Open Access Article
Open Access ArticleIsotope-encoded tetrahedral DNA for multiple SARS-CoV-2 variant diagnosis†
Ziyan
Li
a,
Jing
Zhou
 a,
Chaoqun
Wang
a,
Rui
Liu
a,
Chaoqun
Wang
a,
Rui
Liu
 *b,
Jianyu
Hu
*b,
Jianyu
Hu
 c and
Yi
Lv
c and
Yi
Lv
 *ab
*ab
aAnalytical & Testing Center, Sichuan University, Chengdu 610064, China. E-mail: lvy@scu.edu.cn
bKey Laboratory of Green Chemistry & Technology, Ministry of Education, College of Chemistry, Sichuan University, Chengdu 610064, Sichuan, China. E-mail: liur@scu.edu.cn
cDivision of Analytical and Environmental Toxicology, Faculty of Medicine & Dentistry, University of Alberta, Edmonton T6G 2G3, Alberta, Canada
First published on 25th May 2023
Abstract
The evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has posed an unprecedented demand for accurate and cost-effective diagnostic assays to discriminate between different variants. Whilst many bioassays have been successfully demonstrated for SARS-CoV-2 detection, diagnosis of its variants remains challenging and mainly relies on time-consuming and costly sequencing techniques. Herein, we proposed a triplevalent tetrahedral DNA nanostructure (tTDN) with three overhang isotope probes capable of multiplex simultaneous analysis. HV69/70 del (alpha-specific), K417N (beta-specific) and T478K (delta-specific) and omicron with common mutations above of the SARS-CoV-2 S gene were detected selectively with the aid of the TDN scaffold and MNAzyme system, and a sensitive strategy enabling the screening of four kinds of variants of concern (VOC) was achieved.
Introduction
Multiple variants of SARS-CoV-2 (ref. 1) have shown increased transmissibility and pathogenicity.2 The Centers for Disease Control and Prevention (CDC) has now defined five strains of SARS-CoV-2 alpha/beta/gamma/delta/omicron as VOC,3 with the necessity for strict monitoring. These remarkable mutations, including HV69-70del, K417N, and T478K, all occur in the major transmembrane spike (S) glycoprotein,4–6 enhancing the ability of the virus to invade cells or contribute to the immune escape of the virus.7 Population scale sweeps of multiple SARS-CoV-2 variants brought new challenges for molecular diagnostics and widespread availability of multiplexed, scalable diagnostic screening for the SARS-CoV-2 variant is required to enable active surveillance and focused patient care.8The mainstream for pathogen genomic surveillance is whole genomic sequencing (WGS),9 while it calls for long turnaround times, especially in the screening of multiple variants.10 The quantitative reverse-transcription polymerase chain reaction (qRT-PCR) assays provide the greatest sensitivity and specificity to implement,11 but still shortages of supplies and laboratory personnel exist, and most of these assays either target a single mutation per response or lack sufficient targets for thoroughly characterizing SARS-CoV-2.12 Crucially, tied up by the spectral overlap of fluorescent tags,13 implementations of existing multiplex sequencing14 and PCR15 strategies for SARS-CoV-2 variant diagnosis rely on microarrays14,16 or pre-separations12,17 that are essentially a sum of multiple assays after aliquoting each sample. For key mutations in the virus, a new genotyping method needs to be developed that is more efficient and multiplexable than WGS. Scalable diagnostic tools to efficiently detect multiple variants of SARS-CoV-2 in a single tube are still lacking.
Intriguingly, molecular interpretations encoded by multiple isotopes exhibit high sensitivity, wide dynamic range,18 and, most importantly, no signal overlap compared to most used fluorophore tags.19,20 Efforts have been made in multiplexed assays with respect to nucleic acid21,22 and protein23,24 markers for molecular diagnostics, and mass cytometry (CyTOF),25 spatial single nuclear metabolomics (SEAM)26 have showcasted superior high-dimensional barcoding ability. Overall, isotope-encoding mass spectrometry is ideally suited for multicomponent assays.
To address the need for affordable and accessible multiplexity, a tTDN-magnetic bead (MB) satellite with lanthanide isotope tags at the vertices was constructed for the detection of multi-component character mutations. The TDN provides an oriented scaffold27–31 for subsequent reaction design, which is a ubiquitous method for tracing biological analytes owing to its relatively high sensitivity32,33 and easily fabricated features.34–36 Furthermore, its distinctive multi-vertex structure37–40 makes it an ideal platform for multi-component bioanalysis.40–42 Combining the advantages of TDN and mass spectrometry, a multi-tentacle tTDN with overhang lanthanide isotope probes as MNAzyme substrates for multiplex analysis of HV69/70 del (alpha-specific), K417N (beta-specific) and T478K (delta-specific)3 was established. The tTDN on MBs provides well-controlled nanoscale distance,43 efficiently reducing the non-specific adsorption of the overhang probe DNA. The splicing of the overhang probe by MNAzyme44 could fulfil the re-release of RNA together with isotope tags which are continuously peeled off like satellites in orbit, achieving signal amplification.
The tTDN accelerates the MNAzyme reaction kinetics45 compared to ssDNA probes due to the synergistic contributions of stereo distance and specific reaction orientations, moreover, exhibiting biostability and multiplicity.38 As a proof-of-concept, it is a promising candidate for screening of SARS-CoCV-2 VOC variants.
Experimental
Materials and apparatus
A PerkinElmer NexION 350 inductively coupled plasma mass spectrometer was used throughout the experiments. The ESI† contains a comprehensive list of the parameters of ICPMS (Table S1†), nucleotide sequences (Table S2†), chemicals, reagents, and some of the operational procedures and characterizations studies.Labeling of HV69/70 del, K417N, and T478K-substrates with Tb, Ho and Tm
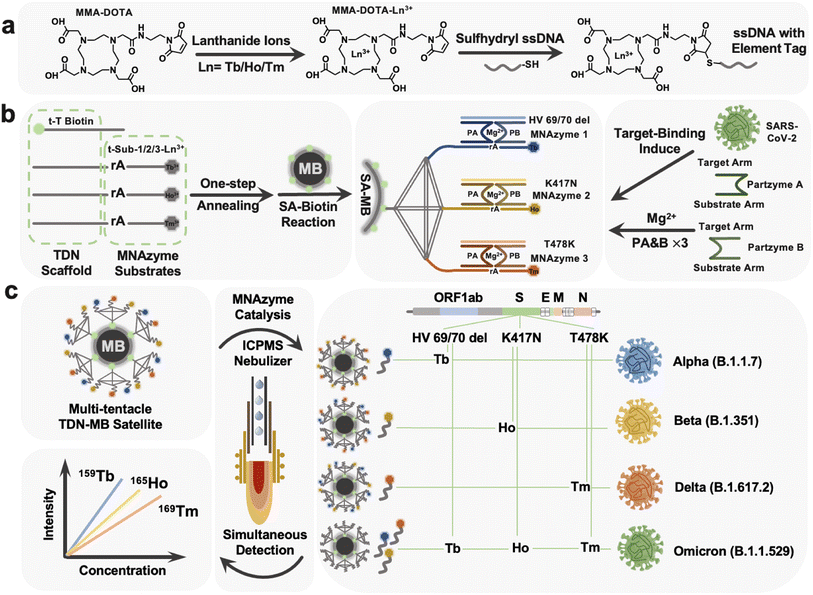
The labeling process was first conducted with protocols adopted from the previous work of Wang's group.46Fig. 1a depicts a diagrammatic sketch of the technique, with individual operations provided in the ESI.† In accordance with that, 1,4,7,10-tetraazacyclodo-decane-1,4,7-tris-acetic acid-10-maleimidoethylacetamide (MMA-DOTA) forms a chelate complex with a lanthanide(III) ion Tb3+/Ho3+/Tm3+ and finally we got the DNA-MMA-DOTA-Tb3+/Ho3+/Tm3+ conformations.Preparation of the tTDN using the thermal annealing protocol
Each of the three isotope-labeled DNA tracks, as well as a biotin-labeled DNA (t-T) as the fourth backbone of the tTDN structure, was diluted to 8 μM. At a final concentration of 2 μM, equal molar quantities of the composition DNA strands were mixed in TM buffer. The mixed strands were heated to 90 °C for 5 minutes before being cooled gradually to 4 °C, remaining for at least 15 min on a K960 Thermal Cycler (Heal Force Inc., China).Preparation of triple probe tTDN–MB satellites
The reactant material employed was a type of commercially available, 1 mm diameter magnetic bead that had the surface of the bead modified with SA (Fig. S5†). 100 μL of SA–MBs were pre-washed with TM buffer before usage and then mixed with 500 μL of prepared 2 M tTDN. The triple probe tTDN–MB satellites were produced when the mixture was incubated for one hour at room temperature with gentle shaking to immobilize DNA on the surface through a particular binding between SA and biotin on tTDN. The tTDN–MB satellites were then washed three more times with TMR buffer for later use after being rinsed with TM buffer to remove extra unreacted DNA. These tTDN–MB satellites in TMR were made up to a final volume of 1 mL in case of usage.Multiplex assay procedure of HV69/70 del, K417N, and T478K based on the tTDN–MNAzyme nanosystem
The multiplex assay procedure of HV69/70 del, K417N, and T478K through tTDN–MB satellites based on the tTDN–MNAzyme nanosystem was made up of several components. Partzyme A and partzyme B for every MNAzyme were previously dissolved in TMR buffer to 1 μM and 5 μL of each was taken; 10 μL of the prepared tTDN–MB satellites was adapted. The addition amount of the analyte samples with different concentrations is unified to 10 μL; finally the volume of the whole tTDN–MNAzyme nanosystem was replenished with TMR to 100 μL. At a temperature of 37 °C, the nanosystem was gently shaken for 3 h to fully react and make the isotope probe entirely sheared.Viral RNA extraction for the standard recovery experiment
Throat swab samples were used to create the real sample matrix for the recovery experiment, which was then purified using the EZ-10 spin column viral total RNA extraction kit. In the ESI,† specific experimental procedures are described.ICPMS measurements
After magnetic separation, the total volume of reactants and products in the supernatant was 100 μL per tube. It was then diluted with 1% HNO3 to a final volume of 1 mL and measured by ICPMS with Tb, Ho, and Tm isotopes in simultaneous detection mode.Results and discussion
Assay overview
Based on the constructed MNAzyme mediated tTDN–MB satellite model, we have created a multiplex detection approach for SARS-CoV-2 variant-related RNA fragments that transforms an RNA molecule into numbers of free lanthanides. The nano-satellite model consists of the MB core and tTDN scaffold with three MNAzyme modules (Fig. 1b). (1) MBs: they offer a platform for and a means of separating heterogeneous biological reactions, as well as a sizable specific surface area to facilitate maximal arrangement of tTDN structures. (2) tTDN: tTDN consists of four chains. One was designed with biotin modification at the 5′ end for connecting to SA–MBs (t-toehold), and at the same time, the toe chain was lengthened with 5 T bases, partially preventing SA–biotin binding from tTDN's steric hindrance. A branched tTDN (Fig. 1b), which exhibits a satellite-like structure on the surface of the MBs and acts as a stiff scaffold for functionalized branch chains to sprout, is made up of both the T-toehold and three other longer chains, t-substrate. Three t-substrates with middle-cleavage site modifications (rA) and 3′ ends modified with various lanthanides to produce appropriate ICPMS signals are present on each branching strand. (3) MNAzyme system: each MNAzyme system is made up of three parts: a pair of partzymes A & B, a substrate from the tTDN branch, and a target for initiating the assembly of the MNAzyme structure. When the analyte is present, the free partzymes A & B are drawn closer by the complementary pairing rule and further allow the other part of partzymes A & B to bind to the substrate.Under the synergistic influence of the Mg2+, the active center can cleave the tTDN branched substrate chain at the rA site. The MNAzyme structure is less sterically hindered by the tTDN structure and more likely to collide with the three t-substrates since there is a gap of 5 T bases between the sequences that participate in the structure and the sequences acting as substrates. Due to thermal instability, the sheared lanthanide-labelled DNA fragments are released into the solution, and the entire MNAzyme system can then be released once more, liberated, and located for the subsequent t-substrate for cleaving to provide isotope signals. As a result, three t-substrates serve as sites for the labelling of lanthanide tags, substrates for MNAzymes, and parts of the tTDN structure.
As triggers for the tTDN–MNAzyme nanosystem, we investigated three characteristic RNA sequences (HV69/70 del, K417N, and T478K) associated with four VOC SARS-CoV-2 variants. Three RNAs were concurrently added to the nanosystem containing tTDN, as seen in Fig. 1c, and this initiated the reaction, releasing significant amounts of the three lanthanides into the supernatant. ICPMS was used to identify the three lanthanide signals. For the purpose of identifying and determining SARS-CoV-2 variations, HV69/70 del is the indicative gene of the alpha variant, K417N is descriptive of the beta variant, and T478K is symptomatic for the gamma variant. It is noteworthy that all the aforementioned mutation sites are present in the recently discovered omicron variants. As a result, we can quantify the characteristic sequence of the variation and identify the relevant SARS-CoV-2 variants based on the combination and intensity of the isotope signals in ICPMS.
Characterization of the tTDN–MB satellites
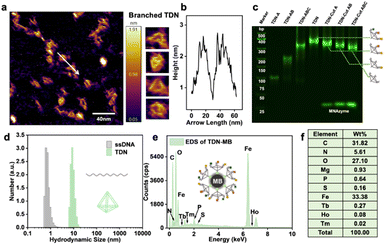
To verify the successful formation of tTDN, we characterized by atomic force microscopy (AFM) (Fig. 2a), PAGE, and dynamic light scattering (DLS) (Fig. 2d and S4†). In PAGE, a single strand of tTDN building blocks, a dimer and a trimer were adapted with the same annealing procedure as tTDN formation. The gradual increase in molecular weight demonstrated the binding of several strands (Fig. 2c). Furthermore, in the AFM image, ssDNA is dispersed and soft, while tTDN has a certain rigidity and maintains a three-dimensional structure (Fig. S3†). In a finer view, the tetrahedral structure can be clearly observed (Fig. 2a). The cross-sectional profile of the tTDN, which was taken along the white line, shows a clear height variation at the location of the tTDN (Fig. 2b). The aqueous ssDNA solution's hydrated particle size is approximately 1 nm, but after combining with tTDN, the hydrated particle size increases to 10 nm. In Fig. 2d and S4,† the particle size distribution indicates that the synthesized tTDN is comparatively pure and homogeneous. Evidence demonstrates the formation of tTDN from a more intuitive perspective.The MB ingredients are Fe3O4 with SA on the surface; tTDNs are immobilized onto the MBs through the SA–biotin linkage. The sphere becomes positively charged overall thanks to SA, and after tTDN was attached, its negative charge (essentially as DNA) replaces the positive charge on the particle surface (Fig. S6†). In isotope analysis by energy dispersive spectroscopy (EDS) (Fig. 2e, f and S7†), MBs contain a large amount of C, N, O, P, S and Fe elements. When the isotope-labeled tTDN was coupled to the MBs, we discovered newly emerged elements: Mg, Tb, Ho and Tm. This not only demonstrates the labeling of the single strand by the lanthanide isotope but also the successful assembly of the four single strands into tTDN and their further immobilization.
To test the feasibility of the developed tTDN–MNAzyme nanosystem, ICPMS detection under different conditions was done (Fig. S14†). In the absence of ingredients, no reaction and release of isotope tags will occur. Only when partzymes A & B exist and accompanied by Mg2+ ions, the tTDN–MB satellite will release the isotope tag under the initiation of the target. And the occurrence of the reaction depends on the type and quantity of the target. In Fig. 2c, the branching t-substrate chains of TDNs were spliced by 1–3 sites when we introduced the same amount of 1–3 kinds of variant RNAs, and the molecular weight of tTDN dropped with the number of splices. This demonstrates the feasibility of the tTDN–MNAzyme nanosystem to operate from single to multiple components.
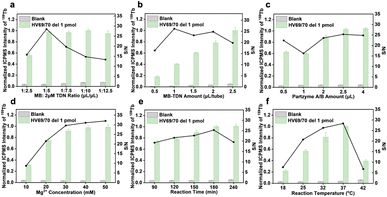
Optimization of experimental conditions
In theory, each type of capture DNA strand accounted for one-third of the total attached capture DNA. Therefore, we took one component as an indicator, optimized the relevant experimental conditions, and extended the results to the three-component analysis. The main contents include the assembly process of tTDN and MBs and the reaction conditions of MNAzyme.Analytical performance
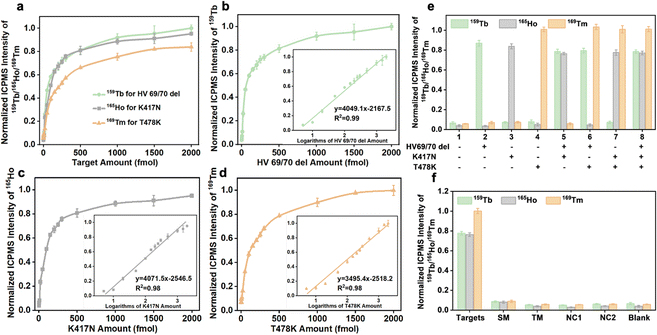
The analytical performance of the established analytical method after optimizing a number of reaction conditions was studied. In the simultaneous detection strategy, the calibration curves were constructed utilizing varying amounts of RNA fragments (HV69/70 del, K417N, and T478K) and ICPMS signal intensities of lanthanide isotopes (159Tb/165Ho/169Tm). Three different target types can be combined and quantified, as seen in Fig. 4a, indicating similar tendencies and performance. After executing logarithmic processing and linear fitting of the data, a good linear relationship for HV69/70 del, K417N, and T478K was found at 5–2000 fmol in the inset figures of Fig. 4b–d, and the correlation equations are shown in Table 1. The limits of detection calculated from 3σ/k (n = 10) for HV69/70 del, K417N, and T478K were 1.2, 1.5, and 1.1 fmol, respectively.| Target | Linear range (fmol) | Linear relationship | R 2 | LOD (fmol) |
|---|---|---|---|---|
| HV69/70 del | 5–2000 |
y = 1758.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(x) − 1881.6 log(x) − 1881.6 |
0.9901 | 1.2 |
| K417N | 5–2000 |
y = 1768.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(x) − 1525.5 log(x) − 1525.5 |
0.9784 | 1.5 |
| T478K | 5–2000 |
y = 1518.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(x) − 977.21 log(x) − 977.21 |
0.9824 | 1.1 |
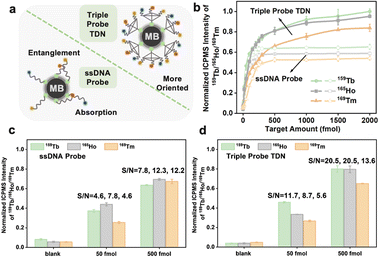
The branched tTDN not only participates in the multi-component reaction but also acts as a rigid molecular scaffold, making the overhang probe more oriented. This is because it spatially keeps the probe away from the MB surface, avoiding the electrostatic adsorption of long ssDNA on the MB surface to a certain extent (Fig. 5a). Obviously, the tTDN-based MNAzyme multiplex strategy possesses better analytical performance than the ssDNA probe. Fig. 5b displays how they performed in the wide range of multi-component detection. We discovered that the isotope signal growth rates of the two are comparable at low analyte concentrations. This is due to their adequate quantity, which facilitates a positive shift in the reaction equilibrium, which is significantly more than the analyte. Above 250 fmol of targets, the ssDNA probes were plainly not enough to handle more targets, and the reaction quickly reached a plateau since it was unable to carry out biological duties as a result of adsorption and entanglement. tTDN's triple probe overhang, however, is supported by a rigid scaffold that is less constrained than ssDNA, allowing it to continue to react at a certain rate, endowing larger incremental for the signal. The tTDN probe strategy in Fig. 5d performed better from a S/N perspective than that of triple ssDNA probes in Fig. 5c, particularly at 500 fmol, where the S/N was noticeably enhanced.
| Sample | Target | Added (fmol) | Found (fmol) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| 1# | HV69/70 del | 0 | Not found | — | 2.9 |
| 50 | 46.8 | 93.6 | 3.8 | ||
| K417N | 0 | Not found | — | 5.6 | |
| 50 | 47.9 | 95.8 | 6.9 | ||
| T478K | 0 | Not found | — | 5.6 | |
| 50 | 45.9 | 91.9 | 7.0 | ||
| 2# | HV69/70 del | 0 | Not found | — | 3.9 |
| 150 | 151.5 | 100.9 | 2.6 | ||
| K417N | 0 | Not found | — | 1.2 | |
| 150 | 152.9 | 101.9 | 2.1 | ||
| T478K | 0 | Not found | — | 2.1 | |
| 150 | 154.8 | 103.2 | 3.9 | ||
| 3# | HV69/70 del | 0 | Not found | — | 0.7 |
| 500 | 493.3 | 98.7 | 0.9 | ||
| K417N | 0 | Not found | — | 1.7 | |
| 500 | 502.8 | 100.6 | 0.8 | ||
| T478K | 0 | Not found | — | 1.4 | |
| 500 | 488.9 | 97.8 | 0.5 |
Conclusions
To address the general problem of the lack of multi-component ability of detection methods for SARS-CoV-2 variants, we have established an isotope encoded tTDN–MNAzyme satellite strategy. Specifically, adopting a multi-component MNAzyme reaction system operated by tTDN with rigid molecular scaffolding and multiple vertex modification properties, tTDN–MBs assembled as a satellite structure, releasing 159Tb, 165Ho, and 169Tm lanthanide isotope tags out of orbit, corresponding to the HV69/70 del, K417N and T478K characteristic gene sites of four VOC SARS-CoV-2 variants. The established method endows a sensitivity of 1.1–1.5 fmol, and a wide quantitative range of 5–2000 fmol. We anticipate that this highly sensitive, multi-component method can be extended to more applications for high throughput detection of biological samples.Data availability
The datasets supporting this article have been uploaded as part of the ESI.† ESI† contains detailed information of apparatus and reagents, detailed experimental protocols of AFM analysis, dynamic light scattering analysis, zeta-potential analysis, isotope labeling, PAGE, and RNA extraction. Operating parameters of ICPMS (Table S1†), nucleotide sequences (Table S2†), ESI-MS of MMA-DOTA-Ln3+ (Fig. S1†), MALDI-TOF-MS of DNA-MMA-DOTA-Ln3+ (Fig. S2†), AFM images (Fig. S3†), DLS of TDN (Fig. S4†), TEM of MBs (Fig. S5†), zeta-potential of MBs (Fig. S6†), EDS of MBs (Fig. S7†), calibration curves of isotopes in ICPMS (Fig. S8†), and optimization of experimental conditions (Fig. S9–S14†).Author contributions
Yi Lv conceptualized the project methodology. Yi Lv and Rui Liu supervised the investigation. Ziyan Li implemented, collected and analyzed the experimental data and wrote the original manuscript. Chaoqun Wang and Jing Zhou performed the validation and sample analysis. Yi Lv, Rui Liu, and Jianyu Hu revised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 22074098 & 22074096), the Science & Technology Department of Sichuan Province (2021ZYD0047) and the Fundamental Research Funds for the Central Universities. We thank Prof. Yunfei Tian, Dr. Xiaobo Xie and Dr. Shuguang Yan of the Analytical & Testing Center of Sichuan University for technical assistance.References
- W. J. Guan, Z. Y. Ni, Y. Hu, W. H. Liang, C. Q. Ou, J. X. He, L. Liu, H. Shan, C. L. Lei, D. S. C. Hui, B. Du, L. J. Li, G. Zeng, K. Y. Yuen, R. C. Chen, C. L. Tang, T. Wang, P. Y. Chen, J. Xiang, S. Y. Li, J. L. Wang, Z. J. Liang, Y. X. Peng, L. Wei, Y. Liu, Y. H. Hu, P. Peng, J. M. Wang, J. Y. Liu, Z. Chen, G. Li, Z. J. Zheng, S. Q. Qiu, J. Luo, C. J. Ye, S. Y. Zhu and N. S. Zhong, N. Engl. J. Med., 2020, 382, 1708–1720 CrossRef CAS PubMed.
- T. P. Peacock, D. H. Goldhill, J. Zhou, L. Baillon, R. Frise, O. C. Swann, R. Kugathasan, R. Penn, J. C. Brown, R. Y. Sanchez-David, L. Braga, M. K. Williamson, J. A. Hassard, E. Staller, B. Hanley, M. Osborn, M. Giacca, A. D. Davidson, D. A. Matthews and W. S. Barclay, Nat. Microbiol., 2021, 6, 899–909 CrossRef CAS PubMed.
- S. Ramesh, M. Govindarajulu, R. S. Parise, L. Neel, T. Shankar, S. Patel, P. Lowery, F. Smith, M. Dhanasekaran and T. Moore, Vaccines, 2021, 9(10), 1195 CrossRef CAS PubMed.
- J. Ou, W. Lan, X. Wu, T. Zhao, B. Duan, P. Yang, Y. Ren, L. Quan, W. Zhao, D. Seto, J. Chodosh, Z. Luo, J. Wu and Q. Zhang, Signal Transduction Targeted Ther., 2022, 7, 138 CrossRef CAS PubMed.
- M. Takeda, Microbiol. Immunol., 2022, 66, 15–23 CrossRef CAS PubMed.
- H. Yousefi, A. Mahmud, D. Chang, J. Das, S. Gomis, J. B. Chen, H. Wang, T. Been, L. Yip, E. Coomes, Z. Li, S. Mubareka, A. McGeer, N. Christie, S. Gray-Owen, A. Cochrane, J. M. Rini, E. H. Sargent and S. O. Kelley, J. Am. Chem. Soc., 2021, 143, 1722–1727 CrossRef CAS PubMed.
- C. B. Jackson, M. Farzan, B. Chen and H. Choe, Nat. Rev. Mol. Cell Biol., 2022, 23, 3–20 CrossRef CAS PubMed.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. C. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. W. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS PubMed.
- M. Gwinn, D. MacCannell and G. L. Armstrong, J. Am. Med. Assoc., 2019, 321, 893–894 CrossRef PubMed.
- K. Karasawa and H. Arakawa, Luminescence, 2022, 37, 822–827 CrossRef CAS PubMed.
- S. A. Byrnes, R. Gallagher, A. Steadman, C. Bennett, R. Rivera, C. Ortega, S. T. Motley, P. Jain, B. H. Weigl and J. T. Connelly, Anal. Chem., 2021, 93, 4160–4165 CrossRef CAS PubMed.
- A. Y. Trick, F. E. Chen, L. B. Chen, P. W. Lee, A. C. Hasnain, H. H. Mostafa, K. C. Carroll and T. H. Wang, Adv. Mater. Technol., 2022, 7(6), 2101013 CrossRef CAS PubMed.
- R. Liu, S. Zhang, C. Wei, Z. Xing, S. Zhang and X. Zhang, Acc. Chem. Res., 2016, 49, 775–783 CrossRef CAS PubMed.
- M. M. Aynaud, J. J. Hernandez, S. Barutcu, U. Braunschweig, K. Chan, J. D. Pearson, D. Trcka, S. L. Prosser, J. Kim, M. Barrios-Rodiles, M. Jen, S. Song, J. Shen, C. Bruce, B. Hazlett, S. Poutanen, L. Attisano, R. Bremner, B. J. Blencowe, T. Mazzulli, H. Han, L. Pelletier and J. L. Wrana, Nat. Commun., 2021, 12, 1405 CrossRef CAS PubMed.
- A. Babiker, C. W. Myers, C. E. Hill and J. Guarner, Am. J. Clin. Pathol., 2020, 153, 706–708 CrossRef CAS PubMed.
- M. M. Hernandez, R. Banu, P. Shrestha, A. S. Gonzalez-Reiche, A. van de Guchte, K. Farrugia, R. Sebra, M. R. Gitman, M. D. Nowak, C. Cordon-Cardo, V. Simon, H. van Bakel, E. M. Sordillo, N. Luna, A. Ramirez, S. A. Castaneda, L. H. Patino, N. Ballesteros, M. Munoz, J. D. Ramirez, A. E. Paniz-Mondolfi and M. S. P. S. Grp, Microbiol. Spectrum, 2022, 10(5), e01736–22 Search PubMed.
- R. J. Dikdan, S. A. E. Marras, A. P. Field, A. Brownlee, A. Cironi, D. A. Hill and S. Tyagi, J. Mol. Diagn., 2022, 24, 309–319 CrossRef CAS PubMed.
- G. Han, S. Zhang, Z. Xing and X. Zhang, Angew. Chem., Int. Ed. Engl., 2013, 52, 1466–1471 CrossRef CAS PubMed.
- J. Hu, R. Liu, J. Zhou and Y. Lv, Chem. Sci., 2021, 12, 13404–13412 RSC.
- X. W. Zhang, M. X. Liu, M. Q. He, S. Chen, Y. L. Yu and J. H. Wang, Anal. Chem., 2021, 93, 6437–6445 CrossRef CAS PubMed.
- Z. Li, X. Chen, Z. Huang, J. Zhou, R. Liu and Y. Lv, Anal. Chem., 2021, 93, 12714–12722 CrossRef CAS PubMed.
- Z. Q. Deng, J. Y. Hu, R. Liu and Y. Lv, At. Spectrosc., 2022, 43, 201–206 CAS.
- Y. Zhou, S. Yuan, K. K. To, X. Xu, H. Li, J. P. Cai, C. Luo, I. F. Hung, K. H. Chan, K. Y. Yuen, Y. F. Li, J. F. Chan and H. Sun, Chem. Sci., 2022, 13, 3216–3226 RSC.
- C. Q. Wang, Z. Q. Deng, H. Zhang, R. Liu and Y. Lv, Chin. Chem. Lett., 2022, 33, 1267–1270 CrossRef CAS.
- G. Han, M. H. Spitzer, S. C. Bendall, W. J. Fantl and G. P. Nolan, Nat. Protoc., 2018, 13, 2121–2148 CrossRef CAS PubMed.
- Z. Y. Yuan, Q. M. Zhou, L. S. Cai, L. Pan, W. L. Sun, S. W. Qumu, S. Yu, J. X. Feng, H. S. Zhao, Y. C. Zheng, M. L. Shi, S. Li, Y. Chen, X. R. Zhang and M. Q. Zhang, Nat. Methods, 2021, 18, 1223 CrossRef CAS PubMed.
- M. Lin, J. Wang, G. Zhou, J. Wang, N. Wu, J. Lu, J. Gao, X. Chen, J. Shi, X. Zuo and C. Fan, Angew. Chem., 2015, 127, 2179–2183 CrossRef.
- J. Wang, D. X. Wang, J. Y. Ma, Y. X. Wang and D. M. Kong, Chem. Sci., 2019, 10, 9758–9767 RSC.
- R. P. Goodman, I. A. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Science, 2005, 310, 1661–1665 CrossRef CAS PubMed.
- J. Yan, Z. Zhang, X. Zhan, K. Chen, Y. Pu, Y. Liang and B. He, Nanoscale, 2021, 13, 9577–9589 RSC.
- J. Li, K. Xun, L. Zheng, X. Peng, L. Qiu and W. Tan, J. Am. Chem. Soc., 2021, 143, 4585–4592 CrossRef CAS PubMed.
- D. Wang, S. Li, Z. Zhao, X. Zhang and W. Tan, Angew. Chem., Int. Ed. Engl., 2021, 60, 15816–15820 CrossRef CAS PubMed.
- X. Chen, J. Huang, S. Zhang, F. Mo, S. Su, Y. Li, L. Fang, J. Deng, H. Huang, Z. Luo and J. Zheng, ACS Appl. Mater. Interfaces, 2019, 11, 3745–3752 CrossRef CAS PubMed.
- H. Pei, X. Zuo, D. Zhu, Q. Huang and C. Fan, Acc. Chem. Res., 2014, 47, 550–559 CrossRef CAS PubMed.
- P. He, W. Han, C. Bi, W. Song, S. Niu, H. Zhou and X. Zhang, ACS Nano, 2021, 15, 6961–6976 CrossRef CAS PubMed.
- F. Li, Y. Liu, Y. Dong, Y. Chu, N. Song and D. Yang, J. Am. Chem. Soc., 2022, 144, 4667–4677 CrossRef CAS PubMed.
- Y. Yang, C. Song, J. Zhang, J. Chao, H. M. Luong, Y. Zhao and L. Wang, Nanoscale, 2022, 14, 4538–4547 RSC.
- Q. Wang, Z. He, H. Zhu, W. Gao, N. Zhang, J. Li, J. Yan, B. He and X. Ye, J. Mater. Chem. B, 2022, 10, 438–449 RSC.
- L. Chen, S. Luo, Z. Ge, C. Fan, Y. Yang, Q. Li and Y. Zhang, Nano Lett., 2022, 22, 1618–1625 CrossRef CAS PubMed.
- R. Peng, X. Zheng, Y. Lyu, L. Xu, X. Zhang, G. Ke, Q. Liu, C. You, S. Huan and W. Tan, J. Am. Chem. Soc., 2018, 140, 9793–9796 CrossRef CAS PubMed.
- M. Lin, P. Song, G. Zhou, X. Zuo, A. Aldalbahi, X. Lou, J. Shi and C. Fan, Nat. Protoc., 2016, 11, 1244–1263 CrossRef CAS PubMed.
- P. Peng, Y. Du, J. Zheng, H. Wang and T. Li, Angew. Chem., Int. Ed. Engl., 2019, 58, 1648–1653 CrossRef CAS PubMed.
- T. Zhang, T. Tian and Y. Lin, Adv. Mater., 2021, e2107820, DOI:10.1002/adma.202107820.
- Q. Kang, M. He, B. Chen, G. Xiao and B. Hu, Anal. Chem., 2021, 93, 737–744 CrossRef CAS PubMed.
- J. Zhuang, J. Tan, C. Wu, J. Zhang, T. Liu, C. Fan, J. Li and Y. Zhang, Nucleic Acids Res., 2020, 48, 8870–8882 CrossRef CAS PubMed.
- X. Yan, L. Yang and Q. Wang, Angew. Chem., Int. Ed. Engl., 2011, 50, 5130–5133 CrossRef CAS PubMed.
- S. Wang, M. Xia, J. Liu, S. Zhang and X. Zhang, ACS Sens., 2017, 2, 735–739 CrossRef CAS PubMed.
- P. Miao, B. Wang, X. Chen, X. Li and Y. Tang, ACS Appl. Mater. Interfaces, 2015, 7, 6238–6243 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc01960h |
| This journal is © The Royal Society of Chemistry 2023 |