Open Access Article
Open Access ArticleExploration of production of C14 and C15 bacillomycin D homologues with enzymatic hydrolysis from maize straws using fed-batch fermentation by Bacillus subtilis NS-174
Shiquan Qian *a,
Xuejin Lib,
Lu Sunc,
Yuanyuan Shena,
Qingyi Rena,
Enjie Diaoa and
Zhaoxin Lud
*a,
Xuejin Lib,
Lu Sunc,
Yuanyuan Shena,
Qingyi Rena,
Enjie Diaoa and
Zhaoxin Lud
aJiangsu Key Laboratory for Eco-Agricultural Biotechnology around Hongze Lake, Jiangsu Collaborative Innovation Center of Regional Modern Agriculture & Environmental Protection, Jiangsu Key Laboratory for Food Safety and Nutrition Function Evaluation, Huaiyin Normal University, Huaian 223300, China. E-mail: qianshq@163.com
bSchool of Biological and Food Engineering, Bengbu University, Bengbu, 233030, China
cSchool of Food and Biological Engineering, Jiangsu University, Zhenjiang, 212000, China
dCollege of Food Science and Technology, Nanjing Agricultural University, 1 Weigang, Nanjing 210095, China
First published on 12th February 2020
Abstract
A strain with strong antifungal activity, Bacillus subtilis NS-174, was identified and the antifungal compounds were purified and structurally analyzed by high performance liquid chromatography-mass spectrometry/mass spectrometry (HPLC-MS/MS). The effects of the enzymatic hydrolysis and the fermentation strategies on production of the antifungal compounds were also investigated. The results showed that the antifungal compounds were characterized to be C14 and C15 bacillomycin D homologues (C14–15BDs). When 400 mL of the cellulase hydrolysates from the maize straws residues (CHMSRs) was fed into 1.0 L of the batch fermentation broth at 60 h, an output of C14–15BDs (OC14–15BDs) was obtained as 44.84 mg gmaize straws−1. Moreover, the fed-batch fermentation was beneficial for the increase of the transcriptional activities of C14–15BDs synthetase genes by up-regulating expression of comA, sigmaH and spo0A, which caused the elevation of C14–15BDs production. The present work provided an effective solution for lipopeptide production in Bacillus subtilis.
1. Introduction
The antifungal lipopeptides produced by bacteria, such as Bacillus subtilis1 and Bacillus amyloliquefaciens,2 were bioactive secondary metabolites that had strong inhibition effects on growth of agricultural fungi.1,3 The biosynthesis of antifungal lipopeptides was mainly regulated by the operon structure module of lipopeptide synthase.4 For example, the synthesis of bacillomycin D, a cyclic lipopeptide from the iturin family, was regulated by bmyA, bmyB, bmyC, bmyD and TE.5 In addition, the biosynthesis of the antifungal lipopeptides was also related to signal genes in Bacillus subtilis, such as ComA, SigmaH and Spo0A.4,5China produces a large amount of maize straws every year and the environmental problems caused by incineration and irrational treatment of maize straws are getting worse and worse.6,7 Therefore, how to rationally make use of maize straws has attracted more and more attention. Enzymatic bioconversion technology provided a reliable way for the utilization of crop straw resources, due to its mild conditions, feasible operations and high selectivities.8 The maize straws were mainly composed of cellulose, hemicellulose and lignin9 and it was difficult for microorganisms to directly utilize crop straws. It was previously reported that maize straws could be efficiently converted into small reducing sugars by enzymatic hydrolysis.10,11 Microorganisms could commendably use these reducing sugars from crop straws as carbon sources to increase cell growth and ferment to useful target substances, such as bioethanol,12 polysaccharides13 and microbial oil.14
In addition, the fed-batch fermentation was widely used in bio-fermentation because of its rapid accumulation of biomass and shortening fermentation cycle.15,16 Use of enzymatic hydrolysates from crop straws as fermentation substrates and optimization fermentation strategies could ulteriorly improve the biomass of microorganisms and the synthesis efficiency of target substances.17–20 However, at present, there were no reports on production of antifungal lipopeptides from microorganisms using enzymatic hydrolysis of crop straws. The present work was aimed at production of antifungal lipopeptides by enzymatic hydrolysis process. Firstly, a strain with antifungal activity was isolated from the soil of Huai'an suburb and the structure of the antifungal compounds was analyzed and elucidated. Then, using maize straws as raw materials, the effects of cellulase enzymatic hydrolysis and fermentation strategies on production of antifungal compounds were studied. Furthermore, the expression of synthase genes and related signal protein genes for biosynthesis of antifungal compounds were also explored.
2. Materials and methods
2.1 Raw materials and enzymes
The raw maize straws were harvested from the suburb of Huaian, China, and cut to a particle size of 1–3 mm after cleaned with tap water. Then, the straws was dried at 60 °C and pulverized to 60 mesh powder. The cellulase in this study was purchased from Sinopharm Chemical Reagent Co., Ltd, China, and the activity of the cellulase was 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U g−1.
000 U g−1.
2.2 Microorganisms and culture mediums
The sample strains isolated from wetland in the suburb of Huaian, China, were cultured in beef extract medium (BEM) at 37 °C. The BEM medium consisted of 3.0 g L−1 beef extract, 10.0 g L−1 peptone and 5.0 g L−1 NaCl. The cellulase enzymatic hydrolyzates medium (CEHM) modified according to the Landy medium21 was used as the fermentation medium for producing C14–15BDs. The CEHM medium consisted of yeast extract 1.0 g L−1, L-glutamic acid 5.0 g L−1, MgSO4·7H2O 0.5 g L−1, KCl 0.5 g L−1, MnSO4 0.05 g L−1, CuSO4·5H2O 0.0016 g L−1, FeSO4·7H2O 0.0015 g L−1, KH2PO4 1.0 g L−1, and 100 mL of enzymatic hydrolyzates. It was maintained at a pH 7.0 and a temperature of 121 °C, and was sterilized for 20 min.2.3 Screening and identification of the strains with antifungal activities
The sample strains were cultured at 37 °C and 180 rpm for 24 h on Luria-Bertani (LB) medium containing 0.5 g L−1 yeast extract, 1.0 g L−1 tryptone, and 1.0 g L−1 NaCl (pH 7.0). The single colonies were stochastically inoculated into 50 mL of BEM medium and shaked at 37 °C and 180 rpm for 16 h. 4% (v/v) of cultures was then fermented into the CEHM medium and cultivated at 33 °C and 180 rpm for 96 h. 20 mL of cultures was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 20 min. The supernatants were filtered through 0.22 μm filters to prepare cell-free supernatants (CFSs). The antifungal activities of CFSs were assayed on a PDA agar, with Aspergillus flavus (CICC 2062) as the indicator strain. The antifungal activities were defined as the diameter of inhibition zone. The CFSs of strains with a significantly large inhibition zone were used as the selected strains. The molecular identification of the isolated strains was performed on basis of the phenotypical analysis.22 In detail, the genomic DNA was extracted according to the procedure of DNA extraction kit (Sangon, China) and its purity was detected on NanoDrop2000 (Thermo Scientific, USA). The universal 16S rRNA PCR primer including 16SF (5′-AGAGTTTGATCMTGGCTCAG-3′) and 16SR (5′-TACGGYTACCTTGTTACGACTT-3′) was used to amplify 16S rRNA. A total volume of 30 μL PCR reaction solution contained 50 ng of genomic DNA, 3.0 μL of each primer, 0.2 μL of Taq DNA polymerase, 3.0 μL of 10 μM PCR buffer, 2.0 μL of 200 μM dNTP and 17.8 μL ddH2O. The PCR program was 95 °C for 5 min, followed by 35 cycles at 95 °C for 3 0 s, 55 °C for 30 s and 72 °C for 60 s, and 72 °C for 10 min.
000g at 4 °C for 20 min. The supernatants were filtered through 0.22 μm filters to prepare cell-free supernatants (CFSs). The antifungal activities of CFSs were assayed on a PDA agar, with Aspergillus flavus (CICC 2062) as the indicator strain. The antifungal activities were defined as the diameter of inhibition zone. The CFSs of strains with a significantly large inhibition zone were used as the selected strains. The molecular identification of the isolated strains was performed on basis of the phenotypical analysis.22 In detail, the genomic DNA was extracted according to the procedure of DNA extraction kit (Sangon, China) and its purity was detected on NanoDrop2000 (Thermo Scientific, USA). The universal 16S rRNA PCR primer including 16SF (5′-AGAGTTTGATCMTGGCTCAG-3′) and 16SR (5′-TACGGYTACCTTGTTACGACTT-3′) was used to amplify 16S rRNA. A total volume of 30 μL PCR reaction solution contained 50 ng of genomic DNA, 3.0 μL of each primer, 0.2 μL of Taq DNA polymerase, 3.0 μL of 10 μM PCR buffer, 2.0 μL of 200 μM dNTP and 17.8 μL ddH2O. The PCR program was 95 °C for 5 min, followed by 35 cycles at 95 °C for 3 0 s, 55 °C for 30 s and 72 °C for 60 s, and 72 °C for 10 min.
The PCR products were purified using Omega Gel Extraction Kits (Omega, USA) and sequenced by BGI (Wuhan, China). The sequence alignment analysis23 was carried out on the website of https://blast.ncbi.nlm.nih.gov/Blast. The phylogenetic tree was constructed and analyzed by the previous method using the software of MEGA5.24
2.4 Isolation and structure characterization of the antifungal compounds
After 5000g of centrifugation, 30 mL of the fermentation broth supernatant was adjusted to a pH of 2.0 with 4 mol L−1 of HCl and stayed at 4 °C for 120 min. After centrifugation, the precipitation was accumulated and extracted by 100% of methanol. The crude antifungal compounds were prepared by 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g of centrifugation. Then, the antifungal compounds was purified by a Sephadex LH-20 column and reverse-phase high performance liquid chromatography (RP-HPLC) (Waters 600, USA) with a C18 column (4.6 × 250 mm, Agilent, USA).The structure of the antifungal compounds and its concentration were analyzed and characterized by high performance liquid chromatography-mass spectrometry/mass spectrometry (HPLC-MS/MS) as described in the previous work.1
000g of centrifugation. Then, the antifungal compounds was purified by a Sephadex LH-20 column and reverse-phase high performance liquid chromatography (RP-HPLC) (Waters 600, USA) with a C18 column (4.6 × 250 mm, Agilent, USA).The structure of the antifungal compounds and its concentration were analyzed and characterized by high performance liquid chromatography-mass spectrometry/mass spectrometry (HPLC-MS/MS) as described in the previous work.1
2.5 Cellulase enzymatic hydrolysis
2 g of dry maize straw powders were sterilized at 121 °C for 20 min and cooled down to room temperature. The enzymatic hydrolysis was performed in a 100 mL conical flask. The sterilized powders were mixed with cellulase in 50 mL of reaction solution and the enzymatic hydrolysis was carried out at 50 °C and 180 rpm. After enzymolysis, the mixture was centrifuged at 8000g for 20 min. The cellulase hydrolysates from the maize straws residues (CHMSRs) were collected to prepare for the fermentation experiments. The maize straws residues were dried at 85 °C to a constant weight and quantified to calculate the consumption of maize straws (CMSs) (g). The reducing sugar content of CHMSRs was determined by the 2,4-dinitrosalicylate method using glucose as the standard.25 The yield of reducing sugar (g g−1) for CHMSRs (YRS-CHMSRs) (g g−1) was defined as the ratio of the amount of reducing sugar in CHMSRs (g) to CMSs (g).2.6 Fermentation process
The strain B. subtilis NS-174 was cultured in 100 mL of BEM medium and shaked at 37 °C and 200 rpm for 20 h to prepare the seed culture. 5% of seed culture was incubated into a 250 mL Erlenmeyer flask which contained 100 mL of CEHM medium to prepare for the flask fermentation. The batch fermentation was performed in a 5.0 L fermenter (GUJZ, Zhenjiang, China) with 1.0 L of CEHM medium. The fed-batch fermentation was carried out by adding 300 mL of CHMSRs into 1.0 L of batch fermentation broth. All fermentations were triplicated and performed at 33 °C and 180 rpm.2.7 RT-qPCR
The fresh cell of B. subtilis NS-174 was cultured in 20 mL of BEM medium at 37 °C and 200 rpm for 36 h. The total RNA was isolated by the method described as the manufacturer's protocol of Trizol Reagent (Sangon, Shanghai, China). Then, the RNA sample was collected and detected by NanoDrop2000 (Thermo Scientific, USA). The cDNA synthesis was performed according to the procedure of HiScript™ 1 st Strand cDNA Synthesis Kit (Vazyme, USA). RT-qPCR was performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems, USA) using SYBR Premix ExTaq™ (TaKaRa, Dalian, China). The primers used for amplification of relative bacillomycin D synthesis genes (bmy A, bmy B, bmy C, bmy D, TE, comA, sigma H and spo0 A) and the PCR program were performed as described in the previous works.26,272.8 Analytical methods
10 mL of fermentation cultures was centrifuged at 8000g for 10 min to obtain wet cells. The wet cells were dried at 80 °C for 6 h and the biomass concentration was defined as the dry cell weight. The concentration of C14–15BDs was determined by HPLC according to Qian et al. (2015).26 The ratio of C14–15BDs/biomass (mg g−1) was defined as the ratio of the concentration of C14–15BDs (mg L−1) to the biomass concentration (g L−1). The output of C14–15BDs (OC14–15BDs) (mg g−1) was defined as the ratio of the amount of C14–15BDs (mg) to CMSs (g).3. Results and discussion
3.1 Identification of the antifungal compounds producing strain
A total of 758 single colonies isolated from wetland were cultured in BEM medium and fermented in Landy medium. The cell-free supernatants (CFSs) of 4 strains exhibited strong antifungal activity against Aspergillus flavus (data not shown) and the strain NS-174 had the highest antifungal activity. Therefore, the strain NS-174 was selected to prepare for the following studies. A 1452 bp 16S rRNA sequence was amplified from the strain NS-174 genomic DNA. The results of sequence alignment and a neighbor-joining phylogenetic tree revealed that the strain NS-174 (accession number MN955309) was 99.72% similar to Bacillus subtilis WX1 (accession number KF624694.1) (Fig. 1). | ||
| Fig. 1 The phylogenetic tree of the strain NS-174. The tree was constructed by the neighbor-joining method using the software MEGA5.0. Bar: sequence divergence of 0.0002. | ||
B. subtilis species are considered to be well known classes for producing antifungal compounds. Many antifungal compounds, such as fengycin, surfactin and iturin, have been gradually found in B. subtilis 28,29. In the present study, the antifungal compounds in CFSs of the strain B. subtilis NS-174 exhibited significant inhibition effects against A. flavus, however, the structure of the antifungal compounds should be further investigated.
3.2 HPLC-MS/MS analysis of the antifungal compounds 1 and 2
The antifungal compounds from CFSs were further purified by Sephadex LH-20 chromatography and RP-HPLC. The results obtained from HPLC profile (Fig. 2a) showed that the antifungal compounds had two active peaks (peaks 1 and 2) with the retention time at 18.34 and 20.93 min, respectively. The two active peaks were further analyzed by MS/MS. The results (Fig. 2b) showed that the isolated peaks 1 and 2 displayed their [M + H]+ ion peaks at m/z 1031.4 and m/z 1045.43. From the results of MS/MS analysis, it was important to note that the isolated compounds individually had the fatty acid chains of C14 and C15 with both contained seven peptide structures (–Asn–Tyr–Asn–Pro–Glu–Ser–Thr–). The similar evidences were found in previous reports.27,30 It was concluded that the antifungal compounds belonged to C14 and C15 of bacillomycin D homologues (C14–15BDs).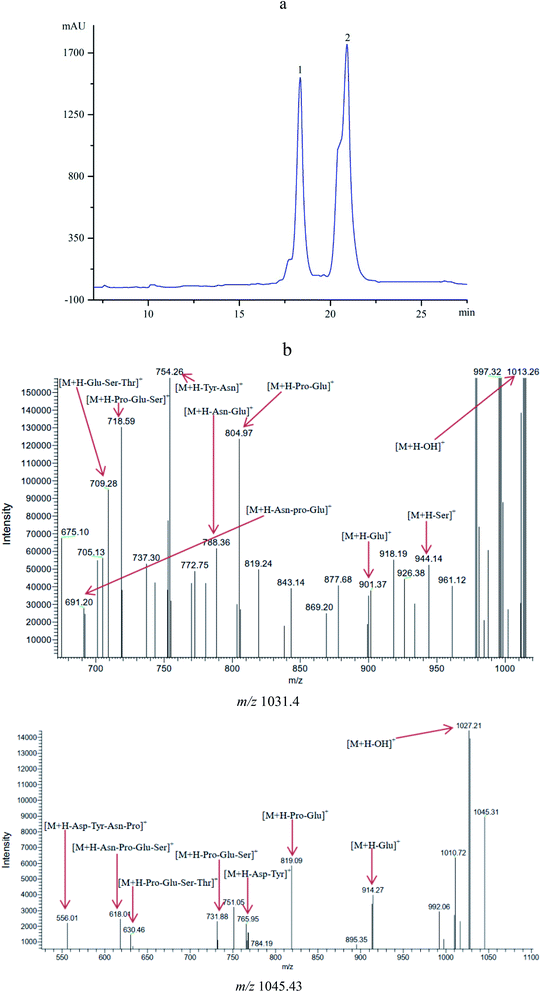 | ||
| Fig. 2 (a) HPLC profile of the antifungal compounds (peaks 1 and 2) with retention time of 18.34 and 20.93 min. (b) MS/MS analysis of the antifungal compounds with m/z 1031.40 and m/z 1045.43. | ||
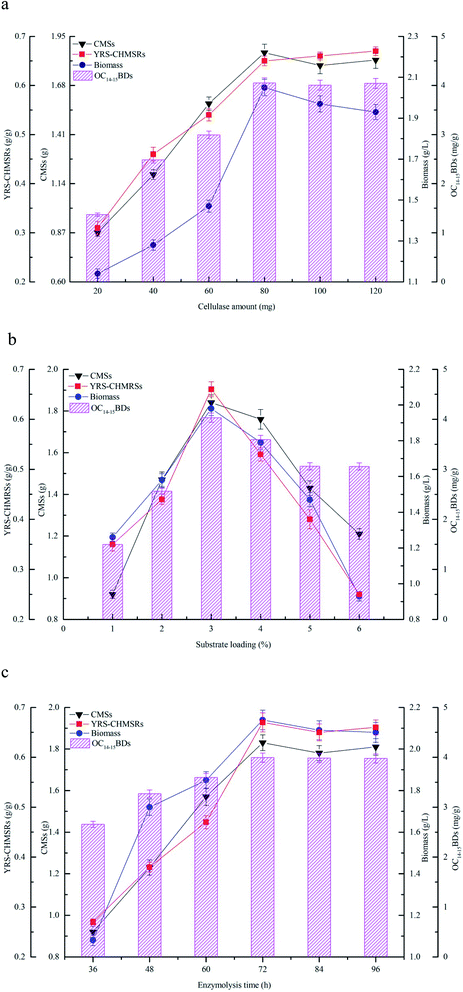
3.3 Effects of cellulase hydrolysis of maize straws on production of C14–15BDs
The effects of cellulase amount, substrate loading, and enzymolysis time on production of reducing sugar and C14–15BDs were investigated using flask fermentations. The results were shown in Fig. 3. Fig. 3a showed that the yield of reducing sugar significantly enhanced along with the consumption of maize straws at the beginning. YRS-CHMRSs was up to 0.65 g g−1 when the cellulase amount was at 80.0 mg, which indicated that cellulase could effectively convert maize straws to reducing sugar within an appropriate range. However, when the cellulase amount continued to mount up, YRS-CHMRSs had no significant increase, demonstrating that an overdose of cellulase was disadvantage to the production of reducing sugar from maize straws. Meanwhile, when the cellulase amount ranged from 20 to 80 mg, the biomass concentration and OC14–15BDs increased from 1.14 to 2.05 g L−1 and 1.37 to 4.05 mg g−1, respectively, which indicated that there was a positive relation between reducing sugar and biomass. The reducing sugar could efficiently help to accumulate more cell growth for C14–15BDs production in the strain B. subtilis NS-174.Fig. 3b profiled the effect of substrate loading on production of reducing sugar and C14–15BDs. It was found that 3.0% of substrate loading was suitable for producing more reducing sugar and YRS-CHMRSs achieved to be 0.68 g g−1. A higher level of substrate loading (over 3.0%) was against the improvement of release of more reducing sugar. An excessive dose of substrate increased the viscosity of the reaction system, which hindered the effective combination of substrate and enzyme and therefore caused an inefficient enzymatic hydrolysis. Furthermore, when the substrate loading ranged from 1% to 3%, the biomass concentration and OC14–15BDs were increased. The biomass concentration and OC14–15BDs were respectively up to 1.98 g L−1 and 4.03 mg g−1 at substrate loading of 3%, which evidenced that OC14–15BDs showed a dose-dependent manner within an appropriate range of substrate loading.
Fig. 3c showed that the release of reducing sugar had an obvious increase when the enzymolysis time was from 36 to 72 h, correlating with the increase of the consumption of maize straws. YRS-CHMRSs was observed to be a maximum value at 72 h. As the enzymolysis time was further extended, YRS-CHMRSs no longer elevated, indicating that an excessive enzymolysis time made little sense of the release of reducing sugar. Therefore, the enzymolysis time was reasonably obtained to be 72 h. At the same time, when the enzymolysis time varied from 36 to 72 h, it was observed that the biomass concentration and C14–15BDs production synchronously increased and their optimal values were achieved at 72 h, which evidenced that the production of C14–15BDs was closely related to the reducing sugar.
The enzymatic hydrolysis was extensively used to produce target active compounds in microorganisms and the previous reports proved that enzymatic hydrolysis was an important tool in bioconversion.31,32 Yu et al. (2017) efficiently used hemicellulose and cellulose from corncob to produce bioethanol by Spathaspora passalidarum U1-58 using the hydrolysis method of cellulase and xylanase.33 Zhao et al. (2019) availably degraded sodium hydroxide pretreated rice straw by three-stage enzymatic hydrolysis to accumulate microbial oil by Mortierella isabellina.14 Maize straws consisted of large molecular carbohydrates including cellulose, hemicellulose and lignin. Enzymatic hydrolysis was beneficial to change these carbohydrates to small reducing sugars,10 which was helpful for bioconversion. Enzyme, substrate and reaction time were considered to be the key factors that were closely relative to digestion efficiency in the process of enzymatic hydrolysis. An appropriate amount of enzymes, substrates and reaction time could promote the combination of enzymes and substrates, thereby improving the efficiency of reducing sugar release and bioconversion of target products.12,34 In the present study, a right dose of cellulase and substrate, and an appropriate enzymolysis time could significantly increase the production of reducing sugar, thus accelerated biomass accumulation and C14–15BDs formation. In general, given the efficiency of cellulase hydrolysis and the production capacity of C14–15BDs, an available cellulase hydrolysis condition was reasonably determined to be the cellulase amount of 80 mg, the substrate loading of 3% and the enzymolysis time of 72 h.
3.4 Evaluation of fermentation strategies for C14–15BDs production
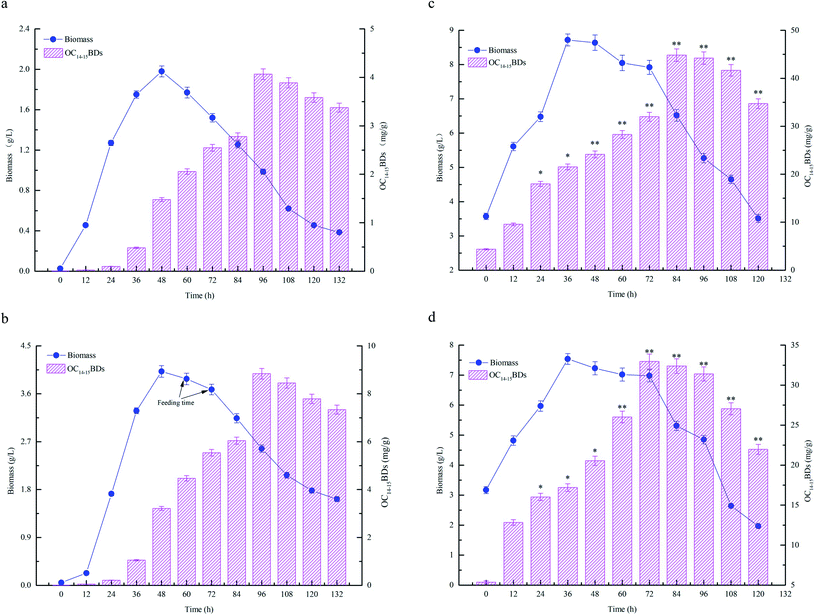
The fermentation characteristics of flask fermentations, batch fermentations and fed-batch fermentations for C14–15BDs production by B. subtilis NS-174 were investigated using hydrolyzates of maize straws as fermentation substrates. The time courses of fermentation curves were illustrated in Fig. 4. The results of flask fermentations in Fig. 4a showed that the biomass concentration apparently increased and remained at a high level when at the fermentation time of 48 h. However, the biomass concentration decreased gradually after 48 h. OC14–15BDs had no significant increase from 0 to 36 h and the strain B. subtilis NS-174 started to produce C14–15BDs after 48 h. The highest OC14–15BDs reached 4.06 mg g−1 at 96 h (Fig. 4a and Table 1). The effects of batch fermentation on C14–15BDs production were shown in Fig. 4b. It was found that the biomass and C14–15BDs production showed the same trend for the flask fermentation and the batch fermentation. The biomass concentration at 48 h and OC14–15BDs at 96 h were 4.03 g L−1 and 8.87 mg g−1, respectively (Fig. 4b and Table 1). While after 48 h, the biomass concentration continued to decrease, due to the lack of carbon sources. B. subtilis NS-174 required ample fermentation substrates to maintain a high level of cell growth for C14–15BDs production. Consequently, the fed-batch fermentation was further performed and the results were shown in Fig. 4c and d. The results demonstrated that fed-batch fermentation effectively improved cell growth and C14–15BDs production. When fed 300 mL of CHMSRs into 1.0 L of batch fermentation broth at 60 h, the maximum biomass concentration, ratio of C14–15BDs/biomass, OC14–15BDs were respectively obtained to be 8.63 g L−1, 70.41 mg g−1 and 44.84 mg g−1. Compared with that of batch fermentation, the biomass concentration, the ratio of C14–15BDs/biomass and OC14–15BDs were individually increased by 114.14%, 430.59% and 405.52%, indicating that the fed-batch fermentation could efficiently promote cell growth and C14–15BDs production of B. subtilis NS-174.| Fermentation process | Maximum biomass (g L−1) | Maximum ratio of C14–15BDs/biomass (mg g−1) | Maximum OC14–15BDs (mg g−1) |
|---|---|---|---|
| a The flask experiment was carried out in a working volume of 100 mL and fermented at 48 h.b The flask experiment was carried out in a working volume of 100 mL and fermented at 96 h.c The batch experiment was carried out in a working volume of 1.0 L and fermented at 48 h.d The batch experiment was carried out in a working volume of 1.0 L and fermented at 84 h.e Feeding 300 mL CHMSRs into 1.0 L batch fermentation broth at 60 h and fermented at 36 h.f Feeding 300 mL CHMSRs into 1.0 L batch fermentation broth at 60 h and fermented at 84 h. | |||
| Flask fermentation | 2.05 ± 0.05a | 5.90 ± 0.15b | 4.06 ± 0.11b |
| Batch fermentation | 4.03 ± 0.11c | 13.27 ± 0.37d | 8.87 ± 0.23d |
| Fed-batch fermentation | 8.63 ± 0.25e | 70.41 ± 1.52f | 44.84 ± 1.31f |
The previous studies confirmed that the reducing sugar formed by enzymatic hydrolysis of maize straws could significantly increase the biomass concentration of microorganisms, thereby improve the yield of target substances.13,14 Bacillomycin D was a cyclic lipopeptide that produced as secondary metabolites by Bacillus spp. The synthesis of cyclic lipopeptides was closely relevant to cell growth of target bacteria and the increase of biomass concentration was advantageous to generate more bacillomycin D.26 From the results of Fig. 4, it was also observed that high level of reducing sugar was beneficial for enhancing the biomass concentration and thus producing more C14–15BDs in B. subtilis NS-174. However, the lack of the reducing sugar supplied in the medium limited cell growth of B. subtilis NS-174, and caused a low yield of C14–15BDs. Therefore, further improvement of fermentation strategy was necessary for the production of C14–15BDs. It was well known that the fed-batch fermentation could significantly improve the biomass concentration of microorganisms and the synthetic ability of target substances.16,35 The similar evidence was also found by Zhong et al. (2014), which significantly increased the production of an antifungal cyclic lipopeptide (jiean-peptide) by Bacillus subtilis using fed-batch fermentation.36 In the present study, according to the evidences of Fig. 4 and Table 1, the production C14–15BDs of fed-batch fermentation significantly enhanced as compared to the production of batch fermentation and flask fermentation.
3.5 Genes expression of C14–15BDs biosynthesis by different fermentation strategies
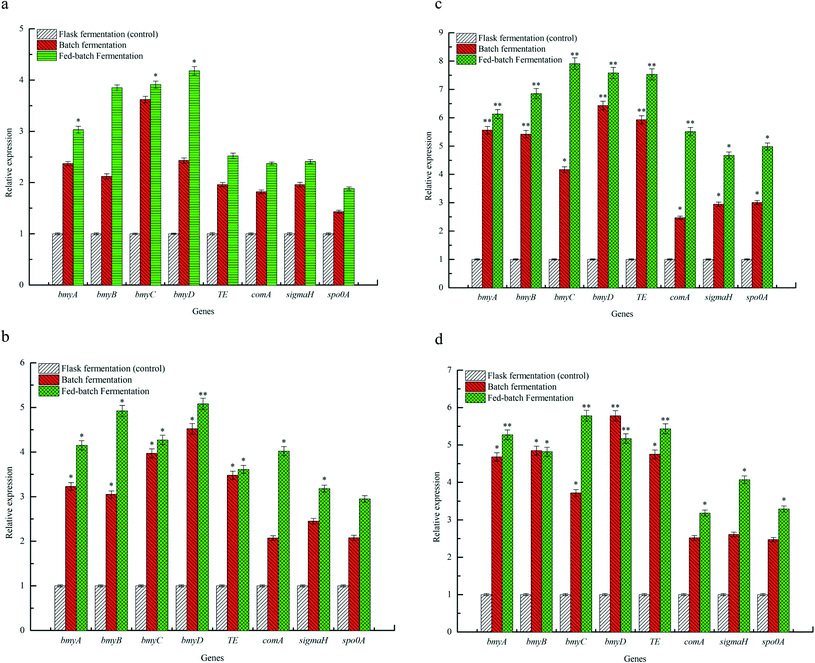
The effects of flask fermentation, batch fermentation and fed-batch fermentation on expression of bmy A, bmy B, bmy C, bmy D, TE, com A, sigma H and spo0 A were presented in Fig. 5. The results showed that expression of bmy A, bmy B, bmy C, bmy D, TE, com A, sigma H and spo0 A was increased when the fermentation time ranged from 12 to 36 h. After 36 h, expression of these eight genes decreased. The expression of bmy A, bmy B, bmy C, bmy D, TE, com A, sigma H and spo0 A by fed-batch fermentation was found to be 6.13 times at 36 h (P < 0.01), 6.85 times at 36 h (P < 0.01), 7.91 times at 36 h (P < 0.01), 7.58 times at 36 h (P < 0.01), 7.53 times at 36 h (P < 0.01), 5.51 times at 36 h (P < 0.01), 4.67 times at 36 h (P < 0.05), 4.98 times at 36 h (P < 0.05) higher than that of batch fermentation. Our evidences showed that the fed-batch fermentation could significantly up-regulate the expression of bmy A, bmy B, bmy C, bmy D, TE, com A, sigma H and spo0 A. The synthesis of a cyclic lipopeptide followed a non-ribosomal enzyme catalysis pattern and was closely correlated to the expression of its synthase.4,37 The biosynthesis of bacillomycin D was regulated by bmyA, bmyB, bmyC, bmyD and TE,5 which catalyzed the formation of cyclic heptapeptides by adenylation, thiolation and concentration of amino acids, the synthesis of fatty acid chains and the cyclization of amino acids. As can be seen from Fig. 5, the fed-batch fermentation significantly promoted expression of bmyA, bmyB, bmyC, bmyD and TE, which contributed to improving the transcriptional activities of synthetase genes of C14–15BDs and caused elevation of C14–15BDs production. In addition, the biosynthesis of cyclic lipopeptides was also associated with signal genes in Bacillus subtilis, such as Com A, Sigma H and Spo0 A.4,5 Expression of ComA, functioned as a transcriptional regulator, was helpful for the formation of spores and the synthesis of cyclic lipopeptide in B. subtilis.38 SigmaH contributed to sporulation and synthetase transcription of cyclic lipopeptides. Spo0A regulated the formation of spores in B. subtilis.39 It was previously evidenced that the increase of expression of Spo0A and number of spores could promote the synthesis of iturin A and bacillomycin D.26,40 From the evidences in Fig. 5, compared with the batch fermentation and the flask fermentation, the fed-batch fermentation significantly increased expression of Com A, Sigma H and Spo0 A, and elevated the transcriptional activity of C14–15BDs synthase genes, thus improved the production of C14–15BDs.4. Conclusions
The antifungal compounds isolated from Bacillus subtilis NS-174 were characterized to be C14 and C15 bacillomycin D homologues (C14–15BDs). The enzymatic hydrolysis of maize straws could be good for noticeably increasing production of C14–15BDs. The fermentation strategies on the production of C14–15BDs were evaluated and the fed-batch process significantly improved the production of C14–15BDs. Feeding CHMSRs could significantly improve transcriptional activities of C14–15BDs synthetase genes through up-regulation expression of comA, sigmaH and spo0A, which contributed to elevating the production of C14–15BDs.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 31571887), Young Talents Project for Jiangsu Collaborative Innovation Center of Regional Modern Agriculture and Environmental Protection (HSXT2-312) and Xiangyu Talents Project for Huaiyin Normal University (31QSQ00).References
- Q. Gong, C. Zhang, F. Lu, H. Zhao, X. Bie and Z. Lu, Food Control, 2014, 36, 8–14 CrossRef CAS.
- J. Yuan, B. Li, N. Zhang, R. Waseem, Q. Shen and Q. Huang, J. Agric. Food Chem., 2012, 60, 2976–2981 CrossRef CAS PubMed.
- S. Qian, H. Lu, J. Sun, C. Zhang, H. Zhao, F. Lu, X. Bie and Z. Lu, Food Control, 2016, 60, 281–288 CrossRef CAS.
- A. L. Moyne, T. E. Cleveland and S. Tuzun, FEMS Microbiol. Lett., 2004, 234, 43–49 CrossRef CAS PubMed.
- X. H. Chen, A. Koumoutsi, R. Scholz, K. Schneider, J. Vater, R. Süssmuth, J. Piel and R. Borriss, J. Biotechnol., 2009, 140, 27–37 CrossRef CAS PubMed.
- H. Liu, X. Ou, J. Yuan and X. Yan, Resour., Conserv. Recycl., 2018, 135, 216–224 CrossRef.
- Q. Yu, R. Liu, K. Li and R. Ma, Resour., Conserv. Recycl., 2019, 107, 51–58 CAS.
- S. H. Ghaffar, M. Fan and B. McVicar, Ind. Crops Prod., 2015, 77, 262–274 CrossRef CAS.
- P. Moniz, H. Pereira, T. Quilhó and F. Carvalheiro, Ind. Crops Prod., 2013, 50, 145–153 CrossRef CAS.
- M. Chen, J. Zhao and L. Xia, Carbohydr. Polym., 2008, 71, 411–415 CrossRef CAS.
- P. Wang, J. Chang, Q. Yin, E. Wang, Q. Zhu, A. Song and F. Lu, Bioresour. Technol., 2015, 194, 165–171 CrossRef CAS PubMed.
- H. Patel, D. Chapla and A. Shah, Renewable Energy, 2017, 109, 323–331 CrossRef CAS.
- X. Xu, P. Wu, T. Wang, L. Yan, M. Lin and C. Chen, Bioresour. Technol., 2019, 278, 43–50 CrossRef CAS PubMed.
- C. Zhao, B. Xie, R. Zhao and H. Fang, Renewable Energy, 2019, 130, 281–289 CrossRef CAS.
- J. Chen, X. Zhang, S. Yan, R. D. Tyagi and P. Drogui, Fuel, 2017, 209, 1–9 CrossRef CAS.
- N. Kumar, S. K. Sudan, R. Garg and G. Sahni, Process Biochem., 2019, 78, 1–7 CrossRef CAS.
- D. Cai, C. Chen, C. Zhang, Y. Wang, H. Wen and P. Qin, Biochem. Eng. J., 2017, 125, 18–22 CrossRef CAS.
- L. Paulová, P. Patáková, M. Rychtera and K. Melzoch, Fuel, 2014, 122, 294–300 CrossRef.
- A. Sawisit, S. Jampatesh, S. S. Jantama and K. Jantama, Bioresour. Technol., 2018, 260, 348–356 CrossRef CAS PubMed.
- J. Soares, M. M. Demeke, M. Van de Velde, M. R. Foulquié-Moreno, D. Kerstens, B. F. Sels, A. Verplaetse, A. A. R. Fernandes, J. M. Thevelein and P. M. B. Fernandes, Bioresour. Technol., 2017, 244, 234–242 CrossRef CAS PubMed.
- M. Landy, G. H. Warren, S. B. Rosenmanm and L. G. Colio, J. Bacteriol., 1947, 54, 24 CAS.
- S. Zhao, J. Han, X. Bie, Z. Lu, C. Zhang and F. Lv, J. Agric. Food Chem., 2016, 64, 2754–2764 CrossRef CAS PubMed.
- S. F. Altschul, J. Mol. Biol., 1990, 215, 403–410 CrossRef CAS PubMed.
- K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei and S. Kumar, Mol. Biol. Evol., 2011, 28, 2731–2739 CrossRef CAS PubMed.
- G. L. Miller, Anal. Biochem., 1959, 31, 426–428 CAS.
- S. Qian, H. Lu, P. Meng, C. Zhang, F. Lv, X. Bie and Z. Lu, Bioresour. Technol., 2015, 179, 260–267 CrossRef CAS PubMed.
- J. Sun, S. Qian, J. Lu, Y. Liu, F. Lu, X. Bie and Z. Lu, J. Agric. Food Chem., 2018, 66, 4422–4430 CrossRef CAS PubMed.
- S. Guo, J. W. Zhang, L. H. Dong, X. Li, M. Asif, Q. G. Guo, W. J. Jiang, P. Ma and L. Q. Zhang, Biol. Control, 2019, 136, 104001 CrossRef CAS.
- F. Siahmoshteh, Z. Hamidi-Esfahani, D. Spadaro, M. Shams-Ghahfarokhi and M. Razzaghi-Abyaneh, Food Control, 2018, 89, 300–307 CrossRef CAS.
- R. Ramachandran, M. Shrivastava, N. N. Narayanan, R. L. Thakur, A. Chakrabarti and U. Roy, Antimicrob. Agents Chemother., 2018, 62, e01457-17 CrossRef PubMed.
- Q. Bei, G. Chen, Y. Liu, Y. Zhang and Z. Wu, J. Funct. Foods, 2018, 47, 512–520 CrossRef CAS.
- Y. Nan, M. Yang, D. Xin, K. Li, S. Kuittinen, A. Pappinen and J. Zhang, Fuel, 2019, 245, 406–412 CrossRef CAS.
- H. Yu, J. Guo, Y. Chen, G. Fu, B. Li, X. Guo and D. Xiao, Bioresour. Technol., 2017, 232, 168–175 CrossRef CAS PubMed.
- L. A. R. Batalha, Q. Han, H. Jameel, H.-m. Chang, J. L. Colodette and F. J. Borges Gomes, Bioresour. Technol., 2015, 180, 97–105 CrossRef CAS PubMed.
- X. Yang, H. S. Choi, J. H. Lee, S. K. Lee, S. O. Han, C. Park and S. W. Kim, Chem. Eng. J., 2018, 349, 25–36 CrossRef CAS.
- J. Zhong, X. Zhang, Y. Ren, J. Yang, H. Tan and J. Zhou, Electron. J. Biotechnol., 2014, 17, 132–136 CrossRef CAS.
- J. S. Lott and T. V. Lee, Structure, 2017, 25, 693–695 CrossRef CAS PubMed.
- L. W. Hamoen, Microbiology, 2003, 149, 9–17 CrossRef CAS PubMed.
- S. D. Seredick and G. B. Spiegelman, J. Mol. Biol., 2007, 366, 19–35 CrossRef CAS PubMed.
- M. S. Rahman, T. Ano and M. Shoda, J. Biotechnol., 2006, 125, 513–515 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |