Self-assembled chromogen-loaded polymeric cocoon for respiratory virus detection†
Indra Memdi
Khoris‡
a,
Akhilesh Babu
Ganganboina‡
 b,
Tetsuro
Suzuki
c and
Enoch Y.
Park
b,
Tetsuro
Suzuki
c and
Enoch Y.
Park
 *ab
*ab
aDepartment of Bioscience, Graduate School of Science and Technology, Shizuoka University, 836 Ohya Suruga-ku, Shizuoka 422-8529, Japan. E-mail: park.enoch@shizuoka.ac.jp; Fax: +81-54-238-4887; Tel: +81-54-238-4887
bResearch Institute of Green Science and Technology, Shizuoka University, 836 Ohya Suruga-ku, Shizuoka 422-8529, Japan
cDepartment of Infectious Diseases, Hamamatsu University School of Medicine, 1-20-1 Higashi-ku, Handa-yama, Hamamatsu 431-3192, Japan
First published on 8th December 2020
Abstract
Inspired by the self-assembly approach, in this work, the chromogen, 3,3′,5,5′-tetramethylbenzidine (TMB), was successfully co-precipitated in aqueous solution to form collective nanoparticles (NPs) of signal molecules (TMB-NPs). Utilizing poly(lactide-co-glycolide) (PLGA) in the molecular delivery approach, the formed emulsion nanovesicle (TMB-NPs@PLGA) exhibits an enrichment of the collective signal molecules in a single antibody–antigen conjugation. A specific antibody-conjugated TMB-NPs@PLGA forms an immunocomplex sandwich structure upon the addition of influenza virus (IV)/A. The addition of dimethyl sulfoxide (DMSO) dissolves the PLGA nanovesicles, releasing the encapsulated TMB-NPs. Sequentially, the TMB-NPs release TMB molecules upon the addition of DMSO. The released TMB is catalytically oxidized by H2O2 with self-assembled protein-inorganic nanoflowers, where copper nanoflowers (CuNFs) acted as the nanozyme. The developed immunoassay demonstrates high sensitivity for IV/A with a limit of detection (LOD) as low as 32.37 fg mL−1 and 54.97 fg mL−1 in buffer and serum, respectively. For practical needs, a clinically isolated IV/A/H3N2 and spike protein of SARS-CoV-2 were detected with the LODs of 17 pfu mL−1 and 143 fg mL−1, respectively. These results show the applicability of the advanced TMB-NPs@PLGA-based colorimetric sensor for the highly sensitive detection of airborne respiratory viruses.
Introduction
In the last decade, immunoassay development has been progressing majorly centering on signal amplification to boost sensitivity and highly selective signals to the target bio-analytes.1–3 The signal measurement used in biosensors is majorly based on colorimetry, fluorescence, and chemiluminescence. The primary drawback of the fluorescence and chemiluminescence principle is physical quenching due to fluorescent dye interaction with the polymeric component and the requirement of instrument measurement. In terms of simplicity, colorimetric detection is favorable for analysis by the naked eye. However, it is difficult to precisely identify small changes occured by low concentration of analyte owing to the low sensitivity,2,4,5 due to the intrinsic limitation of the enzymes.6,7 To overcome these issues while retaining the original advantages, an improved ELISA utilizing an enzyme-loaded nanomaterial,8,9 or a nanomaterial with peroxidase-like activity with high sensitivity, is of significant interest.10–13Recently, nanozymes are being pursued intensively and have experienced rapid expansion in various fields and mechanism studies. However, the nanozymes’ activity and specificity to the substrate are not up to the enzyme's sophistication level.14 To improve the immunoassay for the detection of low concentration analytes, the limitation of the conjugated nanozyme on individually conjugated antibodies lowered the usability to achieve highly sensitive detection and practical requirement. The approach in constructing signal amplification in an immunoassay is being shifted toward the strategy to increase the availability of the nanozyme in single antibody–antigen binding. Several studies have been done to extravagate the signal by facilitating an immunosorbent vesicle-containing nanozyme15,16 or in situ seed growth of the nanozyme.11,17,18 This showed escalating detection signal up to a certain degree of amplification, but the drawback of simplicity needs to be reconsidered.
Recently, the research in immunoassays is being shifted from enzyme-dependent to signal-dependent, pivoting on the signal molecules’ amount corresponding to the antibody–antigen binding. Miao et al.19 and Ren et al.20 reported the hydrophobic interaction of curcumin on MoS2 and π-stacking interaction of thymolphthalein and metal–organic framework (MOF)-polydopamine, respectively, both as signal molecule nanocarriers. These interactions are possibly loosened, leading to leakage of free signal molecules from the nanomaterials. In the signal amplification platforms of biosensors, liposomes are being used as the nano-encapsulation due to their intrinsic hydrophilic and hydrophobic head-and-tail monomer leading to self-assembly encapsulation.16,21 However, the ratio-dependence and their instability limited their practical use in storage and their integrity against physical and chemical conditions.20,22 In our previous work, liposome encapsulation using water-soluble nanozyme hindered the detection of low virus concentration because of the low species incorporation in the liposomal encapsulation in both organic and aqueous phases.16 Polymeric encapsulation or nanovesicles in the nanostructure fabrication comprises proteins or drugs as the core enveloped by a typical polymeric membrane.23 Among the polymeric biomaterials, poly(lactide-co-glycolide) (PLGA) has successfully been implemented in numerous drug delivery formulations due to its synthetic malleability and biocompatibility. Benefited by a rich-carboxylic structure, PLGA can be easily conjugated to biomolecules.24,25 The coupled ligands, such as antibodies conjugated to the surface, could selectively recognize specific antigens that enable the nanovesicle's utilization for biosensor application.
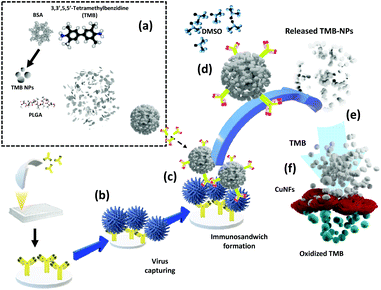
In this work, the chromogen substrate, 3,3′,5,5′-tetramethylbenzidine (TMB), is introduced to the biosensor by being carried within a PLGA encapsulation (TMB-NPs@PLGA). The signal amplification strategy centralized in the enrichment of signal molecules in a single nanovesicle with preceding co-precipitation. TMB-NPs@PLGA was conjugated to antibodies and bound to the captured virus in the microtiter wells. Upon the addition of dimethyl sulfoxide (DMSO), the encapsulated TMB-NPs are released, dissolving the PLGA shell. Simultaneously, the TMB-NPs are also dissolved in the DMSO solution resulting in TMB molecules. The TMB is then catalytically oxidized by self-assembled copper nanoflowers (CuNFs) and H2O2, developing intense blue color as the indicator for a virus's presence. The released TMB-NPs from the captured TMB-NPs@PLGA nanovesicles, even at low virus concentrations, significantly amplify the colorimetric signal according to the quantity of the oxidized TMB. The developed ultrasensitive colorimetric biosensor illustrates its ability to detect respiratory viruses, such as influenza virus (IV) and spike protein of SARS-CoV-2 in a broad range with a low detection limit at the femtogram level due to its profound architecture, easy manipulation, and facile storing of TMB-NPs.
Experimental section
Materials and chemicals
CuSO4·5H2O, ethyl acetate, DMSO, polyvinyl alcohol n = 1500–1800, and TMB were purchased from Dojindo (Osaka, Japan). Bovine serum albumin (BSA) and H2O2 were purchased from Wako Pure Chem Inc. (Osaka, Japan). N-Hydroxysuccinimide (NHS) and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) were obtained from Sigma-Aldrich Co. (St Louis, MO, USA). IV/A/Hongkong/H1N1 was obtained from ProSpec-Tany Technogene, Ltd (East Brunswick, NJ, USA). Recombinant 2019-nCov Spike protein (RBD) (ABL-1-113) and anti-2019-nCov S Protein (RBD) monoclonal antibody (ABN9306) were purchased from Abvigen Inc. (Newark, NJ, USA). Biological agents are described in the ESI.†Preparation of TMB-NPs and TMB-NP-encapsulated PLGA
To increase the water dispersity of small hydrophobic chromogens, such as TMB, the molecules were assembled into nanoparticles (NPs) because of the sudden change in the solvent environment. TMB solution in DMSO (10 mg mL−1) was added into aqueous solution to obtain precipitation with BSA (1 mg mL−1, w/v) in 10 mM PBS (pH 6.5). The mixture was continuously stirred for 12 h at 4 °C, stabilizing the formation of dispersed TMB-NPs. The turbid solution was then centrifuged and washed three times with PBS, followed by freeze-drying for further use.TMB-NPs re-dispersed in DI water were encapsulated in a double emulsion system of PLGA nanovesicles using a previously reported preparation with minor modifications.25 Four hundred microliters of the aqueous solution of TMB-NPs were added to 2 mL of ethyl acetate, and 100 mg of PLGA and the mixture was gently stirred with alternating sonication. After two minutes of emulsification, the PLGA/TMB-NPs and 8 mL of polyvinyl alcohol (5%, w/w, aqueous) were added to the w/o emulsion to form a w/o/w double emulsion and stirred further for 5 min. To solidify the nanoparticles, the organic solvent was extracted by stirring the double emulsion with 200 mL of an aqueous solution of polyvinyl alcohol 0.1% (w/w) for 5 min. The resulting dispersion of nanoparticles was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min and freeze-dried for further use.
000g for 15 min and freeze-dried for further use.
Characterization studies
Scanning electron microscopy (SEM) images were obtained using a scanning electron microscope (SEM, JSM-6510LV, JEOL, Japan). Transmission electron microscopy (TEM, JEM-2100F, JEOL, Ltd, Japan), powder X-ray diffraction (PXRD, RINT ULTIMA XRD, Rigaku Co., Japan), and UV–Vis absorption spectra measurements (UV-1800, Shimadzu, Kyoto, Japan) were performed as described in our previous work.2,16Detection of IV/A/H1N1 and clinically isolated IV/A/H3N2
Briefly, different concentrations of the target virus, IV/H1N1, were added into the wells containing capturing anti-hemagglutinin (HA) antibody (anti-HA Ab) under the optimized amount at 100 ng (Fig. S1a†) and blocking agent, 5% bovine serum albumin. The incubation of antibody-virus binding is optimized up to 1 h (Fig. S1b†). After washing with PBS-0.1%Tween-20, 100 μL of anti-HA Ab-TMB-NPS@PLGA complex was added into the wells and incubated for an additional 1 h. The captured IV/A/H1N1 was bound to the anti-HA Ab-TMB-NPs@PLGA, and the immunocomplex sandwich was formed.The wells were washed, and after the separation, 50 μL of the DMSO solution was added to disrupt the PLGA nanovesicles and release the encapsulated TMB-NPs. One hundred microliters of the prepared substrate solution (H2O2 and CuNFs) were added to the wells, and blue color developed rapidly. After 5 min, 10% H2SO4 was added to stop the reaction, resulting in a change of the blue color to yellow, followed by the absorbance measurement. Clinically isolated IV/A/Yokohama/110/2009/H3N2 and the spike protein of SARS-CoV-2 were applied following the same protocol using virus-specific antibody-conjugated TMB-NPs@PLGA for confirming the versatility of the assay system. For the specificity of the developed immunoassay, six control experiments were performed: 100 pg m−1 of hepatitis E virus-like particles (HEV-LPs) and norovirus-like particles (NoV-LPs), 104 copies per mL white spot syndrome virus (WSSV), 103 pfu mL−1 Zika virus, 20 μg mL−1 norovirus (NoV) and mixture all the viruses.
Results and discussion
Working principle of the encapsulation of TMB-NPs for virus detection
The working principle of the developed TMB-NPs@PLGA-based colorimetric sensing method for the ultrasensitive detection of IV/A/H1N1 is shown in Scheme 1. It comprises TMB-NP-encapsulated PLGA nanovesicles (TMB-NPs@PLGA), which are collectively self-assembled TMB molecules and BSA inside PLGA nanovesicles (Scheme 1a). Initially, the anti-HA Ab is immobilized in the microtiter plate wells via hydrophobic interaction to the polystyrene of the microplate. Different concentrations of the target IV/A are added to the anti-HA Ab immobilized wells (Scheme 1b). Furthermore, anti-HA Ab-conjugated TMB-NPs@PLGA are added to the wells allowing the formation of an immunocomplex sandwich structure. Anti-HA Ab-conjugated TMB-NPs@PLGA and anti-HA Ab formed a sandwich-like structure with IV/A as depicted in Scheme 1c. After incubation, the wells are washed with PBS to remove the excess reagents and unbound anti-HA Ab-conjugated TMB-NPs@PLGA. After that, the successfully separated TMB-NPs@PLGA sandwich nanoconjugates in the wells were treated with DMSO to dissolve the PLGA membrane and release the encapsulated TMB-NPs (Scheme 1d). The dissolving of the TMB-NPs@PLGA represents the first step of releasing the signal molecules in the system.Additionally, the released TMB-NPs are also dissolved by the DMSO solution, acquiring highly concentrated TMB in the solution (Scheme 1e), marking the second step of presenting the signal molecules in the system. The mixture of self-assembled CuNFs as peroxidase-mimicking nanomaterials and H2O2 solution are added to the wells for facilitating the catalytic oxidation of released TMB (Scheme 1f), exhibiting the color signals as proportional to quantification of the captured virus. In the absence of the target virus, the immunocomplex sandwich structure between TMB-NPs@PLGA and the bounded virus will not form. Without the TMB-NPs@PLGA in the system, there is an absence of TMB to be oxidized by the CuNFs and H2O2 mixture for generating a colorimetric signal.
Characterization of the TMB-NPs, TMB-NPs@PLGA, and self-assembled CuNFs
The molecular self-assembly technique for nanoparticle preparation has evolved as a promising strategy to integrate different components for synthesizing desired hybrid nanomaterials spontaneously. The synthesis of TMB-NPs encapsulated within TMB-NPs@PLGA involves two steps, as illustrated in Fig. 1a and b: (a) synthesis of TMB-NPs and (b) encapsulation of the synthesized TMB-NPs inside PLGA nanovesicles. The preparation commences with the self-assembly of TMB molecules using BSA as a template26 with excellent dispersibility. The TEM image of the synthesized TMB-NPs is shown in Fig. 1c. The synthesized TMB-NPs are roughly circular with an average diameter of about 50 nm (Fig. 1d), showing electron density of TMB within the BSA aggregation. The TMB-NP encapsulated PLGA nanovesicles (TMB-NPs@PLGA) are synthesized using a modified double emulsion solvent diffusion method. The formation of TMB-NPs@PLGA is verified using TEM (Fig. 1e and f). TMB-NPs@PLGA is present in the form of collective TMB-NPs clustering together with a diameter of around 200 nm. The TEM image confirms the encapsulation of TMB-NPs in the PLGA nanovesicle (Fig. 1f), which clearly shows TMB-NPs agglomerate as the TMB-NPs are incorporated inside the PLGA nanovesicles with no sign of aggregation. Fig. 1g shows the digital photographs of TMB derived biomaterial dispersibility. TMB molecules were dissolved in an aqueous solution resulting in a light yellowish-white precipitate.In contrast, TMB and BSA can be completely dissolved in DMSO and PBS, respectively, forming a clear solution. Accordingly, the self-assembled TMB-NPs are uniformly dispersed in PBS, attributing to the better aqueous dispersibility obtained from BSA's intrinsic hydrophilic property. TMB-NPs@PLGA are also found to maintain their uniform dispersibility in PBS similar to TMB-NPs due to free carboxyl groups from the lactic acid of PLGA on the surface of TMB-NPs@PLGA.
Inspired by the unique features of protein-inorganic hybrid nanoflowers through self-assembly, BSA-Cu3(PO4)2·3H2O hybrid nanoflowers (CuNFs) were prepared. The SEM image in Fig. 1h shows the flower-like structures of CuNFs with an average size of 400–500 nm, having hierarchical structures with high surface-to-volume ratios. The TEM image of CuNFs (Fig. 1i) clearly shows the uniformly dispersed CuNFs attributed to the loaded hydrophilic BSA molecules. To confirm the structural properties of the CuNFs, XRD analysis was further performed. The XRD patterns of CuNFs (Fig. S2 of ESI†) are in good agreement with the JCPDS card (00-022-0548)27,28 indicating that CuNF crystals are mainly composed of Cu3(PO4)2·3H2O crystals.
Catalytic activity of self-assembled CuNFs
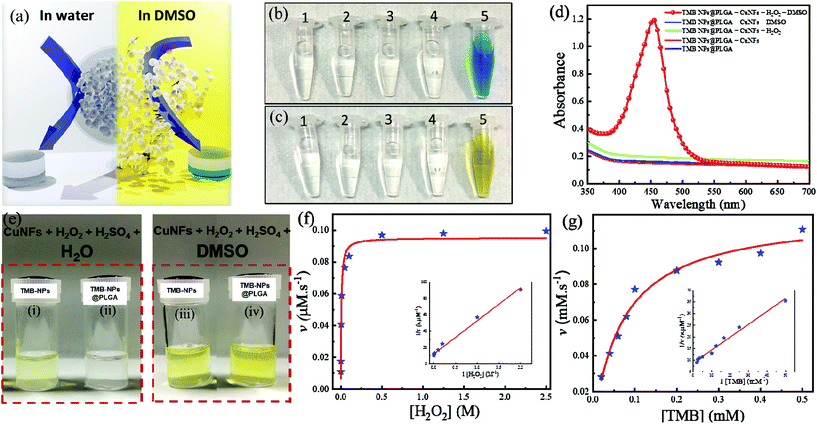
TMB-NPs were successfully encapsulated inside the PLGA nanovesicles. With regard to utilization in immunoassay sensing, these encapsulated TMB-NPs need to be released to exhibit the detection signal required for virus detection. Fig. 2a shows the schematic illustration of the release of TMB-NPs in the presence of water or DMSO. These released TMB-NPs are incubated with H2O2 and CuNFs. As shown in Fig. 2b and c, there is no occurrence of the developed color in the absence of the H2O2 as the co-substrate, CuNFs as the nanozyme, or DMSO as the releasing agent. The blue color changes to yellow after adding the acid solution to stop the TMB oxidation reaction (Fig. 2c). The absorbance of the developed color is shown in Fig. 2d, indicating a single peak in the presence of all components of the assay and the stability of TMB as signal molecules in the encapsulation and dissolving-and-releasing process. Also, the TMB-NPs and TMB-NPs@PLGA are assayed before and after the addition of DMSO. The digital photograph illustrates the difference of TMB-NPs with and without nanoencapsulation of PLGA. It shows a well-dispersed partially transparent solution to TMB-NPs@PLGA in the absence of DMSO (Fig. 2e). Opposite to its counterpart, TMB-NPs without the PLGA nanovesicle develop a slightly yellowish color with a partially white cloudy solution in the presence of H2O2/CuNFs under acidic conditions. These show desirable spatial protection on the TMB-NPs by the PLGA nanovesicle, isolating the TMB-NPs from external stimuli. In DMSO's presence, the higher catalytic oxidation of TMB is observed on both TMB-NPs and TMB-NPs@PLGA, pointing out the dissolving-and-release mechanism of TMB-NPs@PLGA. DMSO dissolves the nanovesicles and follows up with the dissolution of TMB-NPs, releasing the TMB molecules to the H2O2/CuNF solution.The apparent steady-state kinetic parameters are examined to determine the catalytic activity of self-assembled CuNFs. By keeping the concentration of the respective co-substrate constant, the experiments were performed with various concentrations of the substrate and a fixed concentration of the co-substrate. Referring to the typical Michaelis–Menten kinetics, the Lineweaver–Burk double reciprocal plot is generated. The reaction velocity as the function of the respective concentration of the substrate is plotted. It shows excellent linearity indicating the intrinsic properties of CuNFs as a nanozyme (inset of Fig. 2f and g). As listed in Table S1,† the Km value (mM) and maximal velocity (vmax) of CuNFs for both substrates were determined based on the Michelin–Menten graph. The Km values of CuNFs and HRP to TMB are 0.07 mM and 0.43 mM, respectively. This shows that CuNFs exhibit a lower Km value than horseradish peroxidase (HRP), indicating a higher affinity of the nanozyme to the TMB substrate. Besides, the vmax of CuNFs is 0.12 μM s−1 for TMB and 0.02 μM s−1 for H2O2. The CuNFs possess a high surface area and essential electrostatic interaction to the favorable TMB substrate and H2O2, which allow a considerably sufficient peroxidase-like activity fulfilling the prerequisite to develop sensitive nanozyme-based colorimetric immunoassay.
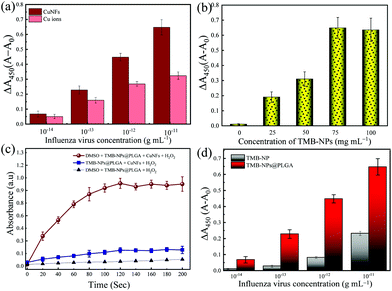
Furthermore, the catalytic activity of CuNFs with the optimum concentration at 50 μg mL−1 (Fig. S1c†) is comparable to copper (Cu) ions for the beneficial utilization in virus detection in the presence of 0.5 M H2O2 (Fig. S1d†). Based on the initial concentration of the CuNFs precursor, 10 mM Cu ions are assayed against the catalytic properties of the CuNFs in the influenza virus detection. Using Cu ions only, the signal is compared to catalytic CuNFs (Fig. 3a) at a low concentration of the influenza virus. At the nanogram level of the influenza virus, the signal by Cu ions is generated down to 50% compared to the CuNFs. The advantage of utilizing the nanozyme implicates the crystalline structure of the copper-phosphate and the catalytic properties of the nanoflower for a better signal generation with a lower baseline signal.
Optimization of TMB-NPs encapsulation in TMB-NPs@PLGA
In this work, the significant points for the signal amplification strategy for immunoassay purposes are the maximal encapsulation of TMB-NPs inside the PLGA nanovesicles and the reaction time required to develop the color change. Firstly, the loading amount of TMB-NPs is optimized to achieve maximal encapsulation inside the PLGA nanovesicles. The as-prepared TMB-NP solutions of 25, 50, 75, and 100 mg mL−1 are used for the PLGA encapsulation with 0 mg mL−1 representing empty PLGA nanovesicles. As shown in Fig. 3b, it is evident that the absorbance signal continuously increased along with the increasing concentrations of TMB-NPs, attributing to the increase of the TMB-NP concentration in the presence of CuNFs and H2O2. To optimize the encapsulated number of TMB-NPs in the PLGA nanovesicles, the assay is performed using various concentrations of TMB-NPs encapsulated in PLGA nanovesicles with and without DMSO. It can be seen that there is no signal amplification with and without the addition of DMSO in empty PLGA nanovesicles, showing the TMB signal solely in the form of TMB-NPs. Comparing the signal generated before and after the DMSO addition, 75 mg mL−1 is determined as the optimal loading concentration in the PLGA nanovesicles.Furthermore, based on the absorbance value, the corresponding TMB molecules’ concentration is calculated corresponding to its molar absorption coefficient. The catalytic oxidized TMB molecules’ concentration is estimated at 43.1 mg mL−1, which indicates that the loading capacity of the PLGA nanovesicle is up to 53.4%. The change of absorbance upon the release of TMB-NPs from PLGA nanovesicles synthesized using 100 mg mL−1 is relatively similar to 75 mg mL−1. The difference in absorbance intensity before and after the release of TMB-NPs upon the addition of DMSO expressed in the change of absorbance is highly desirable for obtaining sensitive and reliable detection. Therefore, TMB-NPs@PLGA nanovesicles, which are synthesized using 75 mg mL−1 of TMB-NPs solution, were used to prepare TMB-NPs@PLGA in further experiments.
Fig. 3c shows the incubation time as the function of the absorbance change in the mixture of TMB-NPs@PLGA and CuNFs/H2O2 with and without DMSO. Before the optimization, the TMB-NPs@PLGA was reacted with only H2O2. Low absorbance was shown due to no involvement of the catalytic reaction by CuNFs and grounding the need for CuNFs as a nanozyme. Although there is an increase in the absorbance response compared to the only H2O2, a slight change of absorbance is observed in TMB-NPs@PLGA incubated with CuNFs/H2O2.
In contrast, the absorbance noticeably and gradually increases after DMSO addition and incubation of TMB-NPs@PLGA with CuNF/H2O2. The absorbance increases rapidly from 0 to 100 s of the incubation time, and the stationary absorbance intensity is observed after 120 s. There is a provision period to maximize the signal, mainly based on the equilibrium of the redox reaction between the TMB/H2O2 system and CuNFs. After the equilibrium, the absorbance is constant until 200 s. Therefore, the reaction time for developing the color is optimized to be 120 s in the developed colorimetric immunoassay for virus detection.
Analytical performance of the sensing system
To achieve a highly sensitive detection limit, the main point of importance is the nanoparticle encapsulation-based signal amplification strategy in this developed colorimetric immunoassay. First, to underline the advantage of the encapsulation of TMB-NPs inside the PLGA nanovesicles in amplifying the signal, the bare TMB-NPs, which are self-assembled with anti-HA Ab and BSA, are used instead of TMB-NPs@PLGA for detecting IV/A. Hypothetically, the TMB-NP can demonstrate a color signal for IV/A detection similar to the TMB-NPs@PLGA. Based on the antigen–antibody interaction, a sole immunocomplex can only be abutted with a single TMB-NP. In contrast, the encapsulation of TMB helps in localization of the signal substrate and enriches the signal generation for individual antibody–antigen binding. The IV detection immunoassay using bare TMB-NPs and TMB-NPs@PLGA is shown in Fig. 3d. Interpreting the difference in the absorbance signal, utilizing the TMB-NPs@PLGA improved 100-fold higher detection sensitivity compared to only TMB-NPs. The encapsulation of the TMB-NPs localizes a higher concentration of TMB molecules even at a low concentration of the virus up to 3.5-fold of localized concentration of the colorimetric signal.Besides the encapsulation of the signal molecules, in juxtaposition to our previous work11,29 using electrostatic interaction as the conjugation force, the antibodies’ chemical conjugation on the nanovesicles upholds the increase of the antibody–antigen by preventing the random confirmation on the antibodies. Moreover, the PLGA nanovesicles provide the conjugation site; instead of the TMB-NPs, the reactivity and morphology of the NPs are well-preserved within the PLGA nanovesicles. Lastly, TMB molecules’ encapsulation instead of the nanozyme like noble metals or metal oxides nanoparticles15,16 shows more effective signal generation. The catalytic oxidation of the TMB molecules as the determining factor is reflected in the low value of the Km of the CuNFs. In relation to the low detection limit, the collective TMB molecules’ reactiveness in the form of NPs within the PLGA nanovesicles is essential for amplifying the signal in the immunoassay format.
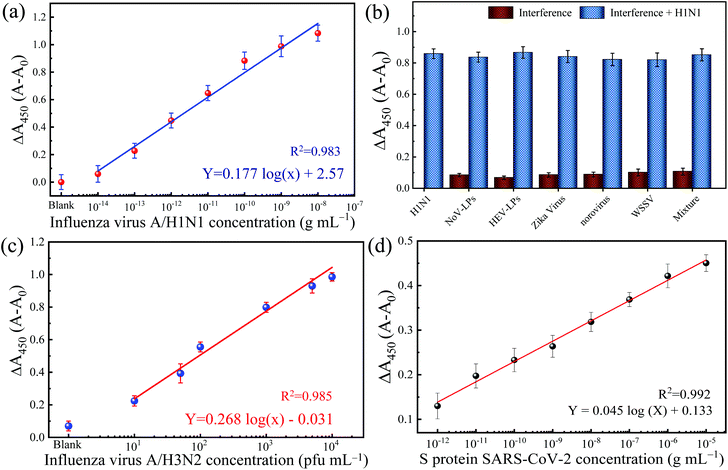
Adopting a typical immunosorbent assay, the colorimetric immunoassay is applied to increasing concentrations of IV/A/H1N1. The corresponding result of blue color shows a gradient pattern from low to intense blue color and changed proportionally to yellow color after the reaction is stopped. The measured absorbance at 450 nm (with 650 nm as the reference absorbance) increased as the function of the concentration of IV/A/H1N1 (Fig. 4a). The calibration line with a correlation coefficient (R2) of 0.983 was obtained with satisfactory linearity from 101–106 fg mL−1. Also, the limit of detection (LOD) is 32.37 fg mL−1, which is determined based on the equation 3.3σ − c/S,30 in which c, S and σ represent the y-intercept and the gradient slope of the calibration line and the standard deviation of the blank signal, respectively. The low LOD can be attributed to two contributing factors: the presence of optimized nanozyme concentration and highly localized TMB molecules in low antibody–antigen conjugation, inclining the increase in the slope of the detection and generating a signal in the low quantity of immuno-conjugation.
To establish a reliable and practical immunoassay, a dilution series of concentrations of IV/A/H1N1 is spiked in the biological human serum matrix. The developed colorimetric immunoassay shows a linear increase of absorbance to the function of the introduced virus in the medium. Fig. S3 in the ESI† shows the calibration line for IV/A/H1N1 detection using 10% human serum as a detection medium. The gradient of the calibration line is slightly flat shifted than the linear calibration from the IV detection in buffer solution up to 15%. However, the responsive linearity of the IV detection confirmed the applicability of the developed colorimetric immunoassay in a sophisticated serum matrix. Prompt to the matrix effect, the slope of the calibration curve is flattened, resulting in an increase of the LOD value to 54.97 fg mL−1, which is comparable to previously reported studies for real applications (Table S2†).31–34
Specificity of the TMB-NPs@PLGA-based immunoassay
The selectivity of the developed TMB-NPs@PLGA based colorimetric immunoassay for IV/A/H1N1 detection is evaluated. The dsDNA virus WSSV, the recombinant expressed NoV-LPs and HEV-LPs, and the RNA virus, Zika virus, and NoV are assayed as interferences (negative analytes) to examine the anti-interference effect and the selectivity of the developed TMB-NPs@PLGA-based immunoassay, which is collated to the response of IV/A/H1N1. Fig. 4b shows the change in absorbance intensity of the developed immunoassay to the mixture of negative analytes and IV/A/H1N1. The intense color developed in the presence of IV/A/H1N1 in the samples, but not in the negative samples containing interferences only. The interfering viruses are unrecognizable by the capturing antibodies and anti-HA antibodies on the conjugated TMB-NPs@PLGA, which are specific to IV/A. As a result, after washing, there is no TMB-NPs@PLGA in the reaction chamber, resulting in no TMB molecules even after the addition of DMSO, the only small increase in the absorbance intensity in the detection of the negative samples justified the superior specificity of TMB-NPs@PLGA-based colorimetric immunoassay towards IV/A detection.Detection of clinical-isolated IV/A/H3N2 and S-protein of SARS-CoV-2
To demonstrate the practicability of the TMB-NPs@PLGA-based colorimetric immunoassay, clinically isolated samples containing IV/A/H3N2 are applied to the developed immunoassay (Fig. 4c). The absorbance intensity increased continuously proportionally to the increasing concentration of IV/A/H3N2 from 101–104 pfu mL−1, developing the proportional gradient of color (Fig. S4†). The detection of the IV/A/H3N2 shows the R2 value of 0.985 with the LOD of 17 pfu mL−1. The developed immunoassay using the TMB-NPs@PLGA nanovesicles proffers a higher sensitivity than the available commercial IV detection kit (Quicknavi-Flu 2, Denka Seiken Co., Ltd, Tokyo, Japan) and the conventional ELISA using HRP and gold nanoparticles (Table S3†).31,32 These results strongly demonstrate that our TMB-NPs@PLGA-based immunoassay possesses a strong anti-interference ability and is highly sensitive for practical use.In response to the recent outbreak of Covid-19, the TMB-NPs@PLGA-based immunoassay was applied to detect the spike protein of SARS-CoV-2. The spike protein is used as the antigen to demonstrate direct coronavirus detection targeting the surface protein. As shown in Fig. 4d, the wide linearity from the picogram to nanogram level is achieved with an excellent linear response depending on the concentration of the spike protein. The LOD is calculated down to 143 fg mL−1 with the R2 value of 0.992, in which the femtogram level detection of the spike protein of SARS-CoV-2 agrees to other biosensors with higher complexity fabrication and analysis.35–37
Conclusions
In this study, we have prepared TMB-NP-encapsulated PLGA nanovesicles using self-assembly and the nano-co-precipitation process. The TMB-NPs@PLGA nanovesicles serve as a signal amplification source for the highly selective and sensitive detection of the IV/A. Self-assembly of TMB-NPs@PLGA integrated the dispersity of hydrophobic molecules by BSA and polymeric encapsulation. The bound TMB-NPs@PLGA recognizing the captured virus could be dissolved with DMSO, releasing encapsulated TMB-NPs. Our results further successfully demonstrated the simultaneous dissolving of the PLGA nanovesicles and TMB-NPs, enriching the signal molecules in low antibody–antigen sandwich structure nanoconjugates. Released TMB-NPs produced an amplified colorimetric signal upon oxidation by self-assembled CuNFs in the presence of H2O2. The developed immunoassay utilizing the TMB-NPs@PLGA nanovesicles and CuNF nanozyme demonstrates the detection of the IV/A using the colorimetric signal readout in a linear response from the 10 fg level to 10 ng level with the LOD of 32 fg mL−1 and a linear range of 101 to 104 pfu mL−1 of clinically isolated IV/A/H3N2 with a LOD of 17 pfu mL−1. Furthermore, concentrated signal molecule nanovesicles in the immunoassay are adopted to target the spike protein of SARS-CoV-2, which shows femtogram level detection. The proposed immunoassay provides a reliable and easy-to-be-adapted ultrasensitive diagnostic platform to detect various respiratory-infecting viruses.Conflicts of interest
There is no conflicts to declare.Acknowledgements
We would like to express our sincere thanks to Dr Tian-Cheng Li of the National Institute of Infectious Diseases, Dr Jun Satoh of the National Research Institute of Aquaculture of Japan Fisheries Research and Education Agency, Professor K. Morita of the Institute of Tropical Medicine Nagasaki University, and Mr F. Abe of the Department of Microbiology, Shizuoka Institute of Environment and Hygiene for providing HEV-LP, WSSV, Zika virus, and NoV, respectively. A. B. G. and I. M. K. sincerely thank the Japan Society for the Promotion of Science (JSPS) for a postdoctoral fellowship (19F19064) and research fellowship-DC (20J22358), respectively.Notes and references
- A. D. Chowdhury, A. B. Ganganboina, Y.-C. Tsai, H.-C. Chiu and R.-A. Doong, Anal. Chim. Acta, 2018, 1027, 109–120 CrossRef
.
- A. D. Chowdhury, A. B. Ganganboina, F. Nasrin, K. Takemura, R.-A. Doong, D. I. S. Utomo, J. Lee, I. M. Khoris and E. Y. Park, Anal. Chem., 2018, 90, 12464–12474 CrossRef
.
- A. B. Ganganboina and R.-a. Doong, Microchim. Acta, 2018, 185, 526 CrossRef
.
- S. Oh, J. Kim, V. T. Tran, D. K. Lee, S. R. Ahmed, J. C. Hong, J. Lee, E. Y. Park and J. Lee, ACS Appl. Mater. Interfaces, 2018, 10, 12534–12543 CrossRef CAS
.
- S. R. Ahmed, J. Kim, T. Suzuki, S. Neethirajan, J. Lee and E. Y. Park, Sci. Rep., 2017, 7, 44495 CrossRef CAS
.
- R. C. Murdock, L. Shen, D. K. Griffin, N. Kelley-Loughnane, I. Papautsky and J. A. Hagen, Anal. Chem., 2013, 85, 11634–11642 CrossRef CAS
.
- L. Yu, C. M. Li, Y. Liu, J. Gao, W. Wang and Y. Gan, Lab Chip, 2009, 9, 1243–1247 RSC
.
- H. Lin, Y. Liu, J. Huo, A. Zhang, Y. Pan, H. Bai, Z. Jiao, T. Fang, X. Wang and Y. Cai, Anal. Chem., 2013, 85, 6228–6232 CrossRef CAS
.
- Y. Zhao, Y. Zheng, R. Kong, L. Xia and F. Qu, Biosens. Bioelectron., 2016, 75, 383–388 CrossRef CAS
.
- C. N. Loynachan, M. R. Thomas, E. R. Gray, D. A. Richards, J. Kim, B. S. Miller, J. C. Brookes, S. Agarwal, V. Chudasama and R. A. McKendry, ACS Nano, 2018, 12, 279–288 CrossRef CAS
.
- I. M. Khoris, A. D. Chowdhury, T.-C. Li, T. Suzuki and E. Y. Park, Anal. Chim. Acta, 2020, 1110, 64–71 CrossRef CAS
.
- N. Cheng, Y. Song, M. M. Zeinhom, Y.-C. Chang, L. Sheng, H. Li, D. Du, L. Li, M.-J. Zhu and Y. Luo, ACS Appl. Mater. Interfaces, 2017, 9, 40671–40680 CrossRef CAS
.
- A. B. Ganganboina and R.-a. Doong, Sens. Actuators, B, 2018, 273, 1179–1186 CrossRef CAS
.
- Y. Zhou, B. Liu, R. Yang and J. Liu, Bioconjugate Chem., 2017, 28, 2903–2909 CrossRef CAS
.
- H. Ye, K. Yang, J. Tao, Y. Liu, Q. Zhang, S. Habibi, Z. Nie and X. Xia, ACS Nano, 2017, 11, 2052–2059 CrossRef CAS
.
- A. B. Ganganboina, A. D. Chowdhury, I. M. Khoris, F. Nasrin, K. Takemura, T. Hara, F. Abe, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2020, 112169 CrossRef CAS
.
- S. Wang, Z. Chen, J. Choo and L. Chen, Anal. Bioanal. Chem., 2016, 408, 1015–1022 CrossRef CAS
.
- I. M. Khoris, K. Takemura, J. Lee, T. Hara, F. Abe, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2019, 126, 425–432 CrossRef CAS
.
- L. Miao, C. Zhu, L. Jiao, H. Li, D. Du, Y. Lin and Q. Wei, Anal. Chem., 2018, 90, 1976–1982 CrossRef CAS
.
- R. Ren, G. Cai, Z. Yu, Y. Zeng and D. Tang, Anal. Chem., 2018, 90, 11099–11105 CrossRef CAS
.
- L. Sercombe, T. Veerati, F. Moheimani, S. Y. Wu, A. K. Sood and S. Hua, Front. Pharmacol., 2015, 6, 286 Search PubMed
.
- Y. Lin, Q. Zhou, Y. Zeng and D. Tang, Microchim. Acta, 2018, 185, 311 CrossRef
.
- U. Nagaich, J. Adv. Pharm. Technol. Res., 2018, 9, 65 CrossRef
.
- P. Kocbek, N. Obermajer, M. Cegnar, J. Kos and J. Kristl, J. Controlled Release, 2007, 120, 18–26 CrossRef CAS
.
- Y. Zhang, H. Wang, S. Stewart, B. Jiang, W. Ou, G. Zhao and X. He, Nano Lett., 2019, 19, 9051–9061 CrossRef CAS
.
- L. Jiao, H. Yan, W. Xu, Y. Wu, W. Gu, H. Li, D. Du, Y. Lin and C. Zhu, Anal. Chem., 2019, 91, 8461–8465 CrossRef CAS
.
- J. Ge, J. Lei and R. N. Zare, Nat. Nanotechnol., 2012, 7, 428–432 CrossRef CAS
.
- M. Li, M. Luo, F. Li, W. Wang, K. Liu, Q. Liu, Y. Wang, Z. Lu and D. Wang, J. Phys. Chem. C, 2016, 120, 17348–17356 CrossRef CAS
.
- S. R. Ahmed, J. Kim, T. Suzuki, J. Lee and E. Y. Park, Biotechnol. Bioeng., 2016, 113, 2298–2303 CrossRef CAS
.
- A. B. Ganganboina and R.-A. Doong, Sci. Rep., 2019, 9, 1–11 CrossRef CAS
.
- K. Takemura, O. Adegoke, N. Takahashi, T. Kato, T.-C. Li, N. Kitamoto, T. Tanaka, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2017, 89, 998–1005 CrossRef CAS
.
- S. R. Ahmed, M. A. Hossain, J. Y. Park, S.-H. Kim, D. Lee, T. Suzuki, J. Lee and E. Y. Park, Biosens. Bioelectron., 2014, 58, 33–39 CrossRef CAS
.
- Y.-F. Chang, S.-F. Wang, J. C. Huang, L.-C. Su, L. Yao, Y.-C. Li, S.-C. Wu, Y.-M. A. Chen, J.-P. Hsieh and C. Chou, Biosens. Bioelectron., 2010, 26, 1068–1073 CrossRef CAS
.
- U. Jarocka, R. Sawicka, A. Góra-Sochacka, A. Sirko, W. Zagórski-Ostoja, J. Radecki and H. Radecka, Sensors, 2014, 14, 15714–15728 CrossRef
.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park and H. G. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS
.
- S. Mavrikou, G. Moschopoulou, V. Tsekouras and S. Kintzios, Sensors, 2020, 20, 3121 CrossRef CAS
.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr06893d |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |