Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Degradable copper(II)-doped starch-based biopolymeric films with antibacterial activity†
Kiryl I.
Trusau
ab,
Paula
Jorge
 cd,
Ana Catarina
Sousa
ae,
Tiago A.
Fernandes
cd,
Ana Catarina
Sousa
ae,
Tiago A.
Fernandes
 a,
Vânia
André
a,
Vânia
André
 a,
Marina V.
Kirillova
a,
Marina V.
Kirillova
 a,
Andrew I.
Usevich
b,
Nuno
Cerca
*cd and
Alexander M.
Kirillov
a,
Andrew I.
Usevich
b,
Nuno
Cerca
*cd and
Alexander M.
Kirillov
 *a
*a
aCentro de Química Estrutural, Institute of Molecular Sciences, Departamento de Engenharia Química, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, 1049-001 Lisboa, Portugal. E-mail: kirillov@tecnico.ulisboa.pt
bDepartment of Oil and Gas Processing and Petroleum Chemistry, Belarusian State Technological University, 13a Sverdlova St., 220006 Minsk, Belarus
cCentre of Biological Engineering, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: nunocerca@ceb.uminho.pt
dLABBELS–Associate Laboratory, Braga/Guimarães, Portugal
eÁrea Departamental de Engenharia Química, ISEL-Instituto Superior de Engenharia de Lisboa, Instituto Politécnico de Lisboa, 1959-007 Lisboa, Portugal
First published on 6th April 2023
Abstract
The search for new bioactive molecules and sustainable materials to address antimicrobial resistance continues to be of significant attention in many research areas. In this work, new copper(II) coordination polymers and complexes containing ammonia and aromatic carboxylate ligands were self-assembled, characterized, and applied as bioactive dopants to produce starch-based biopolymeric films. The structures of [Cu(NH3)2(nca)2] (1) (Hnca = 2-naphthoic acid), [Cu(NH3)2(μ-ndca)]n (2) (H2ndca = 2,6-naphthalenedicarboxylic acid), and [Cu(NH3)2(μ-obba)]n (3) (H2obba = 4,4′-oxybis(benzoic acid)) reveal discrete monocopper(II) units in 1 or 1D coordination polymer chains in 2 and 3. In all compounds, the hexacoordinate Cu(II) centers feature an octahedral {CuN2O4} environment with mutually trans ammonia ligands. The compounds 1–3 were used as bioactive Cu-dopants (5%) to prepare biopolymeric films, 1–3@[PS]n and 1–3@[PS-MCC]n, based on sustainable and low-cost biofeedstocks such as potato starch (PS) or its mixture with microcrystalline cellulose (PS-MCC), respectively. Due to the importance in biomaterial-related infections, the growth inhibition of two clinically significant Gram-positive bacteria species, S. epidermidis and S. aureus, was studied in the presence of the prepared biopolymeric films. The Cu(NH3)2-carboxylates and derived biopolymeric materials showed a pronounced antibacterial activity, with doped films being able to inhibit the growth of 7 out of 8 strains tested, revealing a particularly high performance against the clinical isolates of S. epidermidis. By presenting these novel coordination compounds and biopolymeric films generated from sustainable biofeedstocks, this study combines several research approaches and broadens an antibacterial use of inorganic Cu-based derivatives and related biopolymer materials.
Sustainability spotlightTo address an increasing antimicrobial resistance, the search for new bioactive molecules and sustainable materials is currently in high demand. The present work describes novel antibacterial coordination compounds and degradable biopolymeric films generated from sustainable biofeedstocks (potato starch and cellulose) and simple and low-cost precursors. The obtained antibacterial materials may find prospective use in food processing and packaging, with a possibility to reduce the usage of fossil-fuel plastics. This may lead to sustainability impacts with regard to preventing or combating microbial hazards in the food chain, and to addressing various challenges of the UN 2030 Agenda for Sustainable Development, contributing to the topics on Food Security, Agriculture, Health, Sustainable Production & Consumption, and the respective Goals 2, 3, and 12. |
Introduction
Antimicrobial resistance is a natural phenomenon of microorganisms, such as bacteria, and its existence is ancient.1,2 However, since the introduction of antibiotics, namely penicillin in the 1920s, bacterial antimicrobial resistance has increased significantly. Nowadays, previously treatable infections are becoming life threatening, with a recent estimated burden of ∼5 million deaths in 2019 associated with antimicrobial resistance.3 Complex innate and acquired resistance mechanisms appear more frequently, rendering current therapies ineffective.4,5 Hence, there is a strong need for biomaterials capable of preventing the proliferation of bacteria on their surface.6,7In this respect, bioactive coordination compounds (bioCCs) and coordination polymers (bioCPs) have gained an impetus as potential antibacterial agents.8–12 Such compounds can be generated by merging biocidal metal centers with bioactive linkers, resulting in metal–organic architectures that can also contain antimicrobial guest molecules. Among a variety of metals with recognized antibacterial activity, copper is particularly appealing due to its considerable bioactivity, biocompatibility, and low toxicity to human cells. In fact, copper is one of the essential elements needed for normal function of many processes in the human body, with a necessary daily copper intake of ∼1–2 mg.13 Copper coordination compounds have several pathways of action14,15 that make them important in treating viral infections,16 inflammatory diseases,17 and microbial infections.18–20 In veterinary medicine, a Cu(II) coordination compound based on indomethacin is currently used as an anti-inflammatory agent. Many in vitro experiments have shown that copper has antimicrobial properties against a broad variety of bacteria, viruses, fungi and yeasts.21–24
The coordination environment around copper is critical in stabilizing its different oxidation states. When copper coordination compounds are used, they show extraordinary pharmacological effects that are not seen in the presence of inorganic copper compounds.25 Copper complexes can suppress the growth of different microorganisms and cause their destruction.12,14–16,25–31
For the synthesis of bioactive copper(II) compounds, the selection of commercially available ligands along with low-cost and biocompatible metal nodes plays an important role in targeting the desired properties for biological applications. The inclusion of ammonia as a co-ligand within the Cu-carboxylate compounds was governed by its recognized antimicrobial activity. According to Pinon et al., increased ammonia concentrations promote faster and larger reduction in bacterial populations.32
The principal objectives of this work were to generate novel bioCCs and bioCPs, incorporate them into biopolymers, and evaluate the antibacterial properties of the obtained materials with prospective applications in surface coating, wound healing, or antimicrobial packaging.33–36
For this purpose, we explored starch as a polysaccharide widely used in the production of environmentally friendly biomaterials, and a renewable and commercially available precursor.37 In particular, potato starch (PS) can be employed as a support to immobilize antimicrobial bioCCs or bioCPs to produce novel bioactive materials. In the packaging industry, there is a demand for sustainable materials to replace petrochemically based products with limited biodegradability. Hence, PS might be a suitable feedstock for developing biopolymeric films.38–41 Microcrystalline cellulose (MCC) represents another interesting biomaterial readily available from wood and cotton processing industries. The interest in MCC has also evolved due to its biodegradability and capability of aiding in the degradation of polymers or acting as a reinforcing agent.42,43
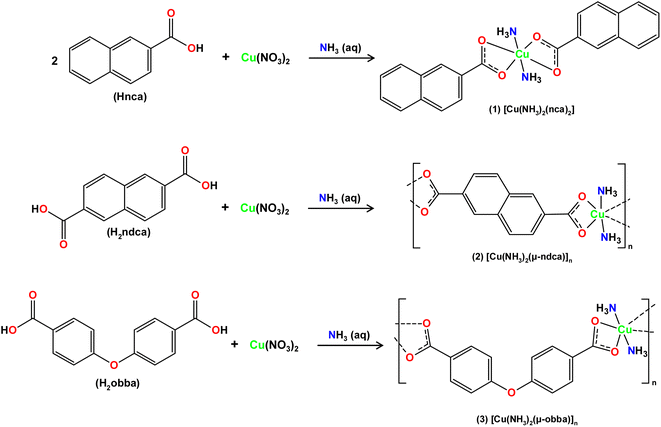
Aiming at the fabrication of new biomaterials and combining our interest in synthetic coordination chemistry, biopolymer materials and their antimicrobial applications,44,45 this study describes the preparation of biologically active copper(II) derivatives, and their use as doping agents for hybrid Cu-doped biopolymers on the basis of [PS]n or [PS-MCC]n matrices. Thus, three new coordination compounds, namely [Cu(NH3)2(nca)2] (1), [Cu(NH3)2(μ-ndca)]n (2), and [Cu(NH3)2(μ-obba)]n (3), were assembled from naphthoic (Hnca), 2,6-naphthalenedicarboxylic (H2ndca), or 4,4′-oxybis(benzoic) (H2obba) acids, in the presence of aqueous ammonia solution, and used to produce hybrid biopolymeric films, which were then tested for antibacterial performance.
The main features of the present study are: (1) the use of an inexpensive and biodegradable starch-based derived biopolymer as a matrix; (2) the preparation of biopolymer films with a low loading of bioactive compounds (5%); (3) the facile synthesis of new Cu(II) coordination compounds from simple and commercially available chemicals; (4) the use of different active building blocks (metal nodes and co-ligands); and (5) the good activity of the obtained biopolymeric films doped with Cu(II) compounds. Thus, the present interdisciplinary work combines various areas of research and broadens the antibacterial applications of Cu-based coordination compounds toward the fabrication of functional biopolymer materials.
Results and discussion
Synthesis of 1–3
Simple reactions in H2O between Cu(NO3)2 with aromatic mono-(Hnca) or dicarboxylic (H2ndca, H2obba) acids and aqueous ammonia resulted in the self-assembly of three new products: [Cu(NH3)2(nca)2] (1), [Cu(NH3)2(μ-ndca)]n (2), and [Cu(NH3)2(μ-obba)]n (3). The crystalline solids of 1–3 were analyzed by standard methods (FTIR spectroscopy, elemental and thermal analyses, powder and single-crystal X-ray diffraction). Products 1, 2, and 3 were obtained as air-stable crystalline solids with a mean particle size of 11, 27, and 24 μm, respectively. The compounds display the resembling [Cu(NH3)2]2+ entities that are linked by terminal (Hnca− in 1) or bridging (μ-ndca2− in 2, μ-obba2− in 3) carboxylate ligands, forming either discrete copper(II) monomers or one-dimensional coordination polymers, respectively. The three compounds (1–3, Scheme 1 and Fig. S1, ESI†) are generally insoluble (2, 3) or barely soluble in water, acetonitrile, ethanol, and methanol (1).Crystal structures of 1–3
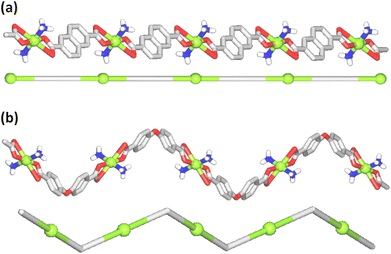
The structure of [Cu(NH3)2(nca)2] (1) reveals a monocopper(II) unit that is composed of a copper atom, two NH3 and two nca− ligands (Fig. 1a). The Cu(II) center is 6-coordinate and possesses an octahedral {CuN2O4} coordination geometry.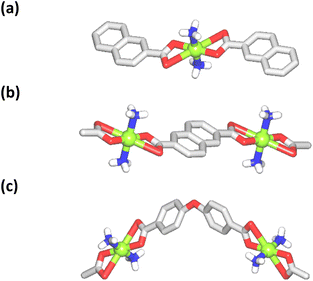 | ||
| Fig. 1 Crystal structure representations of 1 (a), 2 (b), and 3 (c). CH hydrogen atoms are omitted; Cu (green), O (red), N (blue), C (gray), H (white). | ||
It is formed by a pair of ammonia ligands in the axial sites [Cu–N 1.978(3) Å] and two naphthalenecarboxylate(1−) moieties in the equatorial positions [Cu–O 1.958(2) and 2.791(2) Å]. The carboxylate group of nca− shows a terminal bidentate mode with one considerably elongated Cu–O bond. The packing pattern of 1 reveals a 0D → 2D extension of the structure via intermolecular N–H⋯O hydrogen bonds, generating a 2D H-bonded net with an hxl topology.
The compound [Cu(NH3)2(μ-ndca)]n (2) features a linear 1D metal–organic chain structure (Fig. 1b and 2a) composed of the repeating [Cu(NH3)2]2+ entities and the μ-ndca2− linkers. The hexacoordinate Cu(II) centers exhibit an octahedral {CuN2O4} environment, which is formed by two NH3 ligands [Cu–N 1.984(3) Å] in the apical positions and four carboxylate O donors from two μ-ndca2− linkers in the equatorial sites [Cu–O 1.962(2) and 2.711(2) Å]. Both COO− groups of μ-ndca2− are bidentate with one Cu–O bond being considerably elongated. Linear 1D coordination polymer chains (Fig. 2a) show a 2C1 topology and an intrachain Cu⋯Cu separation of 12.966(2) Å. Extensive intermolecular hydrogen bonds (N–H⋯O) sew adjacent 1D chains into a 3D H-bonded network.
The structure of [Cu(NH3)2(μ-obba)]n (3) resembles that of 2 but features a wave-like type of 1D metal–organic chains (Fig. 1c and 2b) due to an angular geometry of the μ-obba2− linkers. These lead to longer Cu⋯Cu separations [14.320(2) Å] within the chain. The bonding parameters in 1–3 are typical for the present type of ligand environment in copper(II) coordination compounds.
TGA and PXRD
Thermogravimetric analysis (TGA) of 1 reveals a release of two NH3 ligands at 235–260 °C (mass loss calcd. 7.7%, exptl. 7.6%), followed by the decomposition of nca− at 260–434 °C (Fig. S2, ESI†). For bioCP 2, two coordinated NH3 moieties are lost within two coinciding thermal effects in the 235–321 °C interval (mass loss calcd. 9.6%, exptl. 9.8%). The next mass loss at 321–428 °C is due to the decomposition of ndca2−. In 3, the NH3 ligands are released during two overlapping steps in the temperature range of 243–315 °C (wt. loss calc. 10.8%, exptl. 10.7%), followed by the degradation of obba2− at 285–428 °C. For all compounds, the major decomposition product is CuO containing some minor amount of carbonous material (mass loss: 1: calcd. 18.2%, exptl. 22.2%; 2: calcd. 25.6%, exptl. 27.1%; 3: calcd. 22.6%, exptl. 25.4%). The experimental powder X-ray diffraction data are in good agreement with the patterns simulated from the CIF files (Fig. S3, ESI†).46Fabrication of hybrid Cu-doped biopolymeric films
These materials were prepared by adding small loadings of compounds 1–3 (5% wt.) to potato starch (PS) or its mixture with microcrystalline cellulose (PS-MCC) and glycerol. Then, the polymerization/curing was performed in Petri dishes to produce films with an ∼1 mm thickness (Scheme 2 and Fig. S4, ESI†). The obtained samples were named as: 1–3@[PS]n or 1–3@[PS-MCC]n (Scheme 2). Besides, the samples of [PS]n and [PS-MCC]n (negative controls) and CuO@[PS]n and CuO@[PS-MCC]n (positive controls) were also produced for comparative purposes. ATR-FTIR (Fig. S5–S13, ESI†) and SEM-EDX (Fig. S14–S16, ESI†) were used for the analysis of the biopolymeric films.Given the structures of starch, MCC, and studied compounds 1–3, the dominant intermolecular interactions between the Cu(II) dopants and biopolymer matrices will be through hydrogen bonding, established between the hydroxyl groups of the α-amylose of starch and the oxygen and nitrogen atoms present in copper(II) compounds. No formation of new covalent bonds is expected. In the course of biopolymer preparation, the gelatinization of the semi-crystalline granular structure (amylopectin) gives rise to a homogeneous and amorphous polymer matrix. The ATR-FTIR spectra of parent biopolymers and their doped versions remain practically the same, showing that the structure of the polymer films is not affected by the addition of the bioactive Cu-dopants.
Water absorption and copper release tests
The stability of biopolymeric films in phosphate buffered saline (PBS) and the absorption of water were examined (Tables S2 and S3, ESI†). After 4 h in PBS, water absorption for Cu-doped [PS]n samples was determined as a weight function, showing a weight increase by 32, 30, and 24% for 1, 2, and 3, respectively. When employing the [PS-MCC]n matrix, the increase in absorption is more pronounced (40% after 4 h for 1@[PS-MCC]n). In PBS medium, both kinds of biopolymers demonstrate increasing disaggregation over time, with a concomitant release of Cu2+ ions. Their amount in PBS after 24 h was measured, resulting in 0.38, 0.32, and 0.32 mg L−1 for 1, 2, and 3@[PS]n, respectively. This corresponds to a release of 28% of all copper present in 1@[PS]n, 15% in 2@[PS]n, and 19% in 3@[PS]n. Copper ion release is faster in biopolymeric films doped with CuO and discrete complex 1, and slower in films containing the coordination polymers 2 and 3 due to their increased stability and insolubility. When using the [PS-MCC]n matrix, the initial release of copper is also slow in samples doped with 1 or CuO. We would like to emphasize that the present coordination compounds do not need to show complete degradation to demonstrate antibacterial activity. The delayed release of copper is what makes these doped biopolymers efficient antibacterial materials, which is why the starch-based biopolymers that easily degrade in water were chosen. The water absorption assays also revealed differences between the films doped with the three Cu compounds, suggesting that there are interactions between the Cu compound and the biopolymer matrix; these interactions are eventually stronger in bioCPs 2 and 3 if compared to a discrete complex 1.Antibacterial activity
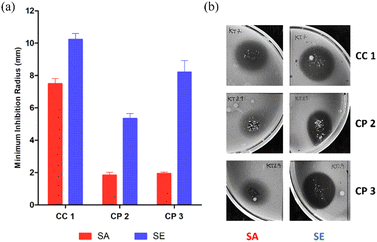
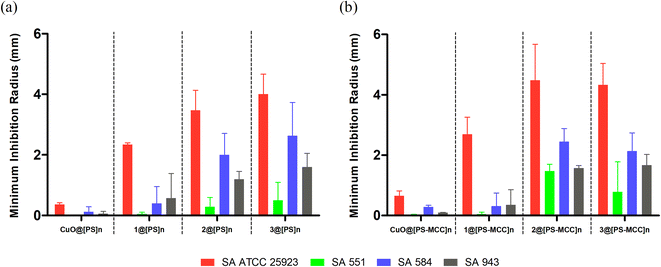
A preliminary assay with bioCC 1 and bioCPs 2 and 3 was performed on the reference strains of each bacterial type (Staphylococcus aureus ATCC 25923 and Staphylococcus epidermidis RP62A). As shown in Fig. 3, all tested compounds demonstrate some level of antimicrobial activity. S. epidermidis is the most susceptible strain as evidenced by the larger inhibition radius. The distinct susceptibilities of both species are especially noticeable for bioCPs 2 and 3, wherein the antimicrobial effect is somewhat minimal for S. aureus. Compound 1 features the highest antibacterial activity against both species, followed by 3 and then 2 for S. epidermidis.Following these preliminary results, we further explored the antimicrobial activity of the obtained Cu(II) compounds using clinical isolates of both species. The [PS]n and [PS-MCC]n biopolymers doped with compounds 2 or 3 present an antibacterial activity against all four strains of S. aureus (Fig. 4).
Not surprisingly, antibacterial effects are strain dependent, with the most susceptible strain being the reference strain, S. aureus ATCC 25923, followed by the clinical isolates 584, 943, and 551. Biopolymers doped with compound 1 are active against the reference strain but virtually inactive against the clinical isolates. While both [PS]n and [PS-MCC]n films that contain 2 and 3 are the most effective, all the films doped with 1–3 disclose higher effectiveness against S. aureus when compared with the control films doped with CuO. Contrary to the previous results in which 1 was the most active compound, films doped with this compound are the least effective among the samples tested. This lower activity of 1 could be related to obstacles in the Cu compound diffusion posed by both biopolymeric films or inferior stability of the complex. Regarding the differences between the two types of films, [PS-MCC]n seems to perform better when doped with 2 against the strains S. aureus ATCC 25923 and 551. Apart from this, both types of films resulted in generally similar outcomes.
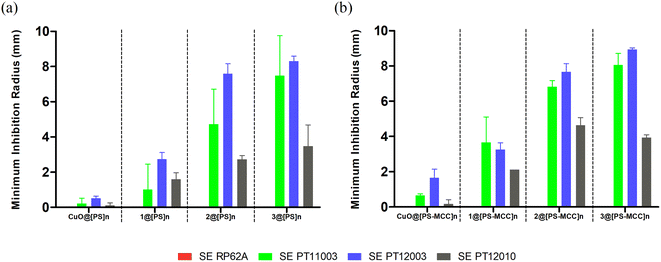
In turn, the antibacterial activity of both types of Cu-doped films against S. epidermidis was visible in three out of four strains (Fig. 5). Similarly to S. aureus, the activity of the films was strain dependent, with clinical isolates of S. epidermidis PT11003 and PT12003 being the most susceptible, followed by PT12010. The reference strain RP62A is not susceptible, contrary to what is observed for bioCPs in their pure form. This lack of activity could be related to obstacles in compound diffusion posed by both types of films. Nevertheless, doped films were overall more effective in inhibiting susceptible S. epidermidis strains compared to S. aureus, given the larger inhibition radius observed, which correlates with the data in Fig. 5. As observed previously, the activity of the CuO-doped control films was much lower compared to those doped with 1–3, thus highlighting an importance of the structures and ligands present in these compounds. Finally, the Cu-doped [PS-MCC]n films seem to perform better, especially for strain PT11003. However, as for S. aureus, this difference does not seem to be significant. A variety of Cu(II) coordination complexes and polymers with Cu–O and/or Cu–N coordination environments were reported as antibacterial compounds with substantial activity against the types of bacteria evaluated in this study (Table S4, ESI†). However, none of the reported examples contains ammonia ligands in their composition. A detailed and quantitative comparison of the observed activities is not possible due to differences in the concentration of antimicrobial components and the use of different assays for evaluating bioactivity.
The antimicrobial activity of copper ions is linked with cell membrane damage, oxidative stress, and DNA degradation, but some questions remain on exact mechanisms by which copper causes these phenomena. Resistance mechanisms to copper action have already been reported,21,47 namely for Gram-positive bacteria, which, along with diffusion issues, could partly explain the lack of susceptibility of some of the tested strains. Gram-positive bacteria can sequester free copper through a chaperone and extrude excess copper via a Cu-exporting ATPase. At high copper concentrations, CopY-like and CsoR-like repressors are released from the DNA to allow for the expression and transcriptions of ATPase and chaperone genes.21,47 Biopolymeric films doped with compounds 1, 2, and 3 revealed a higher effectiveness compared with CuO, probably due to a stronger stability and time-dependent release of copper ions to the medium, as well as a presence of NH3 ligands with potent antibacterial function.32,48 Since the presence of ammonia co-ligands makes bacteria more sensitive to antibiotics, NH3 may have a synergic effect with the copper metallic core of studied coordination compounds 1–3. This improvement in antimicrobial activity is promising and could pave the way for new copper-based antimicrobial biomaterials.
Experimental section
Synthesis and analytical data for 1–3
X-ray crystallography
Monocrystals of 1–3 were mounted in a cryoloop with Fomblin©. X-ray data collection was performed on a BRUKER D8 QUEST diffractometer (Mo Kα, λ = 0.7107 Å), and monitored using APEX3 (ref. 49) software. Corrections for Lorentzian polarization and effects of absorption were undertaken with SAINT50 and SADABS.51 SHELXT 2014/4 (ref. 52) was used for solving structures while SHELXL 2014/7 (ref. 52) was utilized for complete refinement. The software is present in a WINGX-Version 2014.1 program pack53 Anisotropic refinement was applied for all Cu, N, C, and O atoms. Hydrogen atoms bonded to C centers were introduced to idealized locations and refined at the parent carbon atoms. H atoms of NH3 ligands were identified using electron density maps. Crystal data for 1–3 are reported in Table S1 (ESI).† CCDC 2177365–2177367.Bacterial growth conditions and antibacterial assays
Two different types of bacteria (Gram-positive), namely S. epidermidis and S. aureus, were used in this study due to their prominent role in biomaterial related infections.54 A total of four strains were tested for each species, contemplating one collection strain and three clinical isolates (collection of CEB-University of Minho). Growth was performed under aerobic conditions with agitation (120 rpm) at 37 °C. MHA II (Mueller–Hinton agar II) and MHB II (Mueller–Hinton broth II) (Liofilchem) were selected as growth media for antibacterial assays. Antibacterial activity of compounds 1–3 and all biopolymeric films was studied using a soft agar overlay assay, as described previously (for details, see ESI†).43 Normalization of the obtained data for the molar content of copper(II) was performed. For non-normalized results, see Fig. S17–S19 (ESI†).Conclusions
Three new copper(II) coordination compounds 1–3 bearing ammonia and aromatic carboxylate ligands were self-assembled and fully characterized, revealing discrete monocopper(II) units in 1 or 1D coordination polymer chains in 2 and 3 with resembling types of hexacoordinate Cu(II) centers and octahedral {CuN2O4} environments. These Cu(NH3)2-carboxylate compounds were used as antibacterial dopants to produce hybrid Cu-doped biopolymeric films based on potato starch (PS) or its combination with microcellulose (MCC). Their antibacterial properties were investigated against S. aureus and S. epidermidis bacteria, including clinically isolated strains. Overall, all compounds showed a notable antibacterial activity, with doped films inhibiting in varying degrees 7 out of 8 Gram-positive bacterial strains tested. The activity of the bioCPs 2 and 3 was far superior if compared to that of CuO, with 2 and 3 being more effective when incorporated into [PS]n or [PS-MCC]n films. There was no obvious difference between antibacterial activities of matrices based on [PS]n and [PS-MCC]n. As the presence of ammonia, as the co-ligands in the obtained copper(II) compounds, is known to make bacteria more sensitive to antibiotics, the results obtained could indicate a synergic effect between NH3 and Cu.In summary, the present work broadens the types of bioactive copper(II) complexes and coordination polymers,55–57 showing that such compounds can be easily assembled and embedded into degradable biopolymeric matrices, acting as promising antibacterial materials. This work may thus open up new opportunities for designing sustainable biopolymer-based films, coatings, or composites with antibacterial properties.32–35,58,59
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been supported by the Foundation for Science and Technology (FCT) (projects PTDC/QUI-QIN/29697/2017, LISBOA-01-0145-FEDER-029697, PTDC/QUI-QIN/3898/2020, LA/P/0056/2020, UIDB/00100/2020, and UIDP/00100/2020, and contracts CEECIND/02725/2018, CEECIND/00283/2018, CEECIND/00194/2020, and CEECIND/03708/2017), and IPL (IPL/2020/HyBioPol). We thank Eng. Mário Dias (LAIST) for ICP-OES experimental assistance and Sonia Mendes for assistance with graphical design.References
- U. Theuretzbacher, K. Outterson, A. Engel and A. Karlén, The Global Preclinical Antibacterial Pipeline, Nat. Rev. Microbiol., 2020, 18, 275 CrossRef PubMed
.
- J. A. Perry, E. L. Westman and G. D. Wright, The Antibiotic Resistome: What's New?, Curr. Opin. Microbiol., 2014, 21, 45–50 CrossRef CAS PubMed
.
- C. J. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. Robles Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. Kashef Hamadani, E. A. P. Kumaran, B. McManigal, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. De Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, M. Khorana, N. Kissoon, E. Kobeissi, T. Kostyanev, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, N. Mturi, T. Munera-Huertas, P. Musicha, M. M. Mussi-Pinhata, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, A. Y. Peleg, C. Perrone, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, K. E. Rudd, N. Russell, J. Schnall, J. A. G. Scott, M. Shivamallappa, J. Sifuentes-Osornio, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Turner, P. Turner, H. R. van Doorn, S. Velaphi, A. Vongpradith, H. Vu, T. Walsh, S. Waner, T. Wangrangsimakul, T. Wozniak, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Global Burden of Bacterial Antimicrobial Resistance in 2019: a Systematic Analysis, The Lancet, 2022, 399(10325), 629 CrossRef CAS PubMed
.
- P. Jorge, A. P. Magalhães, T. Grainha, D. Alves, A. M. Sousa, S. P. Lopes and M. O. Pereira, Antimicrobial Resistance Three Ways: Healthcare Crisis, Major Concepts and the Relevance of Biofilms, FEMS Microbiol. Ecol., 2019, 95, fiz115 CrossRef CAS PubMed
.
- D. Ş. Karaman, U. K. Ercan, E. Bakay, N. Topaloğlu and J. M. Rosenholm, Evolving Technologies and Strategies for Combating Antibacterial Resistance in the Advent of the Postantibiotic Era, Adv. Funct. Mater., 2020, 30, 1908783 CrossRef
.
- A. Friedlander, S. Nir, M. Reches and M. Shemesh, Preventing Biofilm Formation by Dairy-Associated Bacteria Using Peptide-Coated Surfaces, Front. Microbiol., 2019, 10, 1405 CrossRef PubMed
.
- X. Li, B. Wu, H. Chen, K. Nan, Y. Jin, L. Sun and B. Wang, Recent Developments in Smart Antibacterial Surfaces to Inhibit Biofilm Formation and Bacterial Infections, J. Mater. Chem. B, 2018, 6, 4274 RSC
.
- C. Pettinari, R. Pettinari, C. Di Nicola, A. Tombesi, S. Scuri and F. Marchetti, Antimicrobial MOFs, Coord. Chem. Rev., 2021, 446, 214121 CrossRef CAS
.
- M. Zhang, Z. Y. Gu, M. Bosch, Z. Perry and H. C. Zhou, Biomimicry in metal–organic materials, Coord. Chem. Rev., 2015, 293, 327 CrossRef
.
- G. Wyszogrodzka, B. Marszałek, B. Gil and P. Dorożyński, Metal-organic Frameworks: Mechanisms of Antibacterial Action and Potential Applications, Drug Discovery Today, 2016, 21, 1009 CrossRef CAS PubMed
.
- P. L. Wang, L. H. Xie, E. A. Joseph, J. R. Li, X. O. Su and H. C. Zhou, Metal–organic frameworks for food safety, Chem. Rev., 2019, 119, 10638–10690 CrossRef CAS PubMed
.
- W. Zhuang, D. Yuan, J. R. Li, Z. Luo, H. C. Zhou, S. Bashir and J. Liu, Highly potent bactericidal activity of porous metal-organic frameworks, Adv. Healthcare Mater., 2012, 1, 225 CrossRef CAS
.
-
Agency for Toxic Substances and Disease Registry, Toxicological Profile for Copper, Atlanta, GA, 2004 Search PubMed
.
- M. L. Ermini and V. Voliani, Antimicrobial Nano-Agents: The Copper Age, ACS Nano, 2021, 15, 6008 CrossRef CAS PubMed
.
- S. Zehra, S. Tabassum and F. Arjmand, Biochemical Pathways of Copper Complexes: Progress Over the Past 5 Years, Drug Discovery Today, 2021, 26, 1086 CrossRef CAS PubMed
.
- F. Lebon, N. Boggetto, M. Ledecq, F. Durant, Z. Benatallah, S. Sicsic, R. Lapouyade, O. Kahn, A. Mouithys-Mickalad, G. Deby-Dupont and M. Reboud-Ravaux, Metal-Organic Compounds: A New Approach for Drug Discovery: N1-(4-Methyl-2-Pyridyl)-2,3,6-Trimethoxybenzamide Copper(II) Complex as an Inhibitor of Human Immunodeficiency Virus 1 Protease, Biochem. Pharmacol., 2002, 63, 1863 CrossRef CAS PubMed
.
- G. Psomas, Copper(II) and Zinc(II) Coordination Compounds of Non-Steroidal Anti-Inflammatory Drugs: Structural Features and Antioxidant Activity, Coord. Chem. Rev., 2020, 412, 213259 CrossRef CAS
.
-
(a) M. Claudel, J. V. Schwarte and K. M. Fromm, New Antimicrobial Strategies Based on Metal Complexes, Chemistry, 2020, 2, 849 CrossRef CAS
; (b) H. N. Rubin, B. H. Neufeld and M. M. Reynolds, Surface-Anchored Metal-Organic Framework-Cotton Material for Tunable Antibacterial Copper Delivery, ACS Appl. Mater. Interfaces, 2018, 10, 15189 CrossRef CAS PubMed
; (c) B. H. Neufeld, M. J. Neufeld, A. Lutzke, S. M. Schweickart and M. M. Reynolds, Metal–Organic Framework Material Inhibits Biofilm Formation of Pseudomonas aeruginosa, Adv. Funct. Mater., 2017, 27, 1702255 CrossRef
.
-
(a) D. Mitra, E. T. Kang and K. G. Neoh, Antimicrobial Copper-Based Materials and Coatings: Potential Multifaceted Biomedical Applications, ACS Appl. Mater. Interfaces, 2019, 12, 21159 CrossRef PubMed
; (b) D. Fu, S. Yang, J. Lu, H. Lian and K. Qin, Two Cu(II) Coordination Polymers: Treatment Activity on Spine Surgery Incision Infection by Inhibiting the Staphylococcus aureus Biofilm Formation, J. Cluster Sci., 2022, 33, 529 CrossRef CAS
; (c) S. S. Batool, S. R. Gilani, S. S. Zainab, M. N. Tahir, W. T. A. Harrison, M. Salman Haider, Q. Syed, S. Mazhar and M. Shoaib, Synthesis, crystal structure, thermal studies and antimicrobial activity of a new chelate complex of copper(II) succinate with N,N,N',N'-tetramethylethylenediamine, J. Coord. Chem., 2020, 73, 1778 CrossRef CAS
.
- S. He, Y. Feng, Q. Sun, Z. Xu and W. Zhang, Charge-Switchable CuxO Nanozyme with Peroxidase and Near-Infrared Light Enhanced Photothermal Activity for Wound Antibacterial Application, ACS Appl. Mater. Interfaces, 2022, 14, 25042 CrossRef CAS PubMed
.
- M. Vincent, R. Duval, P. Hartemann and M. Engels-Deutsch, J. Appl. Microbiol., 2018, 124, 1032 CrossRef CAS PubMed
.
- L. P. Arendsen, R. Thakar and A. H. Sultan, The Use of Copper as an Antimicrobial Agent in Health Care, Including Obstetrics and Gynecology, Clin. Microbiol. Rev., 2019, 32, e00125 CrossRef CAS PubMed
.
- Y. Liu, N. Nie, H. Tang, C. Zhang, K. Chen, W. Wang and J. Liu, Effective Antibacterial Activity of Degradable Copper-Doped Phosphate-Based Glass Nanozymes, ACS Appl. Mater. Interfaces, 2021, 13, 11631 CrossRef CAS PubMed
.
- C. Spagnul, L. C. Turner, F. Giuntini, J. Greenman and R. W. Boyle, Synthesis and bactericidal properties of porphyrins immobilized in a polyacrylamide support: influence of metal complexation on photoactivity, J. Mater. Chem. B, 2017, 5, 1834 RSC
.
- J. R. J. Sorenson, Copper-Complexes in Biochemistry and Pharmacology, Chem. Br., 1984, 16, 1110 Search PubMed
.
- A. E. Liberta and D. X. West, Antifungal and Antitumor Activity of Heterocyclic Thiosemicarbazones and their Metal Complexes: Current Status, BioMetals, 1992, 5, 121 CrossRef CAS PubMed
.
- B. Đ. Glišić, I. Aleksic, P. Comba, H. Wadepohl, T. Ilic-Tomic, J. Nikodinovic-Runic and M. I. Djuran, Copper(II) complexes with aromatic nitrogen-containing heterocycles as effective inhibitors of quorum sensing activity in Pseudomonas aeruginosa, RSC Adv., 2016, 6, 86695 RSC
.
- A. H. Totten, C. L. Crawford, A. G. Dalecki, L. Xiao, F. Wolschendorf and T. P. Atkinson, Differential Susceptibility of Mycoplasma and Ureaplasma Species to Compound-Enhanced Copper Toxicity, Front. Microbiol., 2019, 10, 1720 CrossRef PubMed
.
- R. J. Sorenson, Copper Complexes Offer a Physiological Approach to Treatment of Chronic Diseases, Prog. Med. Chem., 1989, 26, 437 CrossRef PubMed
.
-
(a) P. Wang, Y. Yuan, K. Xu, H. Zhong, Y. Yang, S. Jin, K. Yang and X. Qi, Biological Applications of Copper-Containing Materials, Bioact. Mater., 2021, 6, 916 CrossRef CAS PubMed
; (b) A. K. Srivastava, S. K. Singh and A. Srivastava, Spectral, biological antimicrobial activity (in vitro) and effect of solvents on the electrochemical behavior of four newly synthesized copper(II) complexes with 4-hydroxynicotinic acid containing 2,2′-bipyridine/5,5′-Me2-bipy/1,10-phenanthroline/DMP, Chem. Data Collect., 2020, 26, 100357 CrossRef CAS
.
- O. Krasnovskaya, A. Naumov, D. Guk, P. Gorelkin, A. Erofeev, E. Beloglazkina and A. Majouga, Copper Coordination Compounds as Biologically Active Agents, Int. J. Mol. Sci., 2020, 21, 3965 CrossRef CAS PubMed
.
- A. Pinon, S. Decherf, G. Malet, S. Cupferman and M. Vialette, Bactericidal Activity of Ammonia and Monoethanolamine on Pseudomonas Aeruginosa and Staphylococcus Aureus Strains of Various Origins, Int. J. Cosmet. Sci., 2015, 37, 207 CrossRef CAS PubMed
.
- A. Sultana, A. Kathuria and K. K. Gaikwad, Metal–Organic
Frameworks for Active Food Packaging. A Review, Environ. Chem. Lett., 2022, 20, 1479 CrossRef CAS PubMed
.
- J. Salvo and C. Sandoval, Role of copper nanoparticles in wound healing for chronic wounds: literature review, Burns Trauma, 2022, 10, tkab047 CrossRef PubMed
.
- M. Cruz-Romero and J. Kerry, Crop-based Biodegradable Packaging and its Environmental Implications, CAB Rev., 2008, 3, 1 Search PubMed
.
- J. P. Maran, V. Sivakumar, R. Sridhar and V. P. Immanuel, Development of model for mechanical properties of tapioca starch based edible films, Ind. Crops Prod., 2013, 42, 159 CrossRef
.
-
S. Pilla, Engineering Applications of Bioplastics and Biocomposites – An Overview, in Handbook of Bioplastics and Biocomposites Engineering Applications, Scrivener Publishing, LLC, 2011, pp. 1–15 Search PubMed
.
- A. Podshivalov, M. Zakharova, E. Glazacheva and M. Uspenskaya, Gelatin/Potato Starch Edible Biocomposite Films: Correlation Between Morphology and Physical Properties, Carbohydr. Polym., 2017, 157, 1162 CrossRef CAS PubMed
.
- S. Singha, M. Mahmutovic, C. Zamalloa, L. Stragier, W. Verstraete, A. J. Svagan, O. Das and M. S. Hedenqvist, Novel Bioplastic from Single Cell Protein as a Potential Packaging Material, ACS Sustainable Chem. Eng., 2021, 9, 6337 CrossRef CAS
.
- H. M. Fahmy, R. E. S. Eldin, E. S. A. Serea, N. M. Gomaa, G. M. AboElmagd, S. A. Salem, Z. A. Elsayed, A. Edrees, E. Shams-Eldin and A. E. Shalan, Advances in nanotechnology and antibacterial properties of biodegradable food packaging materials, RSC Adv., 2020, 10, 20467 RSC
.
- A. H. D. Abdullah, S. Pudjiraharti, M. Karina, O. D. Putri and R. H. Fauziyyah, Potato Starch-based Bioplastics Plasticized with Glycerol, J. Biol. Sci., 2019, 19, 57 CAS
.
-
G. Moore, Non Wood Fibre Applications in Papermaking, Pira International, London, 1996 Search PubMed
.
- M. N. Waswa, A. S. Merenga and C. M. Migwi, Thermal and Diffusion Characterization of High Density Polyethylene/Cellulose Blend Inoculated with Aspergillus Niger Fungus, Int. J. Sci. Technol., 2014, 3, 320 Search PubMed
.
- T. A. Fernandes, I. F. M. Costa, P. Jorge, A. C. Sousa, V. André, N. Cerca and A. M. Kirillov, ACS Appl. Mater. Interfaces, 2021, 13, 12836 CrossRef CAS PubMed
.
- T. A. Fernandes, I. F. M. Costa, P. Jorge, A. C. Sousa, R. G. Cabral, V. André, N. Cerca and A. M. Kirillov, Hybrid Silver(I)-Doped Soybean Oil and Potato Starch Biopolymer Films to Combat Bacterial Biofilms, ACS Appl. Mater. Interfaces, 2022, 14, 25104 CrossRef CAS PubMed
.
- C. F. Macrae, I. Sovago, S. J. Cottrell, P. T. A. Galek, P. McCabe, E. Pidcock, M. Platings, G. P. Shields, J. S. Stevens, M. Towler and P. A. Wood, Mercury 4.0: From Visualization to Analysis, Design and Prediction, J. Appl. Crystallogr., 2020, 53, 226 CrossRef CAS PubMed
.
- S. Mathews, M. Hans, F. Mücklich and M. Solioz, Contact Killing of Bacteria on Copper is Suppressed if Bacterial-Metal Contact is Prevented and is Induced on Iron by Copper Ions, Appl. Environ. Microbiol., 2013, 79, 2605 CrossRef CAS PubMed
.
- M. Avalos, P. Garbeva, J. M. Raaijmakers and G. P. van Wezel, Production of Ammonia as a Low-Cost and Long-Distance Antibiotic Strategy by Streptomyces Species, ISME J., 2020, 14, 569 CrossRef CAS PubMed
.
-
APEX3, Ver. 2017.3-0, Bruker AXS Inc., Madison, Wisconsin, USA, 2017 Search PubMed
.
-
Bruker AXS: SAINT+, Release 6.22, Bruker Analytical Systems, Madison,WI, 2005 Search PubMed
.
-
Bruker AXS: SADABS, Bruker Analytical Systems, Madison, WI 2005 Search PubMed
.
- G. M. Sheldrick, SHELXT – Integrated Space-Group and Crystal-Structure Determination, Acta Crystallogr., 2015, A71, 3 CrossRef PubMed
.
- L. J. Farrugia, WinGX Suite for Small-Molecule
Single-Crystal Crystallography, J. Appl. Crystallogr., 1999, 32, 837 CrossRef CAS
.
- W. F. Oliveira, P. M. S. Silva, R. C. S. Silva, G. M. M. Silva, G. Machado, L. C. B. B. Coelho and M. T. S. Correia, Staphylococcus Aureus and Staphylococcus Epidermidis Infections on Implants, J. Hosp. Infect., 2018, 98, 111 CrossRef CAS PubMed
.
- J. Deng, C. Peng, L. Hou, Y. Wu, W. Liu, G. Fang, H. Jiang, S. Qin, F. Yang, G. Huang and Y. Gou, Dithiocarbazate-copper complex loaded thermosensitive hydrogel for lung cancer therapy via tumor in situ sustained-release, Inorg. Chem. Front., 2022, 9, 4741 RSC
.
- J. Deng, C. Peng, L. Hou, Y. Wu, W. Liu, G. Fang, H. Jiang, S. Qin, F. Yang, G. Huang and Y. Gou, Dithiocarbazate-copper complex loaded thermosensitive hydrogel for lung cancer therapy via tumor in situ sustained-release, Inorg. Chem. Front., 2022, 9, 6190 RSC
.
- G. Mazzone, E. Sicilia, E. I. Szerb, M. La Deda, L. Ricciardi, E. Furia, B. Sanz Mendiguchia, F. Scarpelli, A. Crispini and I. Aiello, Heteroleptic Cu(II) saccharin complexes: intriguing coordination modes and properties, Inorg. Chem. Front., 2021, 8, 3342 RSC
.
- Z. Huang, D. Zhang, Q. Gu, J. Miao, X. Cen, R. P. Golodok, V. V. Savich, A. P. Ilyushchenko, Z. Zhou and R. Wang, One-step coordination of metal–phenolic networks as antibacterial coatings with sustainable and controllable copper release for urinary catheter applications, RSC Adv., 2022, 12, 15685 RSC
.
- R. Kaur, N. S. Thakur, S. Chandna and J. Bhaumik, Sustainable Lignin-Based Coatings Doped with Titanium Dioxide Nanocomposites Exhibit Synergistic Microbicidal and UV-Blocking Performance toward Personal Protective Equipment, ACS Sustainable Chem. Eng., 2021, 9, 11223 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, antibacterial assays, microscope images of crystalline samples (Fig. S1) and crystal data (Table S1) of 1–3; TGA data (Fig. S2); PXRD patterns (Fig. S3); coupons of [PS]n and [PS-MCC]n biopolymeric films (Fig. S4); ATR-FTIR spectra and discussion (Fig. S5–S13), SEM-EDS data and discussion (Fig. S14–S16); water absorption data (Table S2); Cu release tests (Table S3); additional antibacterial activity data (Fig. S17–S19 and Table S4). CCDC 2177365–2177367. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2su00150k |
| This journal is © The Royal Society of Chemistry 2023 |