brodbelt, Mass Spectrom. Rev., 2013, 32, 247–266 CrossRef CAS PubMed.
- M. Sirajuddin, S. Ali and A. Badshah, J. Photochem. Photobiol., B, 2013, 124, 1–19 CrossRef CAS PubMed.
- K. E. Erkkila, D. T. Odom and J. K. Barton, Chem. Rev., 1999, 99, 2777–2796 CrossRef CAS PubMed.
- L. S. Lerman, J. Mol. Biol., 1961, 3, 18–30 CrossRef CAS PubMed.
- R. Vijayalakshmi, M. Kanthimathi, V. Subramanian and B. U. Nair, Biochem. Biophys. Res. Commun., 2000, 271, 731–734 CrossRef CAS PubMed.
- B.-M. Liu, C.-L. Bai, J. Zhang, Y. Liu, B.-Y. Dong, Y.-T. Zhang and B. Liu, J. Lumin., 2015, 166, 48–53 CrossRef CAS.
- F. Neese, Wiley Interdisciplinary Reviews: Computational Molecular Science, 2018, vol. 8, p. e1327 Search PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- J.-H. Shi, T.-T. Liu, M. Jiang, J. Chen and Q. Wang, J. Photochem. Photobiol., B, 2015, 147, 47–55 CrossRef CAS PubMed.
- M. Purcell, J. F. Neault and H. A. Tajmir-Riahi, Biochim. Biophys. Acta Protein Struct. Mol. Enzymol., 2000, 1478, 61–68 CrossRef CAS.
- M. S. Ali and H. A. Al-Lohedan, J. Mol. Liq., 2017, 236, 232–240 CrossRef CAS.
- Principles of Fluorescence Spectroscopy, ed. J. R. Lakowicz, Springer US, Boston, MA, 2006, pp. 277–330, DOI:10.1007/978-0-387-46312-4_8.
- U. Anand, C. Jash and S. Mukherjee, J. Phys. Chem. B, 2010, 114, 15839–15845 CrossRef CAS PubMed.
- S. M. Ng, M. Koneswaran and R. Narayanaswamy, RSC Adv., 2016, 6, 21624–21661 RSC.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS PubMed.
- S. Naveenraj and S. Anandan, J. Photochem. Photobiol., C, 2013, 14, 53–71 CrossRef CAS.
- Y. Ma, G. Zhang and J. Pan, J. Agric. Food Chem., 2012, 60, 10867–10875 CrossRef CAS PubMed.
- F. A. Tanious, J. M. Veal, H. Buczak, L. S. Ratmeyer and W. D. Wilson, Biochemistry, 1992, 31, 3103–3112 CrossRef CAS PubMed.
- E. Trotta, E. D'Ambrosio, N. Del Grosso, G. Ravagnan, M. Cirilli and M. Paci, J. Biol. Chem., 1993, 268, 3944–3951 CAS.
- E. Trotta, E. D'Ambrosio, G. Ravagnan and M. Paci, Nucleic Acids Res., 1995, 23, 1333–1340 CrossRef CAS PubMed.
- D. Banerjee and S. K. Pal, J. Phys. Chem. B, 2008, 112, 1016–1021 CrossRef CAS PubMed.
- W. D. Wilson, F. A. Tanious, H. J. Barton, R. L. Jones, K. Fox, R. L. Wydra and L. Strekowski, Biochemistry, 1990, 29, 8452–8461 CrossRef CAS PubMed.
- J. Kapuscinski, Biotech. Histochem., 1995, 70, 220–233 CrossRef CAS PubMed.
- X. Cai, P. J. Gray Jr and D. D. Von Hoff, Cancer Treat. Rev., 2009, 35, 437–450 CrossRef CAS PubMed.
- B. S. P. Reddy, S. M. Sondhi and J. W. Lown, Pharmacol. Ther., 1999, 84, 1–111 CrossRef CAS PubMed.
- E. N. Zaitsev and S. C. Kowalczykowski, Nucleic Acids Res., 1998, 26, 650–654 CrossRef CAS PubMed.
- X. Zhou, G. Zhang and J. Pan, Int. J. Biol. Macromol., 2015, 74, 185–194 CrossRef CAS PubMed.
- B. Jana, S. Senapati, D. Ghosh, D. Bose and N. Chattopadhyay, J. Phys. Chem. B, 2012, 116, 639–645 CrossRef CAS PubMed.
- S. Bi, H. Zhang, C. Qiao, Y. Sun and C. Liu, Spectrochim. Acta, Part A, 2008, 69, 123–129 CrossRef PubMed.
- A. Rajendran and B. U. Nair, Biochim. Biophys. Acta Gen. Subj., 2006, 1760, 1794–1801 CrossRef CAS PubMed.
- V. Uma, M. Kanthimathi, T. Weyhermuller and B. U. Nair, J. Inorg. Biochem., 2005, 99, 2299–2307 CrossRef CAS PubMed.
- H. Yang, P. Tang, B. Tang, Y. Huang, X. Xiong and H. Li, RSC Adv., 2017, 7, 10242–10251 RSC.
- S. Satyanarayana, J. C. Dabrowiak and J. B. Chaires, Biochemistry, 1992, 31, 9319–9324 CrossRef CAS PubMed.
- J. M. Kelly, A. B. Tossi, D. J. McConnell and C. OhUigin, Nucleic Acids Res., 1985, 13, 6017–6034 CrossRef CAS PubMed.
- C. Metcalfe, C. Rajput and J. A. Thomas, J. Inorg. Biochem., 2006, 100, 1314–1319 CrossRef CAS PubMed.
- R. Rohs, I. Bloch, H. Sklenar and Z. Shakked, Nucleic Acids Res., 2005, 33, 7048–7057 CrossRef CAS PubMed.
- R. Corradini, S. Sforza, T. Tedeschi and R. Marchelli, Chirality, 2007, 19, 269–294 CrossRef CAS PubMed.
- I. Haq, Arch. Biochem. Biophys., 2002, 403, 1–15 CrossRef CAS PubMed.
- X. Du, Y. Li, Y.-L. Xia, S.-M. Ai, J. Liang, P. Sang, X.-L. Ji and S.-Q. Liu, Int. J. Mol. Sci., 2016, 17, 144 CrossRef PubMed.
- J. Padmanabhan, R. Parthasarathi, V. Subramanian and P. K. Chattaraj, Chem. Res. Toxicol., 2006, 19, 356–364 CrossRef CAS PubMed.
- Y.-Q. Wang and H.-M. Zhang, J. Photochem. Photobiol., B, 2015, 149, 9–20 CrossRef CAS PubMed.
- X.-j. Yu, W. Zhao, Y.-j. Li, F.-x. Li, Z.-j. Liu, H.-l. Xu, L.-y. Lai, R. Xu and S.-y. Xu, Sci. Rep., 2017, 7, 45316 CrossRef CAS PubMed.
- L. Roca-Alonso, L. Pellegrino, L. Castellano and J. Stebbing, Cardiology, 2012, 122, 253–259 CrossRef CAS PubMed.
- C. Carvalho, R. X. Santos, S. Cardoso, S. Correia, P. J. Oliveira, M. S. Santos and P. I. Moreira, Curr. Med. Chem., 2009, 16, 3267–3285 CrossRef CAS PubMed.
- K. Li, J. Yang and X. Han, Int. J. Mol. Sci., 2014, 15, 23519 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra13647a |
| This journal is © The Royal Society of Chemistry 2018 |