Open Access Article
Open Access ArticleHierarchical covalent organic framework-foam for multi-enzyme tandem catalysis†
Satyadip
Paul
 ab,
Mani
Gupta
ab,
Mani
Gupta
 c,
Kaushik
Dey
c,
Kaushik
Dey
 ab,
Ashok Kumar
Mahato
ab,
Ashok Kumar
Mahato
 ab,
Saikat
Bag
ab,
Saikat
Bag
 ab,
Arun
Torris
ab,
Arun
Torris
 d,
E. Bhoje
Gowd
d,
E. Bhoje
Gowd
 e,
Hasnain
Sajid
e,
Hasnain
Sajid
 f,
Matthew A.
Addicoat
f,
Matthew A.
Addicoat
 f,
Supratim
Datta
f,
Supratim
Datta
 *c and
Rahul
Banerjee
*c and
Rahul
Banerjee
 *ab
*ab
aDepartment of Chemical Sciences, Indian Institute of Science Education and Research, Mohanpur, Kolkata, 741246, India
bCentre for Advanced Functional Materials, Indian Institute of Science Education and Research, Mohanpur, Kolkata, 741246, India. E-mail: r.banerjee@iiserkol.ac.in
cDepartment of Biological Sciences, Center for the Climate and Environmental Sciences, Indian Institute of Science Education and Research Kolkata, Mohanpur 741246, India. E-mail: supratim@iiserkol.ac.in
dPolymer Science and Engineering Division, CSIR-National Chemical Laboratory, Dr Homi Bhabha Road, Pune 411008, India
eMaterials Science and Technology Division, CSIR-National Institute for Interdisciplinary Science and Technology, Trivandrum 695 019, Kerala, India
fSchool of Science and Technology, Nottingham Trent University, NG11 8NS Nottingham, UK
First published on 26th May 2023
Abstract
Covalent organic frameworks (COFs) are ideal host matrices for biomolecule immobilization and biocatalysis due to their high porosity, various functionalities, and structural robustness. However, the porosity of COFs is limited to the micropore dimension, which restricts the immobilization of enzymes with large volumes and obstructs substrate flow during enzyme catalysis. A hierarchical 3D nanostructure possessing micro-, meso-, and macroporosity could be a beneficial host matrix for such enzyme catalysis. In this study, we employed an in situ CO2 gas effervescence technique to induce disordered macropores in the ordered 2D COF nanostructure, synthesizing hierarchical TpAzo COF-foam. The resulting TpAzo foam matrix facilitates the immobilization of multiple enzymes with higher immobilization efficiency (approximately 1.5 to 4-fold) than the COF. The immobilized cellulolytic enzymes, namely β-glucosidase (BGL), cellobiohydrolase (CBH), and endoglucanase (EG), remain active inside the TpAzo foam. The immobilized BGL exhibited activity in organic solvents and stability at room temperature (25 °C). The enzyme-immobilized TpAzo foam exhibited significant activity towards the hydrolysis of p-nitrophenyl-β-D-glucopyranoside (BGL@TpAzo-foam: Km and Vmax = 23.5 ± 3.5 mM and 497.7 ± 28.0 μM min−1) and carboxymethylcellulose (CBH@TpAzo-foam: Km and Vmax = 18.3 ± 4.0 mg mL−1 and 85.2 ± 9.6 μM min−1 and EG@TpAzo-foam: Km and Vmax = 13.2 ± 2.0 mg mL−1 and 102.2 ± 7.1 μM min−1). Subsequently, the multi-enzyme immobilized TpAzo foams were utilized to perform a one-pot tandem conversion from carboxymethylcellulose (CMC) to glucose with high recyclability (10 cycles). This work opens up the possibility of synthesizing enzymes immobilized in TpAzo foam for tandem catalysis.
Introduction
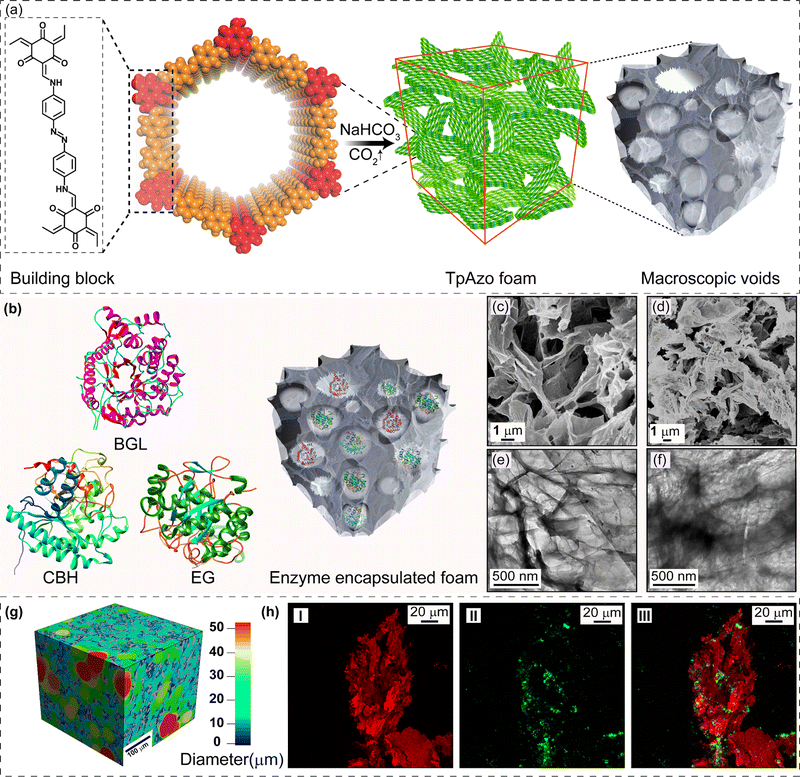
Biocatalysis is becoming an increasingly popular complementary tool to traditional chemosynthesis.1 Multi-enzyme tandem bio-catalysis, which involves sequential steps without isolating intermediates, has garnered attention for its broad range of organic synthesis applications.2,3 However, many naturally occurring enzymes are incompatible with high temperatures, pH, and organic solvents, hindering their industrial applicability.4 Porous materials show promise as enzyme immobilization supports by providing mass transfer channels and conformational constraints. Immobilizing enzymes in porous matrices enhances their stability, simplifies separation, and enables recyclability during multi-enzyme tandem catalysis.5 Porous materials, such as mesoporous silica, metal–organic frameworks (MOFs), hydrogen-bonded organic frameworks (HOFs), and covalent organic frameworks (COFs), have been explored as host matrices for enzyme immobilization.6–8 These crystalline nanostructures must either have sufficient void space or a significant number of defects to enable interactions between large substrates and enzymes. The host matrix's void space size should accommodate multiple enzymes for tandem and synergistic biocatalytic transformations. In the last decade, there has been growing interest in crystalline porous materials, particularly covalent organic frameworks (COFs), due to their high surface area, tunable pore size, stability, and easily tailorable functionality.9 These properties make COFs ideal support for enzyme immobilization. Biomolecules can be anchored in the micro and mesopores of COFs by physical adsorption, and the interconnectivity among the intrinsic micropores facilitates mass transfer at an enhanced rate.10 However, physically adsorbed enzymes may suffer from leaching during the catalytic process, and the uniform microporous nature of COFs restricts the immobilization of enzymes with large volumes. In this study, we propose immobilizing enzymes in a new class of solid-state materials: covalent organic framework foams (COF-foams).11 COF-foams are hierarchical porous structures where micro, meso, and macropores coexist within the ordered COF crystallites. The presence of macropores makes these COF foams more suitable for hosting enzymes with large volumes than conventional COF microcrystals. These COF foams offer a large pore volume, enabling high enzyme adsorption capacity and diffusion through the entire network, enhancing the catalytic efficiency.Results and discussion
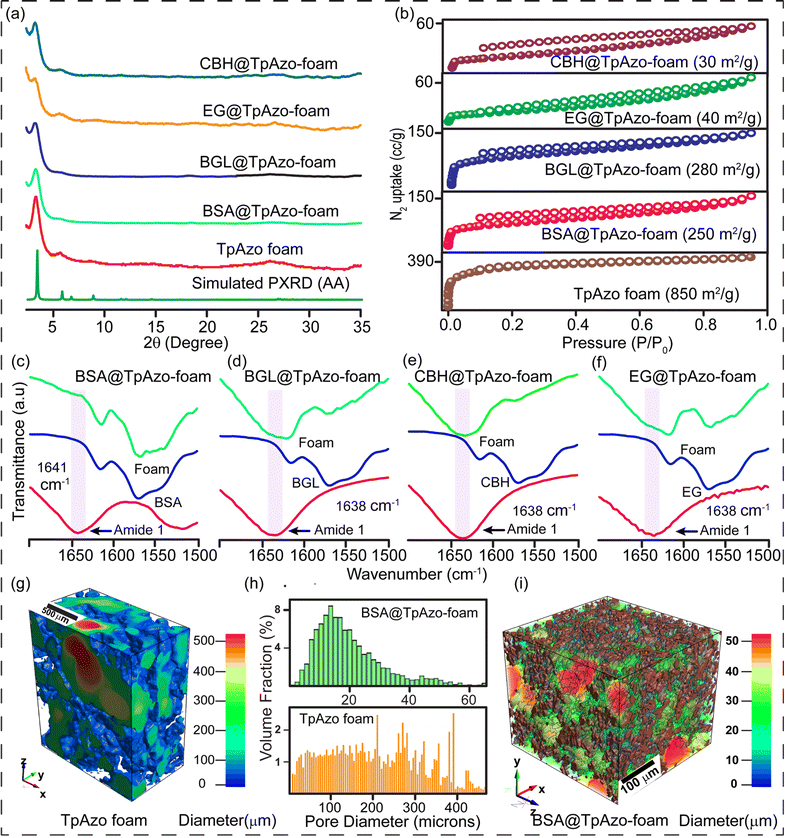
We have synthesized a TpAzo foam using 1,3,5-triformylphloroglucinol (Tp) and 4,4′-azodianiline (Azo), and have immobilized enzymes within the foam using a concentration-dependent adsorption technique. The TpAzo foam was synthesized by adding 0.45 mmol (95.5 mg) of 4,4′-azodianiline (Azo) to 11.6 mmol (2 g) of p-toluenesulfonic acid (PTSA), followed by adding 0.3 mmol (63 mg) of 1,3,5-triformylphloroglucinol (Tp) and 100 μL of water to the mixture. Next, 6 mmol (500 mg) of sodium bicarbonate (NaHCO3) was added to the mixture, resulting in situ CO2 effervescence that turned the paste into a fluffy substance with macropores. The resulting material was freeze-dried, followed by heating at 90 °C for 12 h in a closed condition. The TpAzo foam was washed with dimethylacetamide (DMAC) and water and dried at 120 °C for 12 h. The resulting foam showed a hierarchical nanostructure with macropores and COF nanosheets, as revealed by SEM and TEM images. SEM images reveal that foam is composed of COF nanosheets (1–5 μm) and extrinsic pores ranging from 1–10 μm (Fig. 1c and S8†). TEM images of TpAzo foam also show the random disorder within the architecture where the pores of diameter 100–200 μm are intertwined with the COF crystallites (Fig. 1e and S11†). The immobilization of enzymes onto the TpAzo foam was achieved by a concentration-dependent adsorption technique. The foam was immersed in an enzyme solution of desired concentration, and the mixture was allowed to shake for a specific time to achieve maximum adsorption of enzymes onto the foam. The foam was then washed thoroughly with water to remove unbound enzymes, and the immobilized enzymes were characterized using Fourier-transform infrared spectroscopy (FTIR). The FTIR spectra showed the characteristic peaks of enzymes, indicating successful immobilization onto the TpAzo foam. The immobilized enzymes retained their catalytic activity and stability for up to ten cycles of reuse.The crystallinity of the TpAzo foam was confirmed through powder X-ray diffraction (PXRD), which matched well with the simulated PXRD of the pristine TpAzo COF. The peak observed at 3.3° (2θ) corresponds to a reflection from (100) planes, while the broad peak at a higher angle (2θ = 26.7°) is attributed to the reflections from (001) planes. This also signifies the π–π interaction between the COF layers in the foam matrix (Fig. 2a). To check the microporosity of TpAzo foam, we measured the N2 adsorption isotherm at 77 K, which showed a Type-I adsorption isotherm. The BET surface area and total pore volume of TpAzo foam were found to be 850 m2 g−1 and 0.594 cm3 g−1, respectively. The pore size distribution of the TpAzo foam was checked using non-local density functional theory (NLDFT), indicating an average pore size distribution centered at 2 nm (Fig. 2b and S16†). FT-IR spectra revealed intense peaks at 1619 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1567 (C
O), 1567 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), and 1228 cm−1 (C–N), confirming the β-ketoenamine framework of the foam matrix (Fig. S17†). X-ray micro-computed tomography (micro-CT) was performed to reveal the 3D macrostructure of the TpAzo foam with interconnected pores (Fig. 2g). Within the macro-structure, we were able to isolate the three-dimensional assembly of disordered macropores (5–200 μm) and ordered lamellar COF sheets (up to 200 μm) (Fig. S19 and S20†). The void volume of the foam is approximately 75%. We could control the overall macropore size distribution (up to 400 μm) by controlling the amount of sodium bicarbonate addition. As the macropores were generated stepwise, the pore size distribution exhibited a broad range with a higher volume fraction (Fig. 2h and S21†).
C), and 1228 cm−1 (C–N), confirming the β-ketoenamine framework of the foam matrix (Fig. S17†). X-ray micro-computed tomography (micro-CT) was performed to reveal the 3D macrostructure of the TpAzo foam with interconnected pores (Fig. 2g). Within the macro-structure, we were able to isolate the three-dimensional assembly of disordered macropores (5–200 μm) and ordered lamellar COF sheets (up to 200 μm) (Fig. S19 and S20†). The void volume of the foam is approximately 75%. We could control the overall macropore size distribution (up to 400 μm) by controlling the amount of sodium bicarbonate addition. As the macropores were generated stepwise, the pore size distribution exhibited a broad range with a higher volume fraction (Fig. 2h and S21†).
We performed mass transport simulation in the 3D micro-porous structure to predict the dynamic events occurring during the entrapment of enzymes. A numerical simulator computed digital water flow experiments in all three directions (x, y, and z-axis), using an appropriate solver at 25 °C and 20 Pa pressure drop. The outcome visualized the average flow velocity profiles. The numerical simulation results predict the feasible average water flow profiles through the interconnected pore structure of the foam. The average water flow velocities in the x, y, and z directions were 6.10, 4.62, and 4.89 × 10−3 m s−1, respectively. The significant enhancement of momentum in the x-axis relative to other axes may be attributed to receptivity to flow exerted by the pore walls aligned normal to the x-axis. A higher population of flow velocity streamlines in the x-axis also supports this observation. The results also signify the interconnectivity of the pores in the foam, which should facilitate the transport of enzymes along the x-axis (Fig. S22†).
To investigate the immobilization of enzymes or proteins in TpAzo foam, we used bovine serum albumin (BSA) as a model protein. We determined the loading percentage to be 0.09 mg of BSA per 1 mg of TpAzo foam using UV-vis spectroscopy (Fig. S3†). We rinsed the solid foam 10–15 times with water to remove any BSA that was loosely bound to the foam surface, and we kept a sample for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). We then incubated the BSA@TpAzo-foam samples in water for 24 hours to check for leaching of BSA, and we collected the supernatant after centrifuging the solution. SDS-PAGE showed no band of BSA after the second round of washing of the immobilized foam. The supernatant component of the washed BSA immobilized foam showed no BSA band after being incubated in water for 24 hours (Fig. S6†). These results confirmed the absence of any BSA leaching from the TpAzo foam after a 24 hours incubation in water. We followed a similar method for immobilization of the cellulolytic enzymes β-glucosidase (BGL), cellobiohydrolase (CBH), and endoglucanase (EG). To compute the surface area and volume of the three enzymes, we placed the protein data bank (PDB) structures in a 100 Å vacuum box. We calculated the accessible surface area (ASA) and occupied volume using a 1.86 Å (N2) probe in the Zeo++ software.12 The accessible surface area of BGL, CBH, and EG are 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240, 13
240, 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 565, and 15
565, and 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 489 Å2, respectively. Their accessible volumes are 97
489 Å2, respectively. Their accessible volumes are 97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 880, 74
880, 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 660, and 82
660, and 82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 Å3, respectively.
340 Å3, respectively.
Prior to immobilization in TpAzo foam, BGL, CBH, and EG were purified (Section S-3†). We used HEPES (4-(2-hydroxyethyl)-1-piperazine ethane sulfonic acid), HEPES with 75 mM NaCl, and MES (2-(N-morpholino)ethane sulfonic acid) buffers optimized for the stability of BGL, CBH, and EG, respectively. We placed 1 mg of TpAzo foam in a reaction tube to begin the reaction.
To initiate the reaction, we added 400 μL of freshly purified 4 mg mL−1 BGL in 10 mM HEPES buffer (pH 7) to the tube. The reaction mixture was incubated for 2 hours and then centrifuged. We measured the UV-vis spectrum of the supernatant at 280 nm. The concentration of adsorbed BGL was determined using UV-vis spectroscopy to be 0.14 mg of BGL per 1 mg of foam (Fig. S4†). The BGL-immobilized TpAzo foam samples were washed several times with 10 mM HEPES buffer (pH 7) to remove any loosely bound BGL. SDS-PAGE analyses confirmed the absence of BGL from the second wash onwards. CBH and EG were similarly immobilized in TpAzo foam, and the enzyme-immobilized foams were also washed.
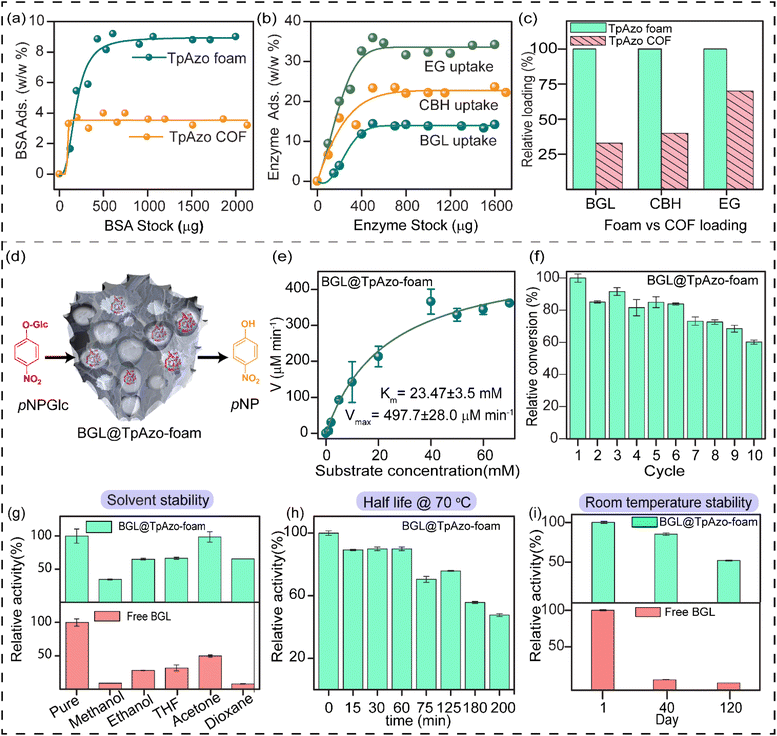
SDS-PAGE experiments were conducted to monitor the leaching of the enzyme from the foam (Fig. S6†). The loading percentage was calculated to be 0.22 mg CBH per 1 mg TpAzo foam and 0.33 mg EG per 1 mg TpAzo foam. SEM (Fig. 1d and S9†) and TEM (Fig. 1F and S12†) images of BGL@TpAzo-foam, CBH@TpAzo-foam, and EG@TpAzo-foam confirmed the retention of their porous morphology after enzyme immobilization. To determine the spatial distribution of the enzymes, we immobilized fluorescein isothiocyanate (FITC)-labeled enzymes in the foam. The FITC-enzyme@TpAzo-foams were excited at 552 nm, and confocal images were collected in a window range from 570 nm to 620 nm for the three enzyme-immobilized foams along the maximum intensity projection (MIP). Confocal images of FITC-EG@TpAzo-foam, FITC-BGL@TpAzo-foam, and FITC-CBH@TpAzo-foam reveal that EG, BGL, and CBH are entrapped inside the foam matrix (Fig. 1h and S14†). The PXRD patterns of the enzyme-immobilized foams are almost identical to the PXRD patterns of the TpAzo foam, indicating that the TpAzo foam retains its crystallinity in the enzymes-immobilized TpAzo foams (Fig. 2a). Micro-computed tomography of the bovine serum albumin (BSA) immobilized foam revealed a wide range of porosity distribution (up to 60 μm), with a macroporous surface area of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 110 m2 m−3. The distribution of macropore diameter of the BSA@TpAzo-foam is significantly lower (∼40 μm) compared to the pristine TpAzo foam (∼400 μm) (Fig. 2h and i), suggesting that immobilized BSA occupies the macropores preferably. The BET surface areas of the BSA@TpAzo-foam, BGL@TpAzo-foam, EG@TpAzo-foam, and CBH@TpAzo-foam dropped to 250, 280, 40, and 30 m2 g−1, respectively. The surface area of the enzyme-loaded TpAzo foam is less than that of the pristine TpAzo-foam, which is attributed to the preferential occupancy of the macropores by the enzymes, as observed through micro-computed tomography. As the macropores and micropores are interconnected, there will also be a blockage in the micropores. This blockage in the macropores led to a drop in porosity and the surface area of enzyme-loaded TpAzo foam. The extent of immobilization of BGL, CBH, and EG in the TpAzo foam is 14% (w/w), 22% (w/w), and 34% (w/w), respectively (Fig. 2b). In this context, a drop in the porosity and surface area is much higher in CBH@TpAzo-foam, and EG@TpAzo-foam followed by BGL@TpAzo-foam. Similarly, the pore volumes of BSA@TpAzo-foam, BGL@TpAzo-foam, CBH@TpAzo-foam, and EG@TpAzo-foam also decrease to 0.209, 0.205, 0.077, and 0.09 cm3 g−1 respectively. NLDFT calculations show that the enzyme-immobilized foams' average micropore size remains unchanged after enzyme immobilization (Fig. S16†). It implies that immobilized enzymes do not occupy the micropores of the foam material. FT-IR spectra of enzyme-immobilized foams exhibit a broad peak of amide-1 at 1635–1642 cm−1, which was absent in the pristine TpAzo foams (Fig. 2c–f, S17 and 18†). A series of volumes of BSA stock of concentration 4 μg μL−1 were added separately in 1 mg of TpAzo foam. The UV-vis spectra of the supernatant were collected after 2 hours of incubation. It was found that the TpAzo foam can encapsulate up to 9.0% (w/w) BSA, whereas TpAzo COF can uptake only 3.6% (w/w) BSA (Fig. 3a). It demonstrates that COF-foam has a greater uptake capability than COF, and macropores in the foam aid in BSA uptake.
110 m2 m−3. The distribution of macropore diameter of the BSA@TpAzo-foam is significantly lower (∼40 μm) compared to the pristine TpAzo foam (∼400 μm) (Fig. 2h and i), suggesting that immobilized BSA occupies the macropores preferably. The BET surface areas of the BSA@TpAzo-foam, BGL@TpAzo-foam, EG@TpAzo-foam, and CBH@TpAzo-foam dropped to 250, 280, 40, and 30 m2 g−1, respectively. The surface area of the enzyme-loaded TpAzo foam is less than that of the pristine TpAzo-foam, which is attributed to the preferential occupancy of the macropores by the enzymes, as observed through micro-computed tomography. As the macropores and micropores are interconnected, there will also be a blockage in the micropores. This blockage in the macropores led to a drop in porosity and the surface area of enzyme-loaded TpAzo foam. The extent of immobilization of BGL, CBH, and EG in the TpAzo foam is 14% (w/w), 22% (w/w), and 34% (w/w), respectively (Fig. 2b). In this context, a drop in the porosity and surface area is much higher in CBH@TpAzo-foam, and EG@TpAzo-foam followed by BGL@TpAzo-foam. Similarly, the pore volumes of BSA@TpAzo-foam, BGL@TpAzo-foam, CBH@TpAzo-foam, and EG@TpAzo-foam also decrease to 0.209, 0.205, 0.077, and 0.09 cm3 g−1 respectively. NLDFT calculations show that the enzyme-immobilized foams' average micropore size remains unchanged after enzyme immobilization (Fig. S16†). It implies that immobilized enzymes do not occupy the micropores of the foam material. FT-IR spectra of enzyme-immobilized foams exhibit a broad peak of amide-1 at 1635–1642 cm−1, which was absent in the pristine TpAzo foams (Fig. 2c–f, S17 and 18†). A series of volumes of BSA stock of concentration 4 μg μL−1 were added separately in 1 mg of TpAzo foam. The UV-vis spectra of the supernatant were collected after 2 hours of incubation. It was found that the TpAzo foam can encapsulate up to 9.0% (w/w) BSA, whereas TpAzo COF can uptake only 3.6% (w/w) BSA (Fig. 3a). It demonstrates that COF-foam has a greater uptake capability than COF, and macropores in the foam aid in BSA uptake.
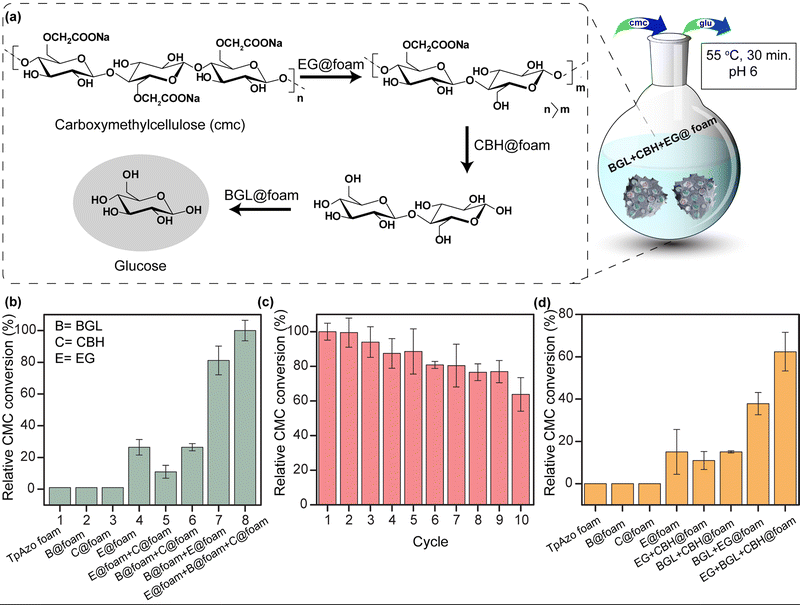
Multi-enzyme tandem catalysis has emerged as an alternative to the tedious process of biocatalytic transformation.13 Cellulose, a readily available material, can be hydrolyzed into glucose by cellulases and further processed to produce biofuels.14 The major components of cellulase are endoglucanase (EG), cellobiohydrolase (CBH), and β-glucosidase (BGL). However, the multi-enzyme catalyzed hydrolysis of carboxymethylcellulose (CMC), a modified form of cellulose, to glucose with high synthetic efficiency is challenging. To address this challenge, we examined the loading of BGL, CBH, and EG with an increasing stock concentration in the foam matrix. The uptake capacity vs. stock concentration graph revealed a saturation plateau in each enzyme immobilization. The total uptake efficiency of the TpAzo foam towards different enzymes differed substantially. BGL, CBH, and EG could be immobilized in the TpAzo foam up to 14% (w/w), 22% (w/w), and 34% (w/w), respectively (Fig. 3b). However, the total uptake efficiency of BGL, CBH, and EG in the TpAzo COF was 2.8% (w/w), 7% (w/w), and 23% (w/w), respectively (Fig. S5†). The relative uptake efficiency of BGL, CBH, and EG in the TpAzo COF compared to TpAzo foam were 27% (w/w), 40% (w/w), and 73% (w/w), respectively (Fig. 3c). Weak non-covalent interactions helped to immobilize these enzymes in the foam.
We undertook molecular dynamics (MD) and hybrid QM/MM simulations to determine the preferable adsorption sites of enzymes on TpAzo foam, along with their non-covalent interaction distances and energies (Section S-15†). Each enzyme was placed directly above a 4-layer, 4 × 4 supercell of the TpAzo foam to model the enzyme-TpAzo foam wall interactions. The bottom two layers of the foam had their positions fixed. To determine the variability of the enzyme-foam binding, each enzyme was placed five times above the TpAzo foam backbone structure in randomly selected orientations. From each starting point, MD simulations were run for 50 ps at 298 K, and QM/MM enzyme-TpAzo foam binding energies were calculated on each final snapshot. The maximum binding energies were: Eint (EG, −667.58 kJ mol−1) > Eint (BGL, −554.47 kJ mol−1) > Eint (CBH, −497.27 kJ mol−1) (Fig. S28–30†). The pore environment and macroscopic pore size dictated the loading of three enzymes into the TpAzo foam. We further checked the close non-covalent interactions between the enzyme and foam by searching for all instances where atoms (including H atoms) from the enzyme and foam approached each other within 3.0 Å. EG interacted more strongly with TpAzo foam due to the significant number of observable H-bonding, followed by BGL, while CBH showed the least number of H-bonding with the TpAzo foam layers (Table S3†).
The substrate chosen for the study was p-nitrophenyl-β-D-glucopyranoside (pNPGlc), and the production of p-nitrophenol (pNP) was monitored at 405 nm using UV-vis spectroscopy to determine β-glucosidase (BGL) activity in the TpAzo foam (Fig. 3d). We added 40 μL of 100 mM pNPGlc as a substrate and 10 μL of BGL@TpAzo-foam (equivalent to 0.5 μg of free BGL) to 50 μL of 10 mM HEPES buffer (pH 7) to make the reaction volume 100 μL (Section S-6†). Hydrolysis of pNPGlc was performed at different temperatures and pH levels. The BGL@TpAzo-foam showed maximum activity at 70 °C and pH 7 (Fig. S23†). The maximum activity of BGL@TpAzo-foam under the optimized condition was set as 100%, and the activities of BGL@TpAzo-foam in other conditions were calculated relatively. The Michaelis–Menten plot of the hydrolysis of pNPGlc to pNP by BGL@TpAzo-foam at the optimized condition revealed that the Km and Vmax were 23.5 ± 3.5 mM and 497.7 ± 28.0 μM min−1, respectively (Fig. 3e), while the Km and Vmax of free BGL were 14.7 ± 0.8 mM and 1116.3 ± 18.9 μM min−1, respectively (Fig. S25†). To confirm the heterogeneity of the reaction, we performed the recyclability of BGL@TpAzo-foam. After centrifugation, we found that BGL@TpAzo-foam is recyclable for up to 10 cycles, as it retained 62% specific activity after the 10th cycle despite the loss of BGL@TpAzo-foam particles in every step during the separation (Fig. 3f). Moreover, there was no change in the morphology of the foam particles after the catalytic cycle (Fig. S10 and S13†).
We also explored the stability of free BGL and BGL@TpAzo-foam in different organic solvents. After incubation in 500 μL of various solvents for 24 hours, we checked their activity. The activity of free BGL and BGL@TpAzo-foam in the optimized buffer was set as 100%. The relative activity of BGL@TpAzo-foam in methanol, ethanol, THF, dioxane, and acetone was 2 to 8-fold higher than that of free BGL due to the confinement of the macropores in the foam matrix (Fig. 3g).15 Additionally, the interaction of BGL with the foam matrix reduces the unfolding tendency of the BGL. To compare the half-life of free BGL (73 minutes)16 with that of BGL immobilized in TpAzo foam, we measured the relative activity of the BGL@TpAzo-foam at different time intervals at 70 °C. The relative activity of the BGL@TpAzo-foam started to decrease with time and became almost half after 200 minutes, implying a higher half-life and thermotolerance of the BGL@TpAzo-foam (Fig. 3h). When free BGL and BGL@TpAzo-foam were kept at room temperature (25 °C), free BGL showed zero activity after 40 days, whereas BGL@TpAzo-foam retained 85% and 51% specific activity after 40 and 120 days, respectively (Fig. 3i). This further demonstrates the stability of BGL inside the foam matrix owing to the confinement effect of the pores.
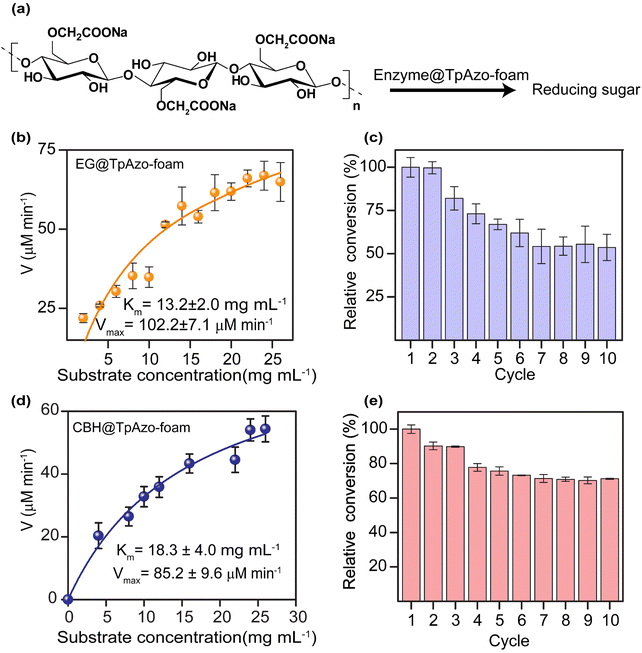
We quantified the generated reducing sugar by UV-vis spectroscopy at 540 nm through DNS assay to determine the activity of endoglucanase (EG) and cellobiohydrolase (CBH) in the foam on carboxymethylcellulose (CMC) (Section S-6†). The maximum activity of EG@TpAzo foam was observed at 55 °C and pH 6 (Fig. S23†). We conducted the hydrolysis of CMC to reducing sugar under different temperatures and pH values. The maximum catalytic activity of EG@TpAzo-foam under the optimized condition was set as 100%, and the activities of EG@TpAzo-foam in other conditions were calculated accordingly. The steady-state kinetics revealed that the Km and Vmax of EG@TpAzo-foam were 13.2 ± 2.0 mg mL−1 and 102.2 ± 7.1 μM min−1, respectively (Fig. 4a), while the Km and Vmax of free EG were 66 mg mL−1 and 545 μM min−1 (Fig. S25†). To confirm the heterogeneity of the reaction, we tested the recyclability of EG@TpAzo-foam, and found that it can be recycled up to 10 cycles. Even though there was an unavoidable loss of EG@TpAzo-foam particles in every step during separation, the relative activity of EG@TpAzo-foam was retained at 53% after the 10th cycle (Fig. 4b). After the catalytic cycle, the morphology of the foam particles remained unchanged (Fig. S10 and S13†).
We also optimized the activity of CBH@TpAzo-foam and found that it showed the highest activity at 55 °C and pH 6 (Fig. S23†). The steady-state kinetics showed that the Km and Vmax of CBH@TpAzo-foam were 18.3 ± 4.0 mg mL−1 and 85.2 ± 9.6 μM min−1, respectively (Fig. 4c), while the Km and Vmax of free CBH were 7.8 mg mL−1 and 327 μM min−1 (Fig. S25†). Similar to EG@TpAzo-foam, CBH@TpAzo-foam was found to be recyclable up to 10 cycles with 71% activity (Fig. 4d). After the catalytic cycle, no changes were observed in the morphology of the foam particles (as shown in Fig. S10 and S13†). The catalytic activity of BGL, CBH, and EG immobilized on TpAzo foam was two to four-fold higher than their activities in TpAzo COF (as demonstrated in Fig. S24†). This lower activity in TpAzo COF can be attributed to the absence of macropores in the structure, which leads to a reduction in the flow velocity of reactants and products.
We simultaneously utilized BGL@TpAzo-foam, CBH@TpAzo-foam, and EG@TpAzo-foam in one pot to convert carboxymethylcellulose to glucose without isolating intermediates (as depicted in Fig. 5a and Section S-6†). No products were observed when the reaction was performed with only TpAzo foam. We added 10 μL each of BGL@TpAzo-foam (equivalent to 0.5 μg of free BGL), CBH@TpAzo-foam (equivalent to 0.5 μg of free CBH), and EG@TpAzo-foam (equivalent to 0.5 μg of free EG), and the conversion of CMC to glucose was monitored by GOD-POD (glucose oxidase-peroxidase) assay. The maximum conversion rate under these conditions was set as 100% to compare the conversion rates in other conditions (as shown in Fig. 5b).
We conducted a control experiment to determine the contribution of each enzyme@TpAzo-foam to glucose conversion. In an equimolar mixture of BGL@TpAzo-foam and EG@TpAzo-foam (equivalent to 0.75 μg of free BGL and 0.75 μg of free EG), the relative conversion dropped to 80%. Similarly, in an equimolar mixture of BGL@TpAzo-foam and CBH@TpAzo-foam (equivalent to 0.75 μg of free BGL and 0.75 μg of free CBH), the relative conversion was 26%. The relative conversion was 10% for an equimolar combination of EG@TpAzo-foam and CBH@TpAzo-foam (equivalent to 0.75 μg of free EG and 0.75 μg of free CBH). In the presence of EG@TpAzo-foam (equivalent to 1.5 μg of free EG), the relative conversion was only 22% (Fig. 5b). The enzymes@TpAzo-foam demonstrated high recyclability (10 cycles) with excellent relative conversions (∼66%) (Fig. 5c). When we immobilized BGL, CBH, and EG simultaneously in the foam (BGL + CBH + EG@TpAzo-foam) in different combinations for CMC conversion, we found that the trend of the relative activity of CMC hydrolysis was similar to that of a mixture of BGL@TpAzo-foam, CBH@TpAzo-foam, and EG@TpAzo-foam (Fig. S26†). However, the relative activity dropped to 62% in the presence of BGL + CBH + EG@TpAzo-foam (Fig. 5d). The different ratios of immobilized enzymes in the same foam (BGL + CBH + EG@TpAzo-foam) compared to the individual mixtures (BGL@TpAzo-foam + CBH@TpAzo-foam + EG@TpAzo-foam) likely caused a drop in relative activity.
EG@TpAzo-foam degrades CMC from the chain end by binding to the substrate until a minimum chain length is achieved. After that, CBH@TpAzo-foam starts to hydrolyze one or a few accessible bonds in the chain from the surface. Finally, BGL@TpAzo-foam hydrolyzes cellobiose and small soluble oligomers to glucose.17 We analyzed the conversion of CMC to glucose by EG@TpAzo-foam + CBH@TpAzo-foam + BGL@TpAzo-foam mixture and EG + CBH + BGL@TpAzo-foam using HRMS. The HRMS value confirmed (observed value M + Na: 203.0553, theoretical value: 203.06) the presence of glucose. Thus, the synergistic action of these three enzymes embedded in foam enables the one-pot production of glucose.
Conclusions
In summary, we have developed and synthesized TpAzo foam with both ordered and disordered pores, which can effectively immobilize various enzymes due to its macroporosity. Compared to TpAzo COF, TpAzo foam exhibited an enhanced BSA uptake capacity of approximately 2.5 times. By immobilizing BGL, CBH, and EG in TpAzo foam through weak non-covalent interactions, we achieved 1.5 to 4 times higher enzyme uptake than TpAzo COF. Furthermore, enzyme@TpAzo-foam demonstrated 2 to 4 times higher activities than enzyme@TpAzo-COF. BGL immobilized in TpAzo foam showed substantial catalytic activity, and stability at room temperature and retained half of its initial activity for up to 120 days. The TpAzo foam can also sustain organic solvents, making it suitable for enzyme catalysis in these conditions. Moreover, we demonstrated the successful one-pot tandem glucose synthesis from CMC with excellent recyclability, which can be used for various industrial applications such as biofuel and chemical production. We believe that this immobilization method can pave the way for developing cost-effective and environmentally friendly biocatalysis-based technologies for diverse industrial applications.Data availability
All experimental and computational data are available in the ESI.†Author contributions
R. Banerjee and S. Datta obtained the funding, and supervised the project. S. Paul designed the project, synthesized the material, performed assays, analyzed the data, and wrote the manuscript. M. Gupta purified the enzymes, designed and performed the assays, analyzed the data. K. Dey verified the manuscript. A. Mahato and S. Bag synthesized the starting material. A. Torris conducted the micro-computed tomography. E. B. Gowd helped to collect the PXRD data. H. Sajid and M. Addicoat calculated the molecular dynamics (MD) simulation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
SP, SB, AKM acknowledges CSIR for Research fellowship. MG acknowledges DST-INSPIRE for a Junior Research Fellowship. KD acknowledges DST-SERB India for a RA fellowship [CRG/2018/000314]. RB acknowledges SERB Core Research Grant [CRG/2018/000314] and SERB SUPRA [SPR/2021/000020] for funding. SD acknowledges SERB Core Research Grant, EMR/2016/003705, and MoE-STARS/STARS-1/643. We acknowledge Mr Amal Raj for the PXRD data collection. We acknowledge Mr Ruturaj and Mr Ritabrata Ghosh for the confocal data collection and Dr Sushant K. Sinha for protein production, purification, and initial assays.Notes and references
- (a) A. Schmid, J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts and B. Witholt, Industrial biocatalysis today and tomorrow, Nature, 2001, 409, 258–268 CrossRef CAS PubMed; (b) C. M. Clouthier and J. N. Pelletier, Expanding the organic toolbox: a guide to integrating biocatalysis in synthesis, Chem. Soc. Rev., 2012, 41, 1585–1605 RSC; (c) R. A. Sheldon, Enzyme Immobilization: The Quest for Optimum Performance, Adv. Synth. Catal., 2007, 349, 1289–1307 CrossRef CAS; (d) R. Ye, J. Zhao, B. B. Wickemeyer, F. D. Toste and G. A. Somorjai, Foundations and strategies of the construction of hybrid catalysts for optimized performances, Nat. Catal., 2018, 1, 318–325 CrossRef.
- (a) P. J. O'Brien and D. Herschlag, Chem. Biol., 1999, 6, 91–105 CrossRef PubMed; (b) J. C. Lewis, P. S. Coelho and F. H. Arnold, Enzymatic functionalization of carbon–hydrogen bonds, Chem. Soc. Rev., 2011, 40, 2003–2021 RSC; (c) S. Simic, E. Zukic, L. Schmermund, K. Faber, C. K. Winkler and W. Kroutil, Shortening Synthetic Routes to Small Molecule Active Pharmaceutical Ingredients Employing Biocatalytic Methods, Chem. Rev., 2022, 122, 1052–1126 CrossRef CAS PubMed; (d) A. J. Straathof, Transformation of biomass into commodity chemicals using enzymes or cells, Chem. Rev., 2014, 114, 1871–1908 CrossRef CAS PubMed.
- (a) E. T. Hwang and S. Lee, Multienzymatic Cascade Reactions via Enzyme Complex by Immobilization, ACS Catal., 2019, 9, 4402–4425 CrossRef CAS; (b) J. Liang, S. Gao, J. Liu, M. Y. B. Zulkifli, J. Xu, J. Scott, V. Chen, J. Shi, A. Rawal and K. Liang, Hierarchically Porous Biocatalytic MOF Microreactor as a Versatile Platform towards Enhanced Multi-enzyme and Cofactor-Dependent Biocatalysis, Angew. Chem., Int. Ed., 2021, 60, 5421–5428 CrossRef CAS PubMed; (c) J. Liang and K. Liang, Multi-enzyme Cascade Reactions in Metal–organic Frameworks, Chem. Rec., 2020, 20, 1100–1116 CrossRef CAS PubMed; (d) Y. Zhang, L. Xu and J. Ge, Multi-enzyme System in Amorphous Metal–Organic Frameworks for Intracellular Lactate Detection, Nano Lett., 2022, 22, 5029–5036 CrossRef CAS PubMed.
- (a) A. S. Bommarius and M. F. Paye, Stabilizing biocatalysts, Chem. Soc. Rev., 2013, 42, 6534–6565 RSC; (b) H. Cui, L. Zhang, L. Eltoukhy, Q. Jiang, S. K. Korkunç, K.-E. Jaeger, U. Schwaneberg and M. D. Davari, Enzyme Hydration Determines Resistance in Organic Cosolvents, ACS Catal., 2020, 10, 14847–14856 CrossRef CAS; (c) R. A. Sheldon and D. Brady, The limits to biocatalysis: pushing the envelope, Chem. Commun., 2018, 54, 6088–60104 RSC; (d) C. Silva, M. Martins, S. Jing, J. Fu and A. Cavaco-Paulo, Practical insights on enzyme stabilization, Crit. Rev. Biotechnol., 2018, 38, 335–350 CrossRef CAS PubMed.
- (a) U. T. Bornscheuer, Immobilizing enzymes: how to create more suitable biocatalysts, Angew. Chem., Int. Ed., 2003, 42, 3336–3337 CrossRef CAS PubMed; (b) R. DiCosimo, J. McAuliffe, A. J. Poulose and G. Bohlmann, Industrial use of immobilized enzymes, Chem. Soc. Rev., 2013, 42, 6437–6474 RSC; (c) M. Dong, Z. Wu, M. Lu, Z. Wang and Z. Li, Combining the physical adsorption approach and the covalent attachment method to prepare a bifunctional bioreactor, Int. J. Mol. Sci., 2012, 13, 11443–11454 CrossRef CAS PubMed; (d) R. Fernandez-Lafuente, Stabilization of multimeric enzymes: strategies to prevent subunit dissociation, Enzyme Microb. Technol., 2009, 45, 405–418 CrossRef CAS; (e) C. Hou, Y. Wang, Q. Ding, L. Jiang, M. Li, W. Zhu, D. Pan, H. Zhu and M. Liu, Facile synthesis of enzyme-embedded magnetic metal–organic frameworks as a reusable mimic multi-enzyme system: mimetic peroxidase properties and colorimetric sensor, Nanoscale, 2015, 7, 18770–18779 RSC; (f) W. Liang, P. Wied, F. Carraro, C. J. Sumby, B. Nidetzky, C. K. Tsung, P. Falcaro and C. J. Doonan, Metal–Organic Framework-Based Enzyme Biocomposites, Chem. Rev., 2021, 121, 1077–1129 CrossRef CAS PubMed; (g) A. Liese and L. Hilterhaus, Evaluation of immobilized enzymes for industrial applications, Chem. Soc. Rev., 2013, 42, 6236–6249 RSC; (h) R. A. Sheldon and S. Van Pelt, Enzyme immobilisation in biocatalysis: why, what and how, Chem. Soc. Rev., 2013, 42, 6223–6235 RSC; (i) V. L. Sirisha, A. Jain and A. Jain, Enzyme Immobilization: An Overview on Methods, Support Material, and Applications of Immobilized Enzymes, Adv. Food Nutr. Res., 2016, 79, 179–211 CAS; (j) M. Vázquez-González, C. Wang and I. Willner, Biocatalytic cascades operating on macromolecular scaffolds and in confined environments, Nat. Catal., 2020, 3, 256–273 CrossRef; (k) J. Zdarta, A. Meyer, T. Jesionowski and M. Pinelo, A General Overview of Support Materials for Enzyme Immobilization: Characteristics, Properties, Practical Utility, Catalysts, 2018, 8, 92 CrossRef; (l) P. Zucca, R. Fernandez-Lafuente and E. Sanjust, Agarose and Its Derivatives as Supports for Enzyme Immobilization, Molecules, 2016, 21, 1577 CrossRef PubMed.
- (a) W. H. Chen, M. Vázquez-González, A. Zoabi, R. Abu-Reziq and I. Willner, Biocatalytic cascades driven by enzymes encapsulated in metal–organic framework nanoparticles, Nat. Catal., 2018, 1, 689–695 CrossRef CAS; (b) Y. Chen, P. Li, H. Noh, C. W. Kung, C. T. Buru, X. Wang, X. Zhang and O. K. Farha, Stabilization of Formate Dehydrogenase in a Metal–Organic Framework for Bioelectrocatalytic Reduction of CO2, Angew. Chem., Int. Ed., 2019, 58, 7682–7686 CrossRef CAS PubMed; (c) Y. Chen, V. Lykourinou, C. Vetromile, T. Hoang, L. J. Ming, R. W. Larsen and S. Ma, How can proteins enter the interior of a MOF? Investigation of cytochrome c translocation into a MOF consisting of mesoporous cages with microporous windows, J. Am. Chem. Soc., 2012, 134, 13188–13191 CrossRef CAS PubMed; (d) R. Greifenstein, T. Ballweg, T. Hashem, E. Gottwald, D. Achauer, F. Kirschhofer, M. Nusser, G. Brenner-Weiss, E. Sedghamiz, W. Wenzel, E. Mittmann, K. S. Rabe, C. M. Niemeyer, M. Franzreb and C. Woll, MOF-Hosted Enzymes for Continuous Flow Catalysis in Aqueous and Organic Solvents, Angew. Chem., Int. Ed., 2022, 61, e202117144 CrossRef CAS PubMed; (e) H. Lyu, O. I. Chen, N. Hanikel, M. I. Hossain, R. W. Flaig, X. Pei, A. Amin, M. D. Doherty, R. K. Impastato, T. G. Glover, D. R. Moore and O. M. Yaghi, Carbon Dioxide Capture Chemistry of Amino Acid Functionalized Metal–Organic Frameworks in Humid Flue Gas, J. Am. Chem. Soc., 2022, 144, 2387–2396 CrossRef CAS PubMed; (f) H. Lyu, H. Li, N. Hanikel, K. Wang and O. M. Yaghi, Covalent Organic Frameworks for Carbon Dioxide Capture from Air, J. Am. Chem. Soc., 2022, 144, 12989–12995 CrossRef CAS PubMed.
- (a) M. Li, S. Qiao, Y. Zheng, Y. H. Andaloussi, X. Li, Z. Zhang, A. Li, P. Cheng, S. Ma and Y. Chen, Fabricating Covalent Organic Framework Capsules with Commodious Microenvironment for Enzymes, J. Am. Chem. Soc., 2020, 142, 6675–6681 CrossRef CAS PubMed; (b) Y. M. Li, J. Yuan, H. Ren, C. Y. Ji, Y. Tao, Y. Wu, L. Y. Chou, Y. B. Zhang and L. Cheng, Fine-Tuning the Micro-Environment to Optimize the Catalytic Activity of Enzymes Immobilized in Multivariate Metal–Organic Frameworks, J. Am. Chem. Soc., 2021, 143, 15378–15390 CrossRef CAS PubMed; (c) K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Biomimetic mineralization of metal–organic frameworks as protective coatings for biomacromolecules, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed; (d) P. Wied, F. Carraro, J. M. Bolivar, C. J. Doonan, P. Falcaro and B. Nidetzky, Combining a Genetically Engineered Oxidase with Hydrogen-Bonded Organic Frameworks (HOFs) for Highly Efficient Biocomposites, Angew. Chem. Int. Ed., 2022, 61, e202117345 CrossRef CAS PubMed; (e) S. Zhang, Y. Zheng, H. An, B. Aguila, C. Yang, Y. Dong, W. Xie, P. Cheng, Z. Zhang, Y. Chen and S. Ma, Covalent Organic Frameworks with Chirality Enriched by Biomolecules for Efficient Chiral Separation, Angew. Chem., Int. Ed., 2018, 57, 16754–16759 CrossRef CAS PubMed.
- (a) Y. Cao, X. Li, J. Xiong, L. Wang, L. T. Yan and J. Ge, Investigating the origin of high efficiency in confined multi-enzyme catalysis, Nanoscale, 2019, 11, 22108–22117 RSC; (b) W. H. Chen, M. Vázquez-González, A. Zoabi, R. Abu-Reziq and I. Willner, Biocatalytic cascades driven by enzymes encapsulated in metal–organic framework nanoparticles, Nat. Catal., 2018, 1, 689–695 CrossRef CAS; (c) T. Man, C. Xu, X. Y. Liu, D. Li, C. K. Tsung, H. Pei, Y. Wan and L. Li, Hierarchically encapsulating enzymes with multi-shelled metal–organic frameworks for tandem biocatalytic reactions, Nat. Commun., 2022, 13, 305 CrossRef CAS PubMed; (d) A. J. Tansell, C. L. Jones and T. L. Easun, MOF the beaten track: unusual structures and uncommon applications of metal–organic frameworks, Chem. Cent. J., 2017, 11, 100 CrossRef CAS PubMed; (e) E. T. Hwang and S. Lee, Multienzymatic Cascade Reactions via Enzyme Complex by Immobilization, ACS Catal., 2019, 9, 4402–4425 CrossRef CAS.
- (a) A. P. Cote, A. I. Benin, N. W. Ockwig, M. O'Keeffe, A. J. Matzger and O. M. Yaghi, Porous, crystalline, covalent organic frameworks, Science, 2005, 310, 1166–1170 CrossRef CAS PubMed; (b) G. Das, F. Benyettou, S. K. Sharama, T. Prakasam, F. Gándara, V. A. De La Peña-O’Shea, N. Saleh, R. Pasricha, R. Jagannathan, M. A. Olson and A. Trabolsi, Covalent organic nanosheets for bioimaging, Chem. Sci., 2018, 9, 8382–8387 RSC; (c) C. S. Diercks and O. M. Yaghi, The atom, the molecule, and the covalent organic framework, Science, 2017, 355, 6328 CrossRef PubMed; (d) S. Kandambeth, K. Dey and R. Banerjee, Covalent Organic Frameworks: Chemistry Beyond the Structure, J. Am. Chem. Soc., 2019, 141, 1807–1822 CrossRef CAS PubMed; (e) S. Kandambeth, A. Mallick, B. Lukose, M. V. Mane, T. Heine and R. Banerjee, Construction of crystalline 2D covalent organic frameworks with remarkable chemical (acid/base) stability via a combined reversible and irreversible route, J. Am. Chem. Soc., 2012, 134, 19524–19527 CrossRef CAS PubMed; (f) S. Karak, S. Kandambeth, B. P. Biswal, H. S. Sasmal, S. Kumar, P. Pachfule and R. Banerjee, Constructing Ultraporous Covalent Organic Frameworks in Seconds via an Organic Terracotta Process, J. Am. Chem. Soc., 2017, 139, 1856–1862 CrossRef CAS PubMed; (g) H. Lyu, C. S. Diercks, C. Zhu and O. M. Yaghi, Porous Crystalline Olefin-Linked Covalent Organic Frameworks, J. Am. Chem. Soc., 2019, 141, 6848–6852 CrossRef CAS PubMed; (h) H. S. Sasmal, S. Bag, B. Chandra, P. Majumder, H. Kuiry, S. Karak, S. Sen Gupta and R. Banerjee, Heterogeneous C–H Functionalization in Water via Porous Covalent Organic Framework Nanofilms: A Case of Catalytic Sphere Transmutation, J. Am. Chem. Soc., 2021, 143, 8426–8436 CrossRef CAS PubMed; (i) V. S. Vyas, M. Vishwakarma, I. Moudrakovski, F. Haase, G. Savasci, C. Ochsenfeld, J. P. Spatz and B. V. Lotsch, Exploiting Non-covalent Interactions in an Imine-Based Covalent Organic Framework for Quercetin Delivery, Adv. Mater., 2016, 28, 8749–8754 CrossRef CAS PubMed; (j) X. Zhao, P. Pachfule, S. Li, T. Langenhahn, M. Ye, C. Schlesiger, S. Praetz, J. Schmidt and A. Thomas, Macro/Microporous Covalent Organic Frameworks for Efficient Electrocatalysis, J. Am. Chem. Soc., 2019, 141, 6623–6630 CrossRef CAS PubMed.
- (a) S. Kandambeth, V. Venkatesh, D. B. Shinde, S. Kumari, A. Halder, S. Verma and R. Banerjee, Self-templated chemically stable hollow spherical covalent organic framework, Nat. Commun., 2015, 6, 6786 CrossRef CAS PubMed; (b) Y. Li, C. Wang, S. Ma, H. Zhang, J. Ou, Y. Wei and M. Ye, Fabrication of Hydrazone-Linked Covalent Organic Frameworks Using Alkyl Amine as Building Block for High Adsorption Capacity of Metal Ions, ACS Appl. Mater. Interfaces, 2019, 11, 11706–11714 CrossRef CAS PubMed; (c) X. Yang, Z. Tan, H. Sun, Y. Tong, X. Huang, J. Ren, C. Wang and W. Shen, Fabrication of a hierarchical nanoreactor based on COFs for cascade enzyme catalysis, Chem. Commun., 2022, 58, 3933–3936 RSC.
- (a) S. Karak, K. Dey, A. Torris, A. Halder, S. Bera, F. Kanheerampockil and R. Banerjee, Inducing Disorder in Order: Hierarchically Porous Covalent Organic Framework Nanostructures for Rapid Removal of Persistent Organic Pollutants, J. Am. Chem. Soc., 2019, 141, 7572–7581 CrossRef CAS PubMed; (b) A. K. Mohammed, S. Usgaonkar, F. Kanheerampockil, S. Karak, A. Halder, M. Tharkar, M. Addicoat, T. G. Ajithkumar and R. Banerjee, Connecting Microscopic Structures, Mesoscale Assemblies, and Macroscopic Architectures in 3D-Printed Hierarchical Porous Covalent Organic Framework Foams, J. Am. Chem. Soc., 2020, 142, 8252–8261 CrossRef CAS PubMed.
- T. F. Willems, C. H. Rycroft, M. Kazi, J. C. Meza and M. Haranczyk, Algorithms and tools for high-throughput geometry-based analysis of crystalline porous materials, Microporous Mesoporous Mater., 2012, 149, 134–141 CrossRef CAS.
- (a) J. H. Schrittwieser, S. Velikogne, M. Hall and W. Kroutil, Artificial Biocatalytic Linear Cascades for Preparation of Organic Molecules, Chem. Rev., 2018, 118, 270–348 CrossRef CAS PubMed; (b) P. A. Santacoloma, G. Sin, K. V. Gernaey and J. M. Woodley, Multienzyme-Catalyzed Processes: Next-Generation Biocatalysis, Org. Process Res. Dev., 2011, 15, 203–212 CrossRef CAS.
- (a) J. Sheehan, L. R. Lynd, M. S. Laser, D. Bransby, B. E. Dale, B. Davison, R. Hamilton, M. Himmel, M. Keller and J. D. McMillan, How biotech can transform biofuels, Nat. Biotechnol., 2008, 26, 169–172 CrossRef PubMed; (b) S. Goswami, N. Gupta and S. Datta, Using the beta-glucosidase catalyzed reaction product glucose to improve the ionic liquid tolerance of beta-glucosidases, Biotechnol. Biofuels, 2016, 9, 72 CrossRef PubMed; (c) D. Klein-Marcuschamer, P. Oleskowicz-Popiel, B. A. Simmons and H. W. Blanch, The challenge of enzyme cost in the production of lignocellulosic biofuels, Biotechnol. Bioeng., 2012, 109, 1083–1087 CrossRef CAS PubMed; (d) S. Aich, S. Datta, Efficient utilization of lignocellulosic biomass: hydrolysis methods for biorefineries, Biorefineries: a step towards renewable and clean energy, Springer, Singapore, 2020, pp. 273–295 Search PubMed.
- F. L. Oliveira, A. S. França, A. M. Castro, R. O. M. Alves de Souza, P. M. Esteves and R. S. B. Gonçalves, Enzyme Immobilization in Covalent Organic Frameworks: Strategies and Applications in Biocatalysis, Chempluschem, 2020, 85, 2051–2066 CrossRef CAS PubMed.
- S. K. Sinha, S. Goswami, S. Das and S. Datta, Exploiting non-conserved residues to improve activity and stability of Halothermothrix orenii beta-glucosidase, Appl. Microbiol. Biotechnol., 2017, 101, 1455–1463 CrossRef CAS PubMed.
- D. Kumar and G. S. Murthy, Development and validation of a stochastic molecular model of cellulose hydrolysis by action of multiple cellulase enzymes, Bioresour. Bioprocess., 2017, 4, 54 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc01367g |
| This journal is © The Royal Society of Chemistry 2023 |