Open Access Article
Open Access ArticleChemical and biological studies on the soft coral Nephthea sp.†
Omnia Hesham Abdelhafez‡
a,
John Refaat Fahim‡ b,
Ramy R. El Masric,
M. Alaraby Salemd,
Samar Yehia Desoukeyb,
Safwat Ahmede,
Mohamed Salah Kamelab,
Sheila Marie Pimentel-Elardof,
Justin R. Nodwell
b,
Ramy R. El Masric,
M. Alaraby Salemd,
Samar Yehia Desoukeyb,
Safwat Ahmede,
Mohamed Salah Kamelab,
Sheila Marie Pimentel-Elardof,
Justin R. Nodwell f and
Usama Ramadan Abdelmohsen
f and
Usama Ramadan Abdelmohsen *ab
*ab
aDepartment of Pharmacognosy, Faculty of Pharmacy, Deraya University, 61111 New Minia, Egypt
bDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, 61519 Minia, Egypt. E-mail: usama.ramadan@mu.edu.eg; Fax: +20-86-2369075; Tel: +20-86-2369075
cDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, October University for Modern Sciences and Arts (MSA), Giza, Egypt
dSchool of Life and Medical Sciences, University of Hertfordshire hosted by Global Academic Foundation, New Administrative Capital, Cairo, Egypt
eDepartment of Pharmacognosy, Faculty of Pharmacy, Suez Canal University, 41522 Ismailia, Egypt
fDepartment of Biochemistry, University of Toronto, MaRS Centre West, Toronto, ON, Canada
First published on 5th July 2021
Abstract
Soft corals belonging to the family Nephtheidae have been appreciated as marine sources of diverse metabolites with promising anticancer potential. In view of that, the current work investigates the anti-proliferative potential of the crude extract, different fractions, and green synthesized silver nanoparticles (AgNPs) of the Red Sea soft coral, Nephthea sp. against a panel of tumor cell lines. The metabolic pool of the soft coral under study was also explored via an LC-HR-ESI-MS metabolomics approach, followed by molecular docking analysis of the characterized metabolites against the target proteins, EGFR, VEGFR, and HER2 (erbB2) that are known to be involved in cancer cell proliferation, growth, and survival. Overall, the n-butanol fraction of Nephthea sp. exhibited the highest inhibitory activities against MCF7 (breast cancer) and A549 (lung cancer) cell lines, with interesting IC50 values of 2.30 ± 0.07 and 3.12 ± 0.10 μg ml−1, respectively, whereas the maximum growth inhibition of HL60 (leukemia) cells was recorded by the total extract (IC50 = 2.78 ± 0.09 μg ml−1). More interestingly, the anti-proliferative potential of the total soft coral extract was evidently improved when packaged in the form of biogenic AgNPs, particularly against A549 and MCF7 tumor cells, showing IC50 values of 0.72 ± 0.06 and 9.32 ± 0.57 μg ml−1, respectively. On the other hand, metabolic profiling of Nephthea sp. resulted in the annotation of structurally diverse terpenoids, some of which displayed considerable binding affinities and molecular interactions with the studied target proteins, suggesting their possible contribution to the anti-proliferative properties of Nephthea sp. via inhibition of tyrosine kinases, especially the EGFR type. Taken together, the present findings highlighted the relevance of Nephthea sp. to future anticancer drug discovery and provided a base for further work on the green synthesis of a range of bioactive NPs from marine soft corals.
1. Introduction
Cancer is one of the major contributing diseases to human morbidity and mortality worldwide.1 About 21 million new cancer cases and 13 million deaths are estimated by 2030, which will exacerbate global health, social, and economic burdens.2 Yet, conventional anticancer therapies, including surgery, chemotherapy, and radiation have not satisfactorily brought the anticipated efficacy, showing both controversial safety and undesirable side effects, e.g. alopecia, liver and kidney dysfunction, anemia, and immunosuppression.3,4 Additionally, a wide variety of malignant tumors has been reported to develop multi-drug resistance, leading to chemotherapy failure, which reflects the unremitting need for other effective and advanced treatment options.5 In this framework, marine ecosystems have been acknowledged as potent supplies of anticancer leads because of their both inconceivable biodiversity and capacity to afford therapeutically useful molecules, some of which are in current clinical use.6,7 Among various marine invertebrates, soft corals belonging to the family Nephtheidae have received attention as special providers of several chemical compounds with antifouling, antibacterial, antiviral, anti-diabetic, and anti-inflammatory activities.8–10 Likewise, a group of assorted metabolites with praiseworthy potential against human cancer cells has been described from various Nephtheidae species,8,9,11–14 which valorizes the promising exploration of these prolific organisms in the realm of nature-inspired anticancer drug discovery.Recent advancements in nanotechnology have blossomed into a myriad of applications that hold the potential to revolutionize modern medicine, encompassing diagnosis, prevention, and remediation of cancer.15 In this respect, nanoparticles (NPs) have gained considerable interest over the last decade thanks to their privileged capacity to refine the compatibility, bioavailability, and efficacy of many synthetic drugs and natural products in the management of chronic disorders, including cancer.16,17 These particles show unique and markedly different chemical, optical, magnetic, mechanical, and biological properties to their larger material counterparts owing to their small sizes (1–100 nm) and high surface-to-volume ratio.18,19 Interestingly, a considerable number of NP-based remedies are now in clinical use, including the chemotherapeutic agents 5-fluorouracil, paclitaxel, and doxorubicin, which endorses the impactful role of this approach in boosting the therapeutic potential of bioactive agents.20 In this connection, metal-based NPs have been introduced as multifunctional tools with valued theranostic abilities and biomedical applications.17,21 This particular type of nanomaterials can act as new therapeutic agents or drug carriers with the advantage of eliciting more targeted activities that help avoid many unwelcome side effects.16,21 Out of various noble metal NPs, silver nanoparticles (AgNPs) have been reported to display remarkable chemical stability, catalytic activity, biocompatibility, and biological activities, including anti-inflammatory, antimicrobial, anti-diabetic, and wound healing properties, among others.18,21,22 They have also been broadly investigated in cancer research because of their notable cytotoxic and antitumor potential.17,21 Among the most striking advantages of AgNPs is their ability to release active ingredients to the anticipated site in a gradual and sustainable manner, leading to boosted efficacies; thus, their production is thought to accelerate in the next few years.18,22
On the other hand, despite their chemical and biological prolificacy, the biogenic synthesis of AgNPs using marine invertebrates has been scarcely explored,23 encompassing our recent report on the anti-inflammatory potential of the biosynthesized AgNPs from the soft coral Nephthea sp., which revealed greater anti-COX-2 properties compared with its bulk extracts.24 Therefore, in continuation of our interest in marine organisms and their bioactive NPs,24–28 the current study investigates the chemical complexity and anti-proliferative activity of the total extract and different fractions of Nephthea sp., along with the green synthesis of potential anti-proliferative AgNPs using the total soft coral extract. Molecular docking analysis was also considered in order to predict the possible binding of the identified metabolites to a number of cancer-related cellular proteins.
2. Materials and methods
2.1. Soft coral material
The animal material used in this work was collected from the Egyptian coasts of the Red Sea at Sharm El-Sheikh using scuba diving at a depth of 10 m by Dr Safwat Ahmed, Professor of Pharmacognosy, Faculty of Pharmacy, Suez Canal University, Ismailia, Egypt. The collected material was immediately frozen and kept at −20 °C until investigation. The soft coral biomass was identified by Dr Tarek Temraz, Marine Science Department, Faculty of Science, Suez Canal University, Ismailia, Egypt. A voucher specimen was deposited in the herbarium section of Pharmacognosy Department, Faculty of Pharmacy, Suez Canal University under the registration number SAA-26.2.2. Chemicals and reagents
Solvents used in this work were of analytical grade and distilled before use. They were obtained from El-Nasr Company for Pharmaceuticals and Chemicals, Egypt. Acetonitrile and methanol of HPLC grade were obtained from SDFCL SD Fine-Chem Limited, Mumbai, India. Both silver nitrate (AgNO3; purity ≥ 99.5%) and ion exchange resin were purchased from Sigma-Aldrich, Germany. Staurosporine (Sigma-Aldrich, Germany) was used as a standard drug in cytotoxicity studies.2.3. Extraction and fractionation
The freeze-dried soft coral was exhaustively extracted with methanol–methylene chloride (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)24,29,30 and the combined extracts were concentrated under reduced pressure until dryness. The obtained extract (24.0 g) was suspended in distilled water and extracted successively with petroleum ether, ethyl acetate, and n-butanol. The organic phase in each step was separately concentrated under vacuum to provide three corresponding fractions: I (10.0 g), II (3.0 g), and III (3.0 g). The remaining mother liquor was desalted using an ion exchange resin, followed by re-extraction with acetone. The latter was also concentrated under vacuum to afford fraction IV (200.0 mg). The crude extract and its fractions were kept at 4 °C for metabolomics and biological analyses.
1)24,29,30 and the combined extracts were concentrated under reduced pressure until dryness. The obtained extract (24.0 g) was suspended in distilled water and extracted successively with petroleum ether, ethyl acetate, and n-butanol. The organic phase in each step was separately concentrated under vacuum to provide three corresponding fractions: I (10.0 g), II (3.0 g), and III (3.0 g). The remaining mother liquor was desalted using an ion exchange resin, followed by re-extraction with acetone. The latter was also concentrated under vacuum to afford fraction IV (200.0 mg). The crude extract and its fractions were kept at 4 °C for metabolomics and biological analyses.
2.4. Metabolic profiling
Metabolic profiling of the total extract and different fractions of Nephthea sp. was carried out according to Abdelmohsen et al.31 using an Acquity Ultra Performance Liquid Chromatography system combined with a Xevo G2S-qToF quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, USA). Chromatographic separation was performed using a reversed phase BEH C18 column (2.1 × 100 mm, 1.7 μm particle size; Waters, Milford, USA) connected to a guard column (2.1 × 5 mm, 1.7 μm particle size; Waters, Milford, USA). The mobile phase used for chromatographic separation was composed of purified water (A) and acetonitrile (B); each containing 0.1% formic acid. Elution was started in the gradient manner at a flow rate of 200 μl min−1 with 10% B linearly increasing to 100% B within 30 min, and then kept isocratic for additional 5 min before linearly returning to 10% B for further 1 min. The injection volume was 2 μl, while the column temperature was adjusted at 40 °C. MSConvert software was employed to convert raw data into separate positive and negative ionization files, which were then exported to the data mining software, MZmine 2.10 for peak picking, deconvolution, deisotoping, alignment, and formula prediction.32 Dereplication of compounds was accomplished by comparison with the Dictionary of Natural Products (DNP), METLIN, and Marinlit databases.33–352.5. Synthesis and characterization of AgNPs
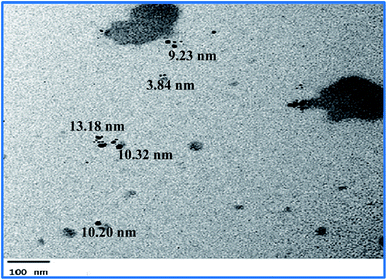
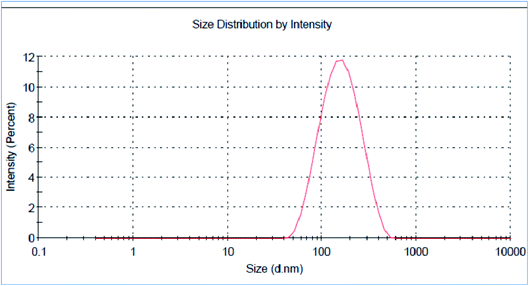
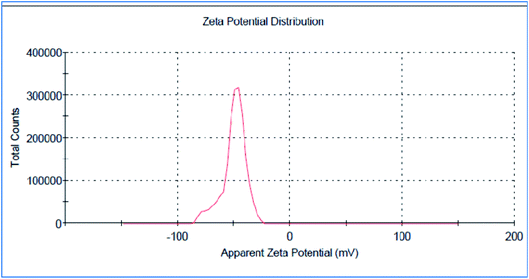
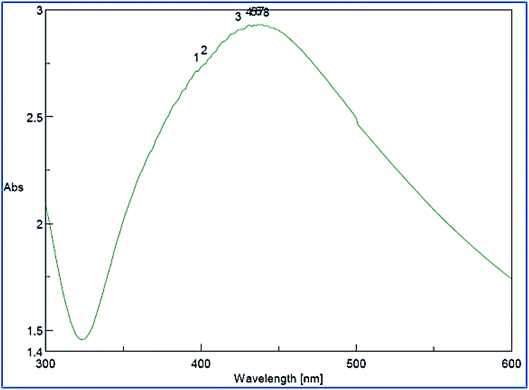
The green synthesized AgNPs were prepared by adding 3 ml of the 0.0005% crude extract of Nephthea sp. in DMSO to 100 ml of 1 mM silver nitrate solution at room temperature. Formation of nanoparticles was observed by the color change from colorless to a brown color and confirmed by measuring the UV-Vis spectrum of the reaction at 200–600 nm using a double beam V-630 spectrophotometer (Jasco, Japan). The shape and size of the resulting AgNPs were determined using a Transmission Electron Microscope (TEM; Jeol model JEM-1010, USA) by adding a drop on a copper grid covered with a carbon support film and left to completely dry at room temperature. Fourier-Transform Infrared Spectroscopy (FT-IR) analysis of AgNPs was performed using a FT-IR-8400S spectrophotometer (IR Prestige-21, IR Affinity-1, Shimadzu, Japan) to determine AgNPs associated biomolecules.24,36 Particle size distribution and polydispersity index (PDI) of the prepared NPs was measured in a disposable cell at 25 °C using a Zeta-sizer Nano ZS (Malvern instruments, UK) and the results were then analyzed by Zeta-sizer 7.01 software, UK.2.6. Anti-proliferative activity
The anti-proliferative potential of the total extract, different fractions, and AgNPs of Nephthea sp. was evaluated against three human tumor cell lines, namely breast cancer (MCF7), lung cancer (A549), and leukemia (HL60) cells via the MTT assay.37 Cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured using DMEM (Invitrogen/Life Technologies, USA) supplemented with 10% FBS (Hyclone, USA), 10 μg ml−1 of insulin (Sigma-Aldrich, Germany), and 1% penicillin-streptomycin. In 96-well plates, cells (at a density of 1.2–1.8 × 104 cells per well) were prepared in a volume of 100 μl per well of each of the complete growth medium and the tested sample for 24 h before testing. The MTT solution was reconstituted with 3 ml of the medium or a balanced salt solution without phenol red or serum and added in an amount equal to 10% of the culture medium volume. Cultures were incubated for 2–4 h depending on cell type and maximum cell density (an incubation period of 2 h was generally adequate, but was lengthened for either low cell densities or cells showing little metabolic activities). After incubation, the resulting formazan crystals were dissolved by adding a volume of DMSO equal to that of the original culture medium. The absorbance of each plate was then measured spectrophotometrically at 570 nm using an ELISA plate reader (Model 550, Bio-Rad, USA). Three independent experiments were performed. IC50 values, representing the concentration responsible for 50% inhibition of cell growth, were calculated by GraphPad Prism 5 (Version 5.01, GraphPad Software, San Diego, CA, USA).2.7. Molecular docking
Three crystal structures were selected to study the potential antitumor activity of the characterized compounds from Nephthea sp., including PDB ID: 1M17 (for the epidermal growth factor receptor (EGFR) tyrosine kinase), PDB ID: 4asd (for the vascular endothelial growth factor receptor (VEGFR) tyrosine kinase), and PDB ID: 3PP0 (for the kinase domain of human HER2 (erbB2)) using erlotinib, sorafenib, and SYR127063 as the co-crystallized ligands, respectively. For all dockings, a grid box with dimensions of 50 grid points and spacing of 0.375 was centered on the given co-crystallized ligand. Docking was performed via Autodock4 implementing 100 steps of genetic algorithm while keeping the default setting provided by Autodock Tools.38 Visualization was achieved using a discovery studio.39 In total, compounds (1–8) were docked into the three active sites in comparison with the three co-crystallized ligands that served as reference molecules.3. Results and discussion
To date, a variety of physical and chemical strategies has been described for the preparation of metal NPs; however, the release of hazardous chemicals and toxic byproducts during these practices have raised several environmental and health concerns.18,22 In contrast, biological methods of synthesis have emerged as less toxic, eco-friendly, and non-destructive techniques, with time- and cost-effective nature. Such green approaches have also efficiently allowed the incorporation of many plant extracts, bacteria, and fungi into various metal NPs in order to explore their biological potential.17,18,22 In this regard, the wide metabolic and chemical diversities of the aforementioned organisms have been reported to mediate the successful reduction, capping, and conversion of metal ions to metallic NPs with particular characteristics.18,22 In recent years, marine flora such as algae, marine bacteria, fungi, finfish, and sponges have been also researched as potential biological precursors for the fabrication of bioactive metal NPs owing to their content of various reductants and stabilizing agents.40,41 Nevertheless, a few reports exist in the literature on the synthesis of biogenic AgNPs using marine invertebrates, which have been mostly studied for their antimicrobial potential, exemplified by oyster (Saccostrea cucullata Born.),42 marine polychaete,23 and sponges, e.g. Acanthella elongate Dendy,43,44 Callyspongia diffusa Ridley,45 Haliclona spp.,46,47 Axinella sinoxia Alvarez & Hooper (Hamed et al., 2017),48 and Amphimedon sp.28 Moreover, we have also earlier reported the first biosynthesis of soft coral-based AgNPs using the petroleum ether and acetone fractions of Nephthea sp., along with their interesting anti-COX-2 activities.24 Inspired by this, the present work considered the green synthesis of AgNPs using the total extract of Nephthea sp., which were further tested for their anti-proliferative potential for the first time among various metal NPs derived from marine invertebrates.3.1. Synthesis and characterization of AgNPs
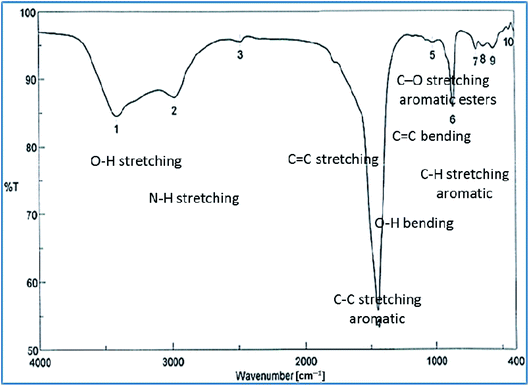
The successful synthesis of AgNPs using the total Nephthea sp. extract was evident by the development of dark brown color, while the control (1 mM AgNO3) solution did not show any color change (ESI Fig. S1†).36 The gradual color change was observed after 24 h of incubation and the fully dark brown color appeared after 48 h. Such change in color is attributable to the excitation of surface plasmon, its vibrations, as well as changes in the metal oxidation state (i.e. the reduction of Ag+ ions to Ag0 by various biomolecules present in the soft coral extract). Therefore, the intensity of the obtained brown color gradually increases as the reaction proceeds.18,22,49![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations of alkene groups. The peak at 1444.42 cm−1 is also assignable to C–C stretching in aromatic rings and O–H bending in carboxylic acids and alcohols, while the stretching of C–H in aromatic rings is responsible for those emerging at 877.452, 694.248, and 647.965 cm−1. Likewise, the peak at 972.912 cm−1 is indicative of C
C stretching vibrations of alkene groups. The peak at 1444.42 cm−1 is also assignable to C–C stretching in aromatic rings and O–H bending in carboxylic acids and alcohols, while the stretching of C–H in aromatic rings is responsible for those emerging at 877.452, 694.248, and 647.965 cm−1. Likewise, the peak at 972.912 cm−1 is indicative of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending of alkenes. Such data reflect the implication of different biomolecules in both the reduction of Ag+ ions and stabilization of the synthesized AgNPs, such as terpenoids, steroids, peptides, proteins, polysaccharides, and pigments.22,24,36,53
C bending of alkenes. Such data reflect the implication of different biomolecules in both the reduction of Ag+ ions and stabilization of the synthesized AgNPs, such as terpenoids, steroids, peptides, proteins, polysaccharides, and pigments.22,24,36,53
3.2. Metabolic profiling
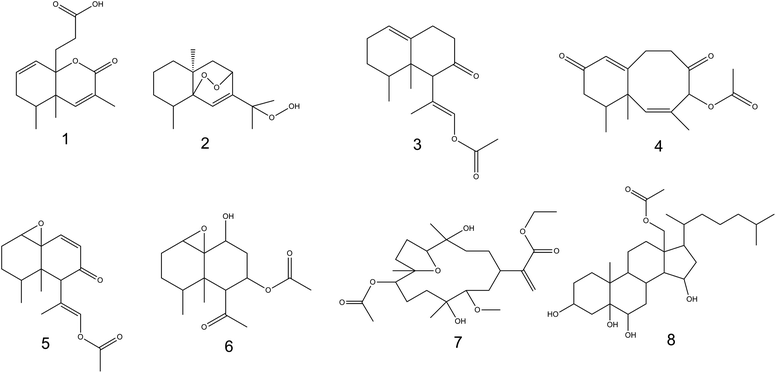
Metabolic profiling of Nephthea sp. using LC-HR-ESI-MS for dereplication purposes led to the annotation of a number of structurally varied secondary metabolites, among which, sesquiterpenoids were noticed to prevail (Fig. 6; ESI Table S1 and Fig. S2†). Identification of these metabolites was achieved via coupling MZmine with some common databases such as DNP, METLIN, and Marinlit. As a result, the mass ion peak at m/z 265.143, corresponding to the molecular formula C15H20O4, was characterized as philippinlin E (1). This 4,5-seconeolemnane sesquiterpene was previously obtained from the soft coral Lemnalia philippinensis that belongs to the family Nephtheidae.54 Moreover, the mass ion peak at m/z 267.159 in consonance with the suggested molecular formula C15H24O4 was annotated as 5,8-epidioxy-11-hydroperoxy-6-eudesmen (2); a sesquiterpenoid that was earlier isolated from Nephthea erecta Kükenthal.55 Another sesquiterpenoid of the nardosinan-type was also identified as paralemnolin L (3) in agreement with both the observed mass ion peak at m/z 276.196 and the molecular formula C17H24O3. Noteworthy, this molecule was previously described among the natural metabolites of the family Nephtheidae, namely the soft coral Paralemnalia thyrosides Ehrenberg.56 In the same context, the mass ion peak at m/z 289.141 was annotated as laevinone A (4) in harmony with the molecular formula C17H22O4. Laevinone A belongs to neolemnane sesquiterpenoids and was formerly reported from Lemnalia laevis Thomson and Dean.57 Furthermore, two nardosinane-type sesquiterpenoids with the molecular formulas C17H22O4 and C16H24O5 were characterized as nardosinanol F (5) and nardosinanol I (6) based on the observed mass ion peaks at m/z 291.158 and 297.151, respectively. Both compounds were identified from some members within the Nephtheidae family, including Paralemnalia clavata Verseveldt and Lemnalia africana May, respectively.58Aside from the abovementioned metabolites, two structurally different terpenoids were also described comprising dendronpholide N (7); a cembrane type-diterpene and the polyhydroxylated sterol, dendronesterol B (8), which were annotated from the mass ion peaks at m/z 469.262 and 494.361 and the corresponding molecular formulas C25H42O8 and C29H50O6, respectively. Both compounds (7) and (8) were earlier reported from the two Nephtheidae soft corals, Dendronephthya sp. and Dendronephthya gigantea Verrill, respectively.59,60 According to the reported literature,8,9 all the characterized metabolites are firstly reported herein from soft corals of the genus Nephthea, except for compound (2).
3.3. Anti-proliferative activity
Marine soft corals are endowed with the ability to accumulate several bioactive molecules that can serve as anticancer drug leads, exemplified by (18S)-18-O-acetyl nephthoacetal; a pentacyclic sterol isolated from Nephthea sp., which showed potent in vitro anti-proliferative activity against human cervical cancer (HeLa) cells (IC50 = 10.1 μg ml−1).13 Sclerosteroid A is another pregnane-type steroid identified from Scleronephthya gracillimum Kükenthal with notable inhibitory potential towards human liver (HepG2) and breast (MDA-MB-231) cancer cells (IC50 = 19.5 and 15.8 μM, respectively).12 Lemnaphilin A was also reported from the Formosan soft coral, Lemnalia philippinensis May as potent cytotoxic sesquiterpenoid against a number of tumor cells, namely HepG2, MDA-MB231, and A549 cell lines, with IC50 values of 15.99, 16.31, and 15.81 μg ml−1, respectively.14 In the same context, members of the genus Nephthea have provided a range of diversified metabolites, mostly dominated by steroids and terpenoids that have been described as the major contributors to their promising anticancer properties.8,61 In view of that, it was of interest to evaluate the anti-proliferative potential of the total extract and different fractions of Nephthea sp. using the MTT assay in comparison with the reference drug, staurosporine. Overall, the tested samples revealed varying in vitro growth inhibitory potencies against MCF7, A549, and HL60 tumor cells, showing IC50 values in the range of 2.30 ± 0.07 to 141.8 ± 0.46 μg ml−1 (Table 1). According to the US NCI (National Cancer Institute) screening guidelines, the majority of the tested extracts from Nephthea sp. exhibited IC50 values less than 20 μg ml−1 (Table 1), indicating their potent anti-proliferative effects.25,54 As depicted in Table 1, the n-butanol fraction exerted the greatest inhibitory activity against MCF7 cells, followed by the ethyl acetate fraction, total extract, and acetone fraction, with interesting IC50 values of 2.30 ± 0.07, 9.96 ± 0.32, 13.95 ± 0.45, and 15.25 ± 0.50 μg ml−1, respectively, which were even more potent than staurosporine (IC50 = 20.30 ± 0.6 μg ml−1). The petroleum ether fraction, on the other hand, showed weaker cytotoxicity against MCF7 cells (IC50 = 27.45 ± 0.90 μg ml−1). Similarly, the n-butanol fraction displayed higher anti-proliferative effects against A549 cells (IC50 = 3.12 ± 0.10 μg ml−1) compared with staurosporine (IC50 = 27.30 ± 0.80 μg ml−1), followed by the acetone and petroleum ether fractions (IC50 = 9.21 ± 0.30 and 11.32 ± 0.30 μg ml−1, respectively), whereas both the ethyl acetate fraction and the total extract had much weaker potential against A549 cells (IC50 = 66.40 ± 2.18 and 84.87 ± 2.70 μg ml−1, respectively). On the other hand, the total soft coral extract exhibited the maximum growth inhibitory potential against HL60 cells (IC50 = 2.78 ± 0.09 μg ml−1), followed by staurosporine (IC50 = 18.17 ± 0.59 μg ml−1), while the ethyl acetate and petroleum ether fractions were moderately active (IC50 = 23.74 ± 0.78 and 31.22 ± 1.02 μg ml−1, respectively). Contrary to this, both the n-butanol and acetone fractions showed very weak activities towards the HL60 cell line (IC50 = 80.45 ± 2.60 and 141.8 ± 0.46 μg ml−1, respectively) (Table 1).| Sample | IC50 (μg ml−1) | ||
|---|---|---|---|
| A549 | MCF7 | HL60 | |
| Total extract | 84.87 ± 2.70 | 13.95 ± 0.45 | 2.78 ± 0.09 |
| Petroleum ether fraction | 11.32 ± 0.30 | 27.45 ± 0.90 | 31.22 ± 1.02 |
| Ethyl acetate fraction | 66.40 ± 2.18 | 9.96 ± 0.32 | 23.74 ± 0.78 |
| n-Butanol fraction | 3.12 ± 0.10 | 2.30 ± 0.07 | 80.45 ± 2.60 |
| Acetone fraction | 9.21 ± 0.30 | 15.25 ± 0.50 | 141.8 ± 0.46 |
| AgNPs of the total extract | 0.72 ± 0.06 | 9.32 ± 0.57 | 10.07 ± 0.71 |
| Staurosporine | 27.30 ± 0.80 | 20.30 ± 0.60 | 18.17 ± 0.59 |
Recent years have witnessed an increasing amount of research on the anticancer perspective of nano-sized natural products, with those packaged as AgNPs revealed a promising potential against various tumor cells both in vitro and in vivo, including cervical, breast, liver, lung, nasopharyngeal, colorectal, and prostate cancers, among others.17,21,62 These natural product-based AgNPs have also been proven to exert advanced pharmacological and cytological effects in comparison with the chemical entities they contain thanks to their superior physicochemical and surface properties, considerable loading ability of natural compounds, and significant cellular interactions, which reflect their possible application in nano-chemoprevention and nano-chemotherapy.17,18 In line with the reported literature,17,21,62 our results unveiled the noteworthy anti-proliferative potential of the synthesized AgNPs from the total extract of Nephthea sp. as inferred from their higher growth inhibitory activities against A549 and MCF7 cell lines compared with the bulk total extract and staurosporine; showing IC50 values of 0.72 ± 0.06 and 9.32 ± 0.57 μg ml−1, respectively (Table 1). These findings therefore imply the possible exceptional role of metallic NPs in augmenting the anticancer potential of Nephthea sp. Thus far, several mechanisms have been proposed to mediate the cytotoxic effects of biogenic AgNPs on cancerous cells, encompassing activation of apoptotic pathways by destroying the ultrastructure of tumor cells, disruption of mitochondrial functions via stimulating the production of reactive oxygen species, and suppression of ATP synthesis, leading to DNA damage. Their ability to target some cellular enzymes and signaling pathways has been also testified in a variety of tumor cells.17,21,62
3.4. Molecular docking
Tyrosine kinases are pivotal mediators of intracellular signaling, showing several key roles in different cellular events, such as growth, differentiation, metabolism, and apoptosis. Though their activity is strictly controlled in normal cells, the over-activation of these entities due to the expression of mutated kinase genes can embolden carcinogenesis.63 Hence, a series of studies have reported the implication of tyrosine kinases in cancer pathogenesis and their potential as propitious anticancer drug targets.64 Members of the EGFR family, such as EGFR and HER2 (erbB2) are currently among the most targeted tyrosine kinases owing to their regulatory roles in some complex signal transduction networks.63,65 Both EGFR and HER2 are also known to be amplified in many cancers, including breast, lung, and colorectal tumors. Such overexpression can halt apoptosis, resulting in unregulated cell cycle, proliferation, invasion, and neovascularization.63,66 In the interim, the proangiogenic factor, VEGF plays an important role in inducing and regulating angiogenesis, which underwrites tumor survival and metastasis. The overexpression of VEGFR tyrosine kinase was also seen to hasten the onset and development of cancer; consequently, blocking of angiogenesis via VEGF suppression represents another attractive target in cancer therapy.67 For this, a range of anticancer agents has been developed to counteract the actions of EGFR, HER2, and VEGF; however, multiple therapeutic downsides and off-target effects have hampered their efficacies.63 In this context, accumulated data have shown the capacity of natural products to modulate or inhibit protein kinases, providing a cornucopia of antitumor drug candidates.68 In light of this as well as the observed anti-proliferative potential of Nephthea sp., the current docking approach was considered to investigate the possible interactions of compounds (1–8) with the above-mentioned cellular proteins in comparison with a number of specific kinase inhibitors. As shown in Table 2, the docked compounds formed considerably stable complexes with 1M17 with markedly comparable binding scores (−6.7 to −7.9 kcal mol−1) to that of the co-crystallized ligand, erlotinib (−7.8 kcal mol−1). Among them, compounds (2, 3, 4, and 5) revealed the highest binding affinities within the active site of 1M17 (Table 2). Erlotinib is an EGFR inhibitor that is used for pancreatic cancer, non-small cell lung cancer, and some other tumor types.69 This drug is known to interact with the gatekeeper Thr766 via a water-bridged H-bond, in addition to hydrogen bonding with the amino acid Met769 in the hinge region of EGFR tyrosine kinase.70 Interestingly, some of the tested metabolites herein displayed analogous interactions to those of erlotinib, exemplified by compounds (3) and (5) that formed hydrogen bonds with Met769. Likewise, some of the hydrophobic contacts exhibited by erlotinib were also reproduced by our compounds, including the interactions of compounds (3) and (5) with Ala719 and those of (3) with Leu764 as shown in Fig. 7. On the other hand, the binding energy scores of compounds (1–8) in case of 3pp0 and 4asd were relatively lower than the co-crystallized ligands, SYR127063 (−10.0 kcal mol−1) and sorafenib (−11.8 kcal mol−1), respectively (Table 2), of which compounds (2) and (4) showed the highest binding aptitudes in terms of their energy scores (−8.3 and −8.7 kcal mol−1 for 3pp0 and −8.1 and −7.7 kcal mol−1 for 4asd, respectively). These results collectively suggest the contribution of the characterized metabolites to the observed anti-proliferative activities of Nephthea sp. that might involve the inhibition of tyrosine kinases, particularly the EGFR type. In this connection, different receptor–ligand interactions displayed by compounds (1–8) could also offer better insights to design alternative anticancer agents with fewer unwanted effects.| Compound | 1M17 | 3pp0 | 4asd |
|---|---|---|---|
| Co-crystallized ligand | −7.8 | −10.0 | −11.8 |
| Philippinlin E (1) | −7.0 | −7.4 | −7.0 |
| 5,8-Epidioxy-11-hydroperoxy-6-eudesmen (2) | −7.7 | −8.3 | −8.1 |
| Paralemnolin L (3) | −7.7 | −7.4 | −7.5 |
| Laevinone A (4) | −7.9 | −8.7 | −7.7 |
| Nardosinanol F (5) | −7.7 | −7.1 | −6.4 |
| Nardosinanol I (6) | −7.1 | −7.3 | −6.6 |
| Dendronpholide N (7) | −7.3 | −7.5 | −7.3 |
| Dendronesterol B (8) | −6.7 | −6.3 | −6.9 |
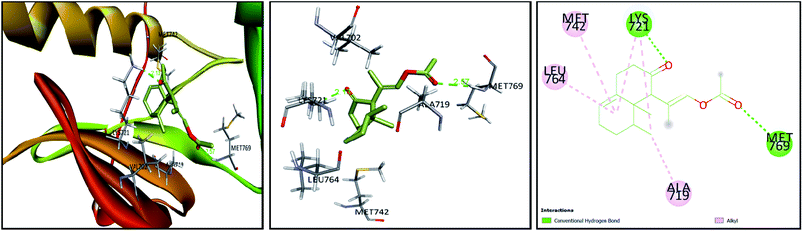 | ||
| Fig. 7 3D and 2D plots of the poses of compound 3 at the active site of EGFR tyrosine kinase (PBD:1M17). | ||
4. Conclusion
The current study highlighted the anti-proliferative potential of the total extract and different fractions of the soft coral Nephthea sp., which showed notable inhibitory activities against MCF7, A549, and HL60 tumor cells. Such effects are likely underlain by the availability of a range of compounds, mostly terpenoids that were mined with the help of LC-MS-based metabolomics. Comparative docking screening of the characterized metabolites also revealed their ability to interact with the active sites of EGFR, HER2, and VEGF, denoting their probable contribution to the anti-proliferative potential of Nephthea sp. as tyrosine kinase inhibitory molecules, particularly compounds (2) and (4). Additionally, our results provided evidence for the interesting role of biogenic AgNPs in enhancing the anticancer properties of Nephthea sp., which were addressed herein for the first time among marine invertebrates. The obtained findings drew attention to the potential of Nephthea sp. to provide the current therapeutic arsenal against cancer with alternative agents of natural origin, offering a good starting point for future research on the development of nanoparticle-based chemotherapies using marine soft corals.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to Dr Khayrya A. Youssif, Department of Pharmacognosy, Faculty of Pharmacy, Modern University for Technology and Information, Cairo, Egypt, for her kind help in the green synthesis of nanoparticles.References
- J. T. McDaniel, K. Nuhu, J. Ruiz and G. Alorbi, Glob. Health Promot., 2019, 26, 41–49 CrossRef.
- L. A. Torre, R. L. Siegel, E. M. Ward and A. Jemal, Cancer Epidemiol. Biomark. Prev., 2016, 25, 16–27 CrossRef.
- V. Schirrmacher, Int. J. Oncol., 2019, 54, 407–419 CAS.
- E. K. Davison and M. A. Brimble, Curr. Opin. Chem. Biol., 2019, 52, 1–8 CrossRef CAS.
- H. Choudhury, M. Pandey, T. H. Yin, T. Kaur, G. W. Jia, S. L. Tan, H. Weijie, E. K. S. Yang, C. G. Keat and S. K. Bhattamishra, Mater. Sci. Eng., C, 2019, 101, 596–613 CrossRef CAS.
- G. Ercolano, P. De Cicco and A. Ianaro, Mar. Drugs, 2019, 17, 31 CrossRef CAS.
- S.-K. Kim, Handbook of Anticancer Drugs from Marine Origin, Springer International Publishing, Switzerland, 2014 Search PubMed.
- O. H. Abdelhafez, J. R. Fahim, S. Y. Desoukey, M. S. Kamel and U. R. Abdelmohsen, Chem. Biodivers., 2019, 16, e1800692 Search PubMed.
- J. Hu, B. Yang, X. Lin, X. Zhou, X. Yang, L. Long and Y. Liu, Chem. Biodivers., 2011, 8, 1011–1032 CrossRef CAS.
- V. T. Sang, T. T. H. Dat, L. B. Vinh, L. C. V. Cuong, P. T. T. Oanh, H. Ha, Y. H. Kim, H. L. T. Anh and S. Y. Yang, Mar. Drugs, 2019, 17, 468 CrossRef CAS PubMed.
- M.-E. Hegazy, A. M. Gamal-Eldeen, T. A. Mohamed, M. A. Alhammady, A. A. Hassanien, M. A. Shreadah, I. I. Abdelgawad, E. M. Elkady and P. W. Paré, Nat. Prod. Res., 2016, 30, 1266–1272 CrossRef CAS PubMed.
- H.-Y. Fang, C.-C. Liaw, C.-H. Chao, Z.-H. Wen, Y.-C. Wu, C.-H. Hsu, C.-F. Dai and J.-H. Sheu, Tetrahedron, 2012, 68, 9694–9700 CrossRef CAS.
- J. Zhang, L.-C. Li, K.-L. Wang, X.-J. Liao, Z. Deng and S.-H. Xu, Bioorg. Med. Chem. Lett., 2013, 23, 1079–1082 CrossRef CAS PubMed.
- Y.-J. Xio, M.Sc. thesis, National Sun Yat-sen University, China, 2011.
- E. S. Kawasaki and A. Player, Nanomed. Nanotechnol. Biol. Med., 2005, 1, 101–109 CrossRef CAS.
- B. Klębowski, J. Depciuch, M. Parlińska-Wojtan and J. Baran, Int. J. Mol. Sci., 2018, 19, 4031 CrossRef PubMed.
- V. Rao, D. Nallappan, K. Madhavi, S. Rahman, L. Jun Wei and S. H. Gan, Oxid. Med. Cell. Longevity, 2016, 2016, 3685671 Search PubMed.
- P. R. M. Hemlata, A. P. Singh and K. K. Tejavath, ACS Omega, 2020, 5, 5520–5528 CrossRef CAS PubMed.
- A. Roy, O. Bulut, S. Some, A. K. Mandal and M. D. Yilmaz, RSC Adv., 2019, 9, 2673–2702 RSC.
- A. C. Anselmo and S. Mitragotri, Bioeng. Transl. Med., 2019, 4, e10143 Search PubMed.
- L. Xu, Y.-Y. Wang, J. Huang, C.-Y. Chen, Z.-X. Wang and H. Xie, Theranostics, 2020, 10, 8996–9031 CrossRef CAS PubMed.
- Y. Singh, S. Kaushal and R. S. Sodhi, Nanoscale Adv., 2020, 2, 3972–3982 RSC.
- R. Singh, S. K. Sahu and M. Thangaraj, J. Nanopart., 2014, 2014, 718240 CrossRef.
- O. H. Abdelhafez, T. F. S. Ali, J. R. Fahim, S. Y. Desoukey, S. Ahmed, F. A. Behery, M. S. Kamel, T. A. Gulder and U. R. Abdelmohsen, Int. J. Nanomed., 2020, 15, 5345–5360 CrossRef CAS PubMed.
- R. F. A. Abdelhameed, E. S. Habib, M. S. Goda, J. R. Fahim, H. A. Hassanean, E. E. Eltamany, A. K. Ibrahim, A. M. AboulMagd, S. Fayez, A. M. A. El-kader, T. Al-Warhi, G. Bringmann, S. A. Ahmed and U. R. Abdelmohsen, Mar. Drugs, 2020, 18, 354 CrossRef CAS PubMed.
- R. F. Abdelhameed, E. S. Habib, N. A. Eltahawy, H. A. Hassanean, A. K. Ibrahim, J. R. Fahim, A. M. Sayed, O. M. Hendawy, U. R. Abdelmohsen and S. A. Ahmed, Tetrahedron Lett., 2021, 72, 152986 CrossRef.
- Y. Elsayed, J. Refaat, U. R. Abdelmohsen, E. M. Othman, H. Stopper and M. A. Fouad, Phytochem. Anal., 2018, 29, 543–548 CrossRef CAS PubMed.
- N. H. Shady, A. R. Khattab, S. Ahmed, M. Liu, R. J. Quinn, M. A. Fouad, M. S. Kamel, A. B. Muhsinah, M. Krischke and M. J. Mueller, Int. J. Nanomed., 2020, 15, 3377–3389 CrossRef CAS PubMed.
- T. H. Hsiao, C. S. Sung, Y. H. Lan, Y. C. Wang, M. C. Lu, Z. H. Wen, Y. C. Wu and P. J. Sung, Mar. Drugs, 2015, 13, 3443–3453 CrossRef CAS PubMed.
- W. A. Tanod, U. Yanuhar, Maftuch, M. Y. Putra and Y. Risjani, Anti-Inflammatory Anti-Allergy Agents Med. Chem., 2019, 18, 126–141 CrossRef CAS PubMed.
- U. Abdelmohsen, C. Cheng, C. Viegelmann, T. Zhang, T. Grkovic, S. Ahmed, R. J. Quinn, U. Hentschel and R. Edrada-Ebel, Mar. Drugs, 2014, 12, 1220–1244 CrossRef CAS.
- Mzmine, http://sourceforge.net/projects/mzmine/, (accessed August 2019) Search PubMed.
- Dictionary of Natural Products, http://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml, (accessed August 2019) Search PubMed.
- METLIN, http://metlin.scripps.edu/index.php, (accessed August 2019) Search PubMed.
- MarinLit, http://pubs.rsc.org/marinlit/, (accessed August 2019) Search PubMed.
- E. G. Haggag, A. M. Elshamy, M. A. Rabeh, N. M. Gabr, M. Salem, K. A. Youssif, A. Samir, A. Bin Muhsinah, A. Alsayari and U. R. Abdelmohsen, Int. J. Nanomed., 2019, 14, 6217–6229 CrossRef CAS PubMed.
- T. Rasheed, M. Bilal, H. M. Iqbal and C. Li, Colloids Surf. B Biointerfaces, 2017, 158, 408–415 CrossRef CAS.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS.
- T. Mohammad, F. I. Khan, K. A. Lobb, A. Islam, F. Ahmad and M. I. Hassan, J. Biomol. Struct. Dyn., 2019, 37, 1813–1829 CrossRef CAS.
- N. Asmathunisha and K. Kathiresan, Colloids Surf. B Biointerfaces, 2013, 103, 283–287 CrossRef CAS PubMed.
- C. R. Singh, K. Kathiresan and S. Anandhan, Afr. J. Biotechnol., 2015, 14, 1525–1532 CrossRef.
- S. Umayaparvathi, M. Arumugam, S. Meenakshi and T. Balasubramanian, Int. J. Sci. Nat., 2013, 4, 199–203 CAS.
- D. Inbakandan, C. Kumar, L. S. Abraham, R. Kirubagaran, R. Venkatesan and S. A. Khan, Colloids Surf. B Biointerfaces, 2013, 111, 636–643 CrossRef CAS.
- D. Inbakandan, G. Sivaleela, D. M. Peter, R. Kiurbagaran, R. Venkatesan and S. A. Khan, Mater. Lett., 2012, 87, 66–68 CrossRef CAS.
- V. Vasanthabharathi, V. Kalaiselvi and S. J. Jayalakshmi, Egypt. Acad. J. Biol. Sci. B. Zoology, 2013, 5, 40–48 CrossRef.
- M. R. Hamed, M. H. Givianrad and A. M. Moradi, Orient. J. Chem., 2015, 31, 125–130 Search PubMed.
- G. Arya, N. Sharma, R. Mankamna and S. Nimesh, Antimicrobial silver nanoparticles: future of nanomaterials, in Microbial Nanobionics, Springer, 2019, pp. 89–119 Search PubMed.
- M. Hamed, M. Givianrad and A. Moradi, Indian J. Geomar. Sci., 2017, 46, 125–130 Search PubMed.
- S. Gurunathan, J. W. Han, V. Eppakayala, M. Jeyaraj and J.-H. Kim, BioMed Res. Int., 2013, 2013, 535796 Search PubMed.
- K. A. Youssif, E. G. Haggag, A. M. Elshamy, M. A. Rabeh, N. M. Gabr, A. Seleem, M. A. Salem, A. S. Hussein, M. Krischke, M. J. Mueller and U. R. Abdelmohsen, PLoS One, 2019, 14, e0223781 CrossRef CAS.
- K. A. Youssif, A. M. Elshamy, M. A. Rabeh, N. Gabr, W. M. Afifi, M. A. Salem, A. Albohy, U. R. Abdelmohsen and E. G. Haggag, ChemistrySelect, 2020, 5, 12278–12286 CrossRef CAS.
- K. Shameli, M. Bin Ahmad, E. A. Jaffar Al-Mulla, N. A. Ibrahim, P. Shabanzadeh, A. Rustaiyan, Y. Abdollahi, S. Bagheri, S. Abdolmohammadi, M. S. Usman and M. Zidan, Molecules, 2012, 17, 8506–8517 CrossRef CAS.
- Y. K. Mohanta, S. K. Panda, R. Jayabalan, N. Sharma, A. K. Bastia and T. K. Mohanta, Front. Mol. Biosci., 2017, 4, 14 CrossRef PubMed.
- Y.-J. Xio, J.-H. Su, Y.-J. Tseng, B.-W. Chen, W. Liu and J.-H. Sheu, Mar. Drugs, 2014, 12, 4495–4503 CrossRef PubMed.
- S.-Y. Cheng, C.-F. Dai and C.-Y. Duh, J. Nat. Prod., 2007, 70, 1449–1453 CrossRef CAS PubMed.
- S.-Y. Cheng, E.-H. Lin, J.-S. Huang, Z.-H. Wen and C.-Y. Duh, Chem. Pharm. Bull., 2010, 58, 381–385 CrossRef CAS PubMed.
- A. A. El-Gamal, E.-P. Chiu, C.-H. Li, S.-Y. Cheng, C.-F. Dai and C.-Y. Duh, J. Nat. Prod., 2005, 68, 1749–1753 CrossRef CAS.
- A. Bishara, D. Yeffet, M. Sisso, G. Shmul, M. Schleyer, Y. Benayahu, A. Rudi and Y. Kashman, J. Nat. Prod., 2008, 71, 375–380 CrossRef CAS PubMed.
- A. Ma, Z. Deng, L. van Ofwegen, M. Bayer, P. Proksch and W. Lin, J. Nat. Prod., 2008, 71, 1152–1160 CrossRef CAS PubMed.
- K. Yoshikawa, S. Kanekuni, M. Hanahusa, S. Arihara and T. Ohta, J. Nat. Prod., 2000, 63, 670–672 CrossRef CAS.
- Y.-S. Lee, T.-H. Duh, S.-S. Siao, R.-C. Chang, S.-K. Wang and C.-Y. Duh, Mar. Drugs, 2017, 15, 392–401 CrossRef.
- K. C. Hembram, R. Kumar, L. Kandha, P. K. Parhi, C. N. Kundu and B. K. Bindhani, Artif. Cells, Nanomed., Biotechnol., 2018, 46, S38–S51 CrossRef PubMed.
- M. K. Paul and A. K. Mukhopadhyay, Int. J. Med. Sci., 2004, 1, 101–115 CrossRef CAS PubMed.
- P. S. Sharma, R. Sharma and T. Tyagi, Curr. Pharm. Des., 2009, 15, 758–776 CrossRef CAS.
- R. J. Craven, H. Lightfoot and W. G. Cance, Surg. Oncol., 2003, 12, 39–49 CrossRef.
- C. T. Guy, R. D. Cardiff and W. J. Muller, J. Biol. Chem., 1996, 271, 7673–7678 CrossRef CAS PubMed.
- I. F. Nerini, M. Cesca, F. Bizzaro and R. Giavazzi, Chin. J. Canc., 2016, 35, 61 CrossRef.
- M. S. A. Gill, H. Saleem and N. Ahemad, Curr. Top. Med. Chem., 2020, 20, 1093–1104 CrossRef.
- C. Schettino, M. A. Bareschino, V. Ricci and F. Ciardiello, Expet Rev. Respir. Med., 2008, 2, 167–178 CrossRef CAS.
- K. Choowongkomon, O. Sawatdichaikul, N. Songtawee and J. Limtrakul, Molecules, 2010, 15, 4041–4054 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra03045k |
| ‡ Authors have equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2021 |