Preparation of one- to four-branch silver nanostructures of various sizes by metallization of hybrid DNA–protein assemblies†
Sergii Rudiukabc,
Anna Venancio-Marquesabc,
Géraldine Hallaisabc and
Damien Baigl*abc
aDepartment of Chemistry, Ecole Normale Supérieure, 24 rue Lhomond, 75005 Paris, France. E-mail: damien.baigl@ens.fr; Web: http://www.baigllab.com/ Tel: +33 1 4432 2405
bUniversité Pierre et Marie Curie Paris 6, 4 place Jussieu, 75005 Paris, France
cUMR 8640, CNRS, France
First published on 1st May 2013
Abstract
We exploit the versatility of DNA–protein assemblies to generate branched metal nanostructures, referred to as nanoshurikens, of various sizes and degrees of branching. Branched silver nanostructures are prepared by metallization of star-shaped DNA–protein templates composed of monobiotinylated DNA molecules surrounding a single streptavidin protein core. DNA–protein templates are prepared by direct assembly that results in a mixture of 1-, 2-, 3- and 4-branch structures that can be separated by gel electrophoresis. A one-pot, bulk metallization is then performed in situ by successive addition of silver nitrate and sodium borohydride. This results in branched metal nanostructures with one to four branches of a well-defined length that is about 3-fold shorter than that of the template. We show that it is possible to tune two structural parameters: (i) the degree of branching by varying the concentration of streptavidin and (ii) the branch length (from 21 ± 5 nm to 107 ± 22 nm) by using biotinylated DNA molecules of different sizes.
Introduction
Reduction of various metal ions in the presence of anionic templates (so-called metallization process) is a widely used strategy to prepare various kinds of metal nanostructures of well-defined size and shape.1–3 While biological templates, such as proteins4–9 and bacteria,10 have been investigated, the remarkable versatility of DNA templates for metallization was clearly established.11–35 Self-organized structures obtained by DNA compaction36 were also used to prepare metal nanorings.37 With recent development of DNA nanotechnology,38 for instance, a large number of 2D and 3D nanostructures can be obtained through Watson–Crick base pairing39–41 or DNA–protein conjugation.42 However, the metallization of such highly organized nanotemplates has been only partially explored for the generation of well-defined metal nanostructures. In particular, there is no available methodology for preparing branched metal nanostructures with an easily variable and controllable degree of branching. Recently, remarkable 3-branch silver nanoobjects have been produced by using T-shaped DNA origamis as templates,43,44 although the authors did not describe higher degrees of branching. DNA origami is a powerful technology but it requires specific templates and large libraries of DNA fragments of precise sequence.39 Metallization have also been applied with 3-branch RecA–DNA templates,45 2-branch DNA–gold nanoparticle conjugates46 and DNA networks combed on a mica substrate,17 but each of these methods provided a fixed degree of branching. Note also that in all reported approaches, the metallization was performed on templates already adsorbed on a surface. In this article, we describe a one-pot strategy for the preparation of nanostructures with various branch sizes and degrees of branching through bulk metallization. We used well-defined branched templates obtained by conjugating biotinylated DNA branches to a single streptavidin core, where the branch length was controlled by the initial DNA size, and the degree of branching was tuned by varying the DNA and streptavidin concentrations.Materials and methods
Materials
Streptavidin from S. avidinii was purchased from Life Technologies. It was dissolved in water to give 1 mg mL−1 stock solution and stored at 4 °C. PCR primers, silver nitrate AgNO3 (99.9999%), sodium chloride NaCl and agarose were purchased from Sigma-Aldrich. Sodium borohydride NaBH4 (≥99%) was purchased from Fluka. Deionized water (Millipore, 18 MΩ cm) was used for all experiments.Preparation of monobiotinylated DNA
Three monobiotinylated DNA of 206 bp (B-DNA206), 541 bp (B-DNA541) and 978 bp (B-DNA978) were prepared by PCR using pAG001 plasmid as the template (a gift from A. Gautier, ENS), the same biotinylated forward primer (Biotin-TTACGGTAAACTGCCCACTTG) and three different non-biotinylated reverse primers (CCACGCCCATTGATGTACTGC, CTCAGCCTCTCGATCTCGTACC and TCAATGAGACAAGTGATGTGCAGG respectively). 50 μL reaction solutions were placed on ice with the following final composition: 0.2 mM dNTPs, 50 ng of DNA template, 0.5 μM primers and 0.5 μL (for B-DNA206 and B-DNA541) or 0.25 μL (for B-DNA978) of 2000 U mL−1 Phusion® High-Fidelity DNA polymerase in HF buffer (New England BioLabs Inc.). After the first step at 98 °C for 30 s, 25 cycles (98 °C for 10 s, 66 °C for 20 s and 72 °C for 10, 15 or 20 s for B-DNA206, B-DNA541 and B-DNA978, respectively) were performed followed by the last step at 72 °C for 5 min. Length and purity of amplified DNA were checked by agarose gel electrophoresis. Finally, amplified B-DNA206, B-DNA541 and B-DNA978 were purified using a QIAquick PCR Purification Kit (QIAGEN). The concentrations of the resulting monobiotinylated DNA, as determined by absorption using a BioPhotometer plus spectrophotometer (Eppendorf), were 55 μg mL−1 (i.e., 400 nM in DNA molecules) for B-DNA206, 70 μg mL−1 (i.e., 196 nM in DNA molecules) for B-DNA541, and 25 μg mL−1 (i.e., 38 nM in DNA molecules) for B-DNA978.Preparation of DNA nanostar templates
Solutions of monobiotinylated DNA and streptavidin were mixed to reach different target concentrations in 0.2 M NaCl for a final volume of 4 μL. Resulting solutions were mixed by pipetting up and down prior to overnight incubation at 28 °C.Metallization
Within one day after preparation of the solution containing DNA nanostar templates, water, AgNO3 and a freshly prepared NaBH4 solution were added in this order with the following final concentrations: [DNA] = 0.5 μM in phosphate groups (regardless of monobiotinylated DNA size); [AgNO3] = 50 μM; [NaBH4] = 50 μM. The final volumes were 50 μL, 100 μL and 200 μL for B-DNA978, B-DNA541 and B-DNA206, respectively. The reaction mixture was immediately mixed by pipetting for several minutes.Atomic force microscopy (AFM)
A freshly cleaved mica surface (potassium aluminosilicate (Muscovite Mica) from Goodfellow) was first treated with 0.2 mM spermine solution for 1 hour, rinsed with an excess of water and stored under a drop of water for up to a few hours. Right after removing water with filter paper (Whatman), a 40 μL drop of a diluted sample solution (0.5 μM in DNA phosphate groups), before or after metallization, is then placed on the mica slide. After 1 hour of incubation in wet atmosphere at room temperature, the mica plate was rinsed with an excess of water, dried with compressed argon and left under vacuum for 1 hour. AFM measurements were performed using a 5100 Atomic Force Microscope (Agilent Technologies-Molecular Imaging) operated in a dynamic tip deflection mode (Acoustic Alternating Current mode, AAC). All AFM experiments were performed using silicon probes (Applied NanoStructures-FORT) in the tapping mode with a spring constant of 3 N m−1 at 69 kHz. The images were scanned in topography mode. To establish the branch length distributions, we performed analysis of the AFM images using the freehand line tool provided by ImageJ software. For DNA–streptavidin nanostar templates, the length of each DNA branch was measured by calculating the length of a freehand line drawn along the DNA from the edge of the streptavidin to the DNA extremity. For silver nanostructures, we performed the same procedure but starting from the center of the objects. The radius of streptavidin, determined on non-metallized samples (8.5 ± 1.5 nm), was then subtracted to give the estimated branch length. Each distribution was established on 50–150 individual branches.Agarose gel electrophoresis
Electrophoresis analysis was performed using a 1.5% agarose gel containing GRGreen DNA stain (Excellgen), with a constant electric field of 100 V for 1 h. Three 6 μL solutions were loaded into the gel (Fig. 3):- lane 1: 100 bp DNA ladder (New England BioLabs);
- lane 2: biotinylated 541 bp DNA solution without conjugation (16.3 nM in molecules);
- lane 3: DNA nanostar template solution obtained by mixing streptavidin (16.6 nM) and B-DNA541 (49 nM) and diluted 3-fold prior to gel loading.
After migration, the gel was analyzed using a G:BOX imaging system (Syngene).
Results and discussion
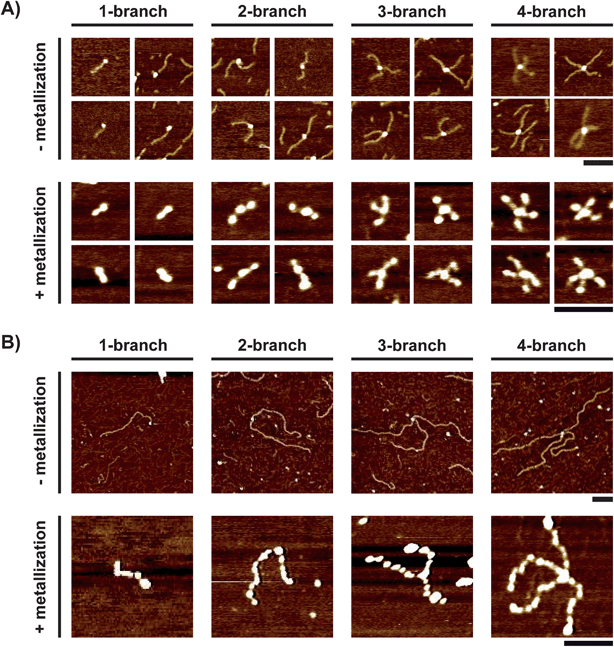
Fig. 1A shows our one-pot, two-step synthesis of multibranched silver nanoobjects. In the first step, monobiotinylated DNA was mixed with streptavidin to give structures with one to four branches. Interestingly, the salt concentration proved to be a critical parameter, as a high salt concentration (0.2 M) was necessary at this step to obtain a high yield of conjugation as well as a significant fraction of 3- and 4-branch DNA–protein structures (ESI, Fig. S1†). In contrast, at a low salt concentration, a large amount of non-conjugated DNA as well as very few 4-branch structures were observed (ESI, Fig. S1†), in agreement with a previous report.47 The presence of salt probably enables the formation of structures with a high degree of branching by screening the electrostatic repulsions between DNA branches and thus allowing the approach of biotinylated DNA to the streptavidin core once two or three DNA branches have already been conjugated. In the second step, these templates were diluted in pure water and metallized by adding AgNO3 and a reducing agent (NaBH4). The dilution was necessary to decrease the ionic strength and improve the efficiency of templated metallization. This procedure resulted in silver nanostructures with one to four branches referred to as “nanoshurikens” for their shapes similar to ancient ninja throwing weapons. We used atomic force microscopy (AFM) as it allowed us to characterize the size and morphology of both templates and resulting metal nanostructures. Fig. 1B shows typical AFM large-scale images of the templates obtained with a 541 bp monobiotinylated DNA (B-DNA541) and Fig. 1C shows the resulting nanoshurikens after metallization. Before metallization, we observed a large number of branched structures composed of DNA chains (height: 0.37 ± 0.08 nm) surrounding a central part of larger height (1.1 ± 0.3 nm) attributed to a single streptavidin core (Fig. 1B).47–49 A small number of unconjugated DNA molecules were also detected. After metallization, we observed branched structures with a greater height (1.6 ± 0.6 nm), which indicated successful silver deposition (Fig. 1C). A large number of such silver nanoshurikens were detected together with some individual spherical particles attributed to non-templated silver reduction. | ||
| Fig. 1 (A) A monobiotinylated 541bp DNA (B-DNA541, 49 nM) is assembled with streptavidin (16.3 nM) in NaCl solution (0.2 M) prior to 100-fold dilution in water and addition of AgNO3 (50 μM) and NaBH4 (50 μM). Representative AFM images of the DNA nanostar templates (B) and resulting silver nanostructures after metallization (C). All scale bars are 200 nm. | ||
By analyzing AFM images, four DNA–protein structures could be distinguished before metallization. They consisted of a single streptavidin surrounded by one to four DNA branches (Fig. 2A). Fig. 3 shows agarose gel electrophoresis analysis of B-DNA541 before and after conjugation to streptavidin. Before conjugation, a single band was observed corresponding to the size of monobiotinylated 541 bp DNA. Notably, after conjugation, 4 well-defined bands were observed. The lower one was at the same position as that of the unconjugated DNA and was attributed to a mixture of free DNA and 1-branch template. The three upper bands were attributed to 2-, 3- and 4-branch nanostar templates, respectively. All these results show that only four well-defined protein–DNA hybrid structures existed in the solution.
 | ||
| Fig. 2 AFM images of the four kinds of branched nanostructures before (A) and after (B) silver metallization prepared under the same conditions as in Fig. 1. All scale bars are 100 nm. The height scale is the same as that in Fig. 1B and C. (C) Branch length distributions before (blue) and after (red) metallization for each degree of branching. n shows the number of measured branches. | ||
 | ||
| Fig. 3 Agarose gel electrophoresis analysis of B-DNA541 before and after conjugation to streptavidin. | ||
By systematic analysis of AFM images on a large number of templates, we measured the DNA branch length and established the distributions shown in Fig. 2C. These measurements showed that all templates had a narrow branch length distribution centered around a similar average length, regardless of the number of branches (180.6 ± 15.6 nm; 181.4 ± 12.5 nm; 173.6 ± 14.5 nm and 175.3 ± 15.1 nm for one-, two-, three- and four-branch template, respectively). The narrow size distributions and the average values close to the expected contour length of fully stretched 541 bp DNA (184 nm) show that no detectable DNA damage occurred during the template preparation. After metallization, silver nanoshurikens with one to four branches were clearly distinguished (Fig. 2B). To our knowledge, this was the first time that such metal nanostructures with different degrees of branching had been obtained by templated metallization. By AFM image analysis, we also established the branch length distributions and found that all nanoshurikens had a similar branch length distribution (Fig. 3C). Notably, although the distribution width was similar to that of the corresponding template, a marked change in branch length was observed upon metallization. The average branch length of the nanoshurikens (67 ± 17 nm; 64 ± 17 nm; 60 ± 18 nm and 68 ± 14 nm for one-, two-, three- and four-branch structures, respectively) was approximately 2.7-fold shorter than the branch size of the corresponding template. This effect is probably specific to the bulk metallization process since such a shrinking has never been reported on adsorbed DNA templates. It could be due to the reorganization of DNA to optimize packing of silver atoms upon reduction and/or to the local accumulation of cationic charges during the metal growth leading to partial DNA compaction.50 All these results show for the first time that the metallization of DNA nanostar templates obtained by streptavidin–biotin assembly results in metal nanostructures with homogeneous branch sizes and degrees of branching varying from one to four.
Our assembly method generates a mixture of templates with different degrees of connectivity. To evaluate the possibility to control the branching levels, we studied the influence of streptavidin concentration at a fixed B-DNA541 concentration (49 nM) on the distribution of DNA nanostar templates. For each streptavidin concentration, AFM analysis showed that DNA molecules were either unconjugated or conjugated to a single streptavidin to give the same four kinds of branched nanostructures shown in Fig. 2A. However, the proportion of each nanostructure was strongly dependent on streptavidin concentration. By systematic AFM analysis of hundreds of DNA molecules, we established the distribution of free DNA, 1-, 2-, 3- and 4-branch templates as a function of streptavidin concentration (Fig. 4). At a low streptavidin concentration ([strep] ≤ 16 nM), DNA was mainly in the unconjugated form, which was due to the excess of DNA with respect to streptavidin. For intermediate concentrations (16 nM < [strep] ≤ 33 nM), a majority of 4-branch and 3-branch structures were observed while the fraction of unconjugated DNA continuously decreased and only minor fractions of 2-branch and 1-branch structures were observed. For higher streptavidin concentrations ([strep] > 33 nM), unconjugated, 4-branch and 3-branch structures decreased while increasing fractions of 2-branch and 1-branch conjugates were observed. These results show that the streptavidin concentration can be used to tune the distribution in degrees of branching during the preparation of templates. Since these templates are well separated by electrophoresis (Fig. 3), it could be possible to separate a given template using agarose gel electrophoresis purification.48
![Effect of streptavidin concentration on the distribution of DNA nanostar templates. A fixed concentration of B-DNA541 (49 nM) was mixed with a streptavidin concentration ([strep]) varying from 10 to 100 nM in 0.2 M NaCl. For each sample, the mixture was diluted 100-fold, deposited on the mica surface and analyzed by AFM. The observed structures were categorized into 5 types: free DNA, 1-, 2-, 3-, or 4-branch conjugates. The graph shows the fraction of DNA molecules incorporated in each type as a function of streptavidin concentration.](/image/article/2013/SM/c3sm50710f/c3sm50710f-f4.gif) | ||
| Fig. 4 Effect of streptavidin concentration on the distribution of DNA nanostar templates. A fixed concentration of B-DNA541 (49 nM) was mixed with a streptavidin concentration ([strep]) varying from 10 to 100 nM in 0.2 M NaCl. For each sample, the mixture was diluted 100-fold, deposited on the mica surface and analyzed by AFM. The observed structures were categorized into 5 types: free DNA, 1-, 2-, 3-, or 4-branch conjugates. The graph shows the fraction of DNA molecules incorporated in each type as a function of streptavidin concentration. | ||
We then investigated the possibility to control the branch size of the silver nanostructures. For this purpose, we followed the procedure for the preparation of the nanostar templates shown in Fig. 1A and studied the effect of using monobiotinylated DNA of various lengths. In addition to B-DNA541 that has hitherto been described, we also used monobiotinylated DNA with 206 bp (B-DNA206) and 978 bp (B-DNA978). We prepared the nanostar templates under the same conditions as with B-DNA541 and we observed the resulting structures by AFM. Regardless of DNA size, the same four characteristic conjugates could be distinguished having a single streptavidin core surrounded by one to four DNA branches (Fig. 5A and 5B top). With both B-DNA206 and B-DNA978, the presence of salt (0.2 M NaCl) and a high DNA over streptavidin ratio was necessary to get nanostructures having a high degree of branching, which confirms the observations made with B-DNA541. Interestingly, the branch length distribution determined by AFM was homogenous and did not depend on the degree of branching. The average branch size (70 ± 8 nm for B-DNA206 and 366 ± 23 nm for B-DNA978) corresponded well to the expected contour lengths of fully stretched 206 bp and 978 bp DNA (70 nm and 333 nm, respectively). After metallization, four silver nanostructures, similar to the four kinds of nanoshurikens observed with B-DNA541, were distinguished by AFM for both B-DNA206 (Fig. 5A bottom) and B-DNA978 (Fig. 5B bottom). The height did not depend on the size of template branches nor on the degree of branching, which indicates that silver deposition was not affected by the template size and shape. In contrast, the branch length strongly depended on the template branch size (21 ± 5 nm and 107 ± 22 nm for B-DNA206 and B-DNA978 bp, respectively). This corresponds to respectively 3.3- and 3.4-fold decrease of branch length upon silver metallization. To analyze how this bulk metallization method enables the control over the length of resulting metal branches, we compared the average length of DNA branches before and after silver metallization as a function of the initial number of base pairs of biotinylated DNA (Fig. 6). Before metallization, the average branch length of the template seems to linearly increase as a function of biotinylated DNA size, with a measured slope equaled to 0.36 nm bp−1, a value close to the length of the DNA base pair (0.34 nm). After metallization, the average branch size of the sliver nanoshurikens showed also an apparent linear behavior but with a much smaller slope (0.11 nm bp−1). There is thus a direct correlation between the final average branch size of the silver nanoshurikens and the initial length of the biotinylated templates used for the preparation of the templates (inset of Fig. 6). The bulk metallization thus induces a decrease in the branch size but this effect seems to be reproducible enough (about 3-fold shrinking regardless of the template size and the degree of branching) to predict the final branch length of the resulting silver nanoshurikens.
 | ||
| Fig. 5 AFM images of the four kinds of branched nanostructures before (top) and after (bottom) silver metallization obtained using a monobiotinylated DNA having (A) 206 bp (B-DNA206, 220 nM) and (B) 978 bp (B-DNA978, 21 nM). Streptavidin concentrations are 40 nM (A) and 3 nM (B). All scale bars are 100 nm. The height scale is the same as that in Fig. 1B and C. | ||
 | ||
| Fig. 6 Measured length of DNA branches before (−Ag, blue diamonds) and after (+Ag, red squares) silver metallization of nanostar templates as a function of the number of base pairs of the biotinylated DNA used for template preparation. The inset shows the branch length after metallization as a function of the branch length before metallization. Symbols and error bars are mean ± SD values determined by AFM. The slopes of the linear fits (solid lines) are 0.36 nm bp−1 (−Ag, blue line) and 0.11 nm bp−1 (+Ag, red line). | ||
Conclusions
We described a one-pot preparation of branched metal nanostructures called nanoshurikens with well-defined and controllable branch length, and a number of branches varying between one and four. We used self-assembled DNA–protein complexes as versatile templates for metallization. This strategy based on the metallization of star-shaped templates obtained by DNA–protein assembly offers several advantages: (i) a large variety of template shapes and sizes can be generated by DNA–protein assembly;42 (ii) DNA of different lengths can be used, from a few bases to several kbp; (iii) while we made branched structures with only one connection point, more complex networks can be devised, e.g. by using dibiotinylated DNA.47 Used as such or combined with other templating approaches, this method can be readily extended to various other templates to give a broad range of single or interconnected nanostructures of different metals.Acknowledgements
We thank A. Gautier for experimental support and discussion. The research leading to these results has received funding from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007–2013)/ERC Grant agreement no. 258782 and from Institut Universitaire de France. S.R. and A.V.-M. received fellowships from ENS (Paris) and ENS de Lyon, respectively.References
- T. K. Sau and A. L. Rogach, Complex-shaped Metal Nanoparticles, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2012 Search PubMed
.
- S. Sotiropoulou, Y. Sierra-Sastre, S. S. Mark and C. A. Batt, Chem. Mater., 2008, 20, 821–834 CrossRef CAS
.
- A. A. Zinchenko, Polym. Sci., Ser. C, 2012, 54, 80–87 CrossRef CAS
.
- F. Patolsky, Y. Weizmann and I. Willner, Nat. Mater., 2004, 3, 692–695 CrossRef CAS
.
- T. Scheibel, R. Parthasarathy, G. Sawicki, X.-M. Lin, H. Jaeger and S. L. Lindquist, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4527–4532 CrossRef CAS
.
- G. Wei, J. Reichert and K. D. Jandt, Chem. Commun., 2008, 3903–3905 RSC
.
- R. Kirsch, M. Mertig, W. Pompe, R. Wahl, G. Sadowski, K. J. Böhm and E. Unger, Thin Solid Films, 1997, 305, 248–253 CrossRef CAS
.
- M. Mertig, R. Kirsch and W. Pompe, Appl. Phys. A: Mater. Sci. Process., 1998, 66, S723–S727 CrossRef CAS
.
- W. Habicht, S. Behrens, K. Böhm and E. Dinjus, J. Phys.: Conf. Ser., 2007, 61, 374–378 CrossRef CAS
.
- R. Mogul, J. J. Getz Kelly, M. L. Cable and A. F. Hebard, Mater. Lett., 2006, 60, 19–22 CrossRef CAS
.
- E. Braun, Y. Eichen, U. Sivan and G. Ben-Yoseph, Nature, 1998, 391, 775–778 CrossRef CAS
.
- W. E. Ford, O. Harnack, A. Yasuda and J. M. Wessels, Adv. Mater., 2001, 13, 1793–1797 CrossRef CAS
.
- M. Mertig, L. Colombi Ciacchi, R. Seidel, W. Pompe and A. De Vita, Nano Lett., 2002, 2, 841–844 CrossRef CAS
.
- O. Harnack, W. E. Ford, A. Yasuda and J. M. Wessels, Nano Lett., 2002, 2, 919–923 CrossRef CAS
.
- H. Yan, S. H. Park, G. Finkelstein, J. H. Reif and T. H. LaBean, Science, 2003, 301, 1882–1884 CrossRef CAS
.
- K. Keren, R. S. Berman, E. Buchstab, U. Sivan and E. Braun, Science, 2003, 302, 1380–1382 CrossRef CAS
.
- Z. Deng and C. Mao, Nano Lett., 2003, 3, 1545–1548 CrossRef CAS
.
- C. F. Monson and A. T. Woolley, Nano Lett., 2003, 3, 359–363 CrossRef CAS
.
- K. Keren, R. S. Berman and E. Braun, Nano Lett., 2004, 4, 323–326 CrossRef CAS
.
- H. A. Becerril, R. M. Stoltenberg, C. F. Monson and A. T. Woolley, J. Mater. Chem., 2004, 14, 611–616 RSC
.
- R. Seidel, L. Colombi Ciacchi, M. Weigel, W. Pompe and M. Mertig, J. Phys. Chem. B, 2004, 108, 10801–10811 CrossRef CAS
.
- H. Li, S. H. Park, J. H. Reif, T. H. LaBean and H. Yan, J. Am. Chem. Soc., 2004, 126, 418–419 CrossRef CAS
.
- G. Braun, K. Inagaki, R. A. Estabrook, D. K. Wood, E. Levy, A. N. Cleland, G. F. Strouse and N. O. Reich, Langmuir, 2005, 21, 10699–10701 CrossRef CAS
.
- L. Berti, A. Alessandrini and P. Facci, J. Am. Chem. Soc., 2005, 127, 11216–11217 CrossRef CAS
.
- H. A. Becerril, P. Ludtke, B. M. Willardson and A. T. Woolley, Langmuir, 2006, 22, 10140–10144 CrossRef CAS
.
- J. Lund, J. Dong, Z. Deng, C. Mao and B. A. Parviz, Nanotechnology, 2006, 17, 2752–2757 CrossRef CAS
.
- S. H. Park, M. W. Prior, T. H. LaBean and G. Finkelstein, Appl. Phys. Lett., 2006, 89, 033901 CrossRef
.
- D. Aherne, A. Satti and D. Fitzmaurice, Nanotechnology, 2007, 18, 125205 CrossRef
.
- M. Fischler, U. Simon, H. Nir, Y. Eichen, G. A. Burley, J. Gierlich, P. M. E. Gramlich and T. Carell, Small, 2007, 3, 1049–1055 CrossRef CAS
.
- Q. Gu and D. T. Haynie, Mater. Lett., 2008, 62, 3047–3050 CrossRef CAS
.
- S. Kundu, V. Maheshwari and R. F. Saraf, Langmuir, 2008, 24, 551–555 CrossRef CAS
.
- S. Kundu and H. Liang, Langmuir, 2008, 24, 9668–9674 CrossRef CAS
.
- S. Kundu, K. Wang, D. Huitink and H. Liang, Langmuir, 2009, 25, 10146–10152 CrossRef CAS
.
- P. Gao and Y. Cai, ACS Nano, 2009, 3, 3475–3484 CrossRef CAS
.
- J. Timper, K. Gutsmiedl, C. Wirges, J. Broda, M. Noyong, J. Mayer, T. Carell and U. Simon, Angew. Chem., Int. Ed., 2012, 51, 7586–7588 CrossRef CAS
.
- A. Estévez-Torres and D. Baigl, Soft Matter, 2011, 7, 6746 RSC
.
- A. A. Zinchenko, K. Yoshikawa and D. Baigl, Adv. Mater., 2005, 17, 2820–2823 CrossRef CAS
.
- N. C. Seeman, Annu. Rev. Biochem., 2010, 79, 65–87 CrossRef CAS
.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS
.
- B. Saccà and C. M. Niemeyer, Angew. Chem., Int. Ed., 2012, 51, 58–66 CrossRef
.
- H. Li, T. H. LaBean and K. W. Leong, Interface Focus, 2011, 1, 702–724 CrossRef
.
- C. M. Niemeyer, Angew. Chem., Int. Ed., 2010, 49, 1200–1216 CrossRef CAS
.
- J. Liu, Y. Geng, E. Pound, S. Gyawali, J. R. Ashton, J. Hickey, A. T. Woolley and J. N. Harb, ACS Nano, 2011, 5, 2240–2247 CrossRef CAS
.
- Y. Geng, J. Liu, E. Pound, S. Gyawali, J. N. Harb and A. T. Woolley, J. Mater. Chem., 2011, 21, 12126–12131 RSC
.
- T. Nishinaka, A. Takano, Y. Doi, M. Hashimoto, A. Nakamura, Y. Matsushita, J. Kumaki and E. Yashima, J. Am. Chem. Soc., 2005, 127, 8120–8125 CrossRef CAS
.
- G. Wang, A. Ishikawa, A. Eguchi, Y. Suzuki, S. Tanaka, Y. Matsuo, K. Niikura and K. Ijiro, ChemPlusChem, 2012, 77, 592–597 CrossRef CAS
.
- C. M. Niemeyer, M. Adler, B. Pignataro, S. Lenhert, S. Gao, L. Chi, H. Fuchs and D. Blohm, Nucleic Acids Res., 1999, 27, 4553–4561 CrossRef CAS
.
- Y. Tian, Y. He, A. E. Ribbe and C. Mao, Org. Biomol. Chem., 2006, 4, 3404–3405 CAS
.
- S. Rudiuk, A. Venancio-Marques and D. Baigl, Angew. Chem., Int. Ed., 2012, 51, 12694–12698 CrossRef CAS
.
- A. A. Zinchenko, D. Baigl, N. Chen, O. Pyshkina, K. Endo, V. G. Sergeyev and K. Yoshikawa, Biomacromolecules, 2008, 9, 1981–1987 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: Sequences of B-DNA206, B-DNA541 and B-DNA978; effect of salt on the distribution of DNA nanostar templates (Fig. S1). See DOI: 10.1039/c3sm50710f |
| This journal is © The Royal Society of Chemistry 2013 |
