Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An electrochemical SARS-CoV-2 biosensor inspired by glucose test strip manufacturing processes†
Vincent J.
Vezza
 *a,
Adrian
Butterworth
*a,
Adrian
Butterworth
 a,
Perrine
Lasserre
a,
Perrine
Lasserre
 a,
Ewen O.
Blair
a,
Alexander
MacDonald
a,
Ewen O.
Blair
a,
Alexander
MacDonald
 a,
Stuart
Hannah
a,
Stuart
Hannah
 a,
Christopher
Rinaldi
b,
Paul A.
Hoskisson
a,
Christopher
Rinaldi
b,
Paul A.
Hoskisson
 c,
Andrew C.
Ward
c,
Andrew C.
Ward
 d,
Alistair
Longmuir
d,
Alistair
Longmuir
 e,
Steven
Setford
e,
Eoghan C. W.
Farmer
f,
Michael E.
Murphy
fg and
Damion K.
Corrigan
e,
Steven
Setford
e,
Eoghan C. W.
Farmer
f,
Michael E.
Murphy
fg and
Damion K.
Corrigan
 a
a
aBiomedical Engineering, University of Strathclyde, Glasgow, G1 1XP, UK. E-mail: vincent.vezza@strath.ac.uk
bPure & Applied Chemistry, University of Strathclyde, Glasgow, G1 1XP, UK
cStrathclyde Institute of Pharmacy and biomedical Sciences (SIPBS), University of Strathclyde, Glasgow, G1 1XP, UK
dCivil & Environmental Engineering, University of Strathclyde, Glasgow, G1 1XP, UK
eLifeScan Scotland Ltd, Beechwood Park North, Inverness, IV2 3ED, UK
fNHS GGC, Glasgow Royal Infirmary, Department of Microbiology, NEW Lister Building, Glasgow, G31 2ER, UK
gSchool of Medicine, Dentistry & Nursing, College of Medical Veterinary & Life Sciences, University of Glasgow, G12 8QQ, UK
First published on 12th March 2021
Abstract
Accurate and rapid diagnostic tests are critical to reducing the impact of SARS-CoV-2. This study presents early, but promising measurements of SARS-CoV-2 using the ACE2 enzyme as the recognition element to achieve clinically relevant detection. The test provides a scalable route to sensitive, specific, rapid and low cost mass testing.
SARS-CoV-2 came to the attention of health authorities during late 2019, shortly followed by declaration of a “public health emergency of international concern” on 30th of January 2020. SARS-CoV-2 quickly spread around the globe and was declared a pandemic by the World Health Organisation on 11th of March 2020. The virus, SARS-CoV-2, is the aetiological agent of coronavirus disease (Covid-19). Diagnostics have been a major priority and challenge thus far in the pandemic with assay quantity, reagent costs and time to result being of prime concern. SARS-CoV-2 has four major structural proteins,1 with the spike protein known to bind to the surface of cells expressing angiotensin converting enzyme 2 (ACE2) on their surface. The affinity between ACE2 and the spike protein has been shown to be in the low nanomolar range2 giving a similar level of affinity to an antibody-antigen interaction. The enzyme represents an important candidate molecule for construction of a biosensor because of the high affinity between the spike protein and ACE2 and the fact that a limited number of coronaviruses utilise ACE2 for entry (SARS-CoV-1, SARS-CoV-2 and HCoV-NL63) HCoV-NL63 causes infection primarily in young children, has a lower overall affinity for ACE2 (3–10 times lower)3 and is a pathogen responsible for only mild/moderate childhood disease. It is therefore possible for ACE2 to be deployed as a selective receptor in various biosensor formats for this crucial category of human respiratory pathogen, either to allow definitive diagnosis of SARS-CoV-2 in adults or as a screening tool for identifying positive cases, which then receive confirmatory lab testing. Finally, the ACE2 enzyme is a carboxypeptidase responsible for the hydrolytic cleavage of angiotensin II to angiotensin [1,7] liberating phenylalanine in the process. The active site is positioned away from SARS-CoV-2 binding site meaning that enzyme activity and binding from other proteins or small molecules in human samples is likely to be insignificant when compared to binding of an entire SARS-CoV-2 viral particle (50–200 nm) and the resulting effect on electrochemical signal. A critical feature of the ACE2 enzyme is the hydrophobic region which normally facilitates insertion into cell membranes4 and allows ACE2 insertion into a synthetically made amphiphobic structures resembling cell membranes.
The fluorous effect is a well-known and well described tendency for fluorine atoms to avoid unfavoured interactions with other elements.5 Formation of fluorous self-assembled monolayers (SAMs) have been utilised in organic electronics to reduce biological fouling of surfaces.6 Here, a perfluorocarbon SAM formed of 1H,1H,2H,2H-Perfluorodecanethiol (PFDT) is deployed on a gold sensor surface to form a layer of amphiphobic character which facilitates straightforward insertion of ACE2 via its hydrophobic tail7 (ordinarily employed for membrane insertion in vivo). Use of a perfluorocarbon SAM with its unique properties provided a robust combination of anti-biofouling behaviour and enzyme insertion, allowing straightforward sensor preparation.
Electrochemical biosensors offer a very attractive route to sensitive and low cost detection of biological analytes. The most notable example of this is the glucose biosensor, operating via the glucose oxidase enzyme which is in widespread use each day by diabetic patients for routine determination of blood glucose levels. As a result of demand, facilities exist with very high volume manufacturing capability (e.g. several million sensors per day) and very well characterised chemical layer and enzyme deposition protocols. Due to the volume of sales in the glucose market (similar demand would exist for Covid-19 screening) unit costs of 20 UK pence8 can be achieved using established manufacturing processes. Very few diagnostic technologies, including lateral flow assays, can currently compete from a cost and volume perspective (e.g. current lateral flow Covid-19 assays cost £5–20 per test in the UK). In contrast, the sensor presented here requires a simple two stage preparation procedure of (1) PFDT deposition on a low cost PCB electrode and (2) ACE2 functionalisation through physisorbtion into the PFDT (Scheme 1). These steps are compatible with current manufacturing practices for glucose biosensors and importantly provide opportunity to rapidly translate the approach described here for SARS-CoV-2 detection into a glucose test strip production environment. This unlocks potential to achieve an extremely low cost Covid-19 assay, which utilises already CE marked substrates and approved assay readers, thus minimising the regulatory burden. Additionally, the assay is mutation proof as it exploits the interaction between SARS-CoV-2 spike protein and ACE2, has a degree of in-built surface orientation through the ability of ACE2 to insert via its hydrophobic region and offers the opportunity to develop similar tests for other respiratory viruses entering cells via membrane bound surface proteins.
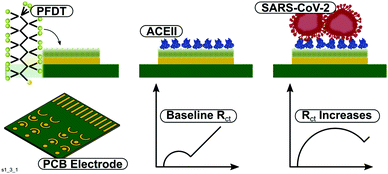 | ||
| Scheme 1 Top – Two stage functionalisation process of the PFDT-ACE sensor and virus binding. Bottom – The electrode chip and the resulting RCT increase upon virus recognition. | ||
PFDT-SAM and ACE2 functionalisation experiments were performed on a PCB sensor array bearing eight gold working electrodes. Such PCB arrays are low cost and allow high throughput assay development and represent an excellent test bed for this assay. When functionalised with PFDT and ACE2, the charge transfer resistance (RCT) of the [Fe(CN)6]−3/−4 redox reaction, a signal parameter commonly used in impedimetric biosensor measurements, increased as expected (Fig. S1, ESI†). The PFDT SAM layer caused an increase in RCT and a further increase was noted when ACE2 was physisorbed into the PFDT layer, confirming successful insertion of the enzyme. This was in contrast to experiments on facile ACE2 insertion with thiolated-hydrocarbon layers, where ACE2 insertion behaviour and sensor performance was not reproducible (Fig. S2, ESI†).
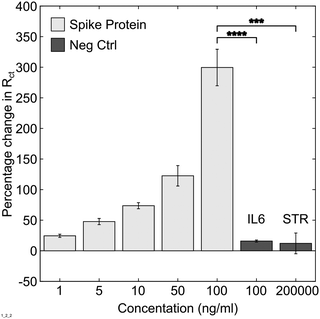
To characterise the sensitivity and specificity of the sensor, experiments to assess dose response behaviour and determine specificity of the ACE2 spike protein interaction were conducted using recombinant spike protein, streptavidin and IL-6. IL-6, which is found at elevated levels in Covid-19 patients, and streptavidin, widely used in bioassays, were employed as negative controls. A clear dose response effect was found for recombinant spike-protein binding to ACE2 (Fig. 1). Fitting the dose response curve gave a limit of detection for recombinant spike protein of 1.68 ng mL−1. The binding between ACE2 and spike protein at 100 ng mL−1 was shown to be significantly different to IL-6 (100 ng mL−1, P = 3.158E-7) and streptavidin (200 μg mL−1, P = 3.981E-4) binding. This suggests it was possible to construct a sensitive and selective sensor operating on the principle of ACE2-spike glycoprotein interactions. Note – example Nyquist plots, equivalent circuit and representative values are shown in Fig. S3 (ESI†).
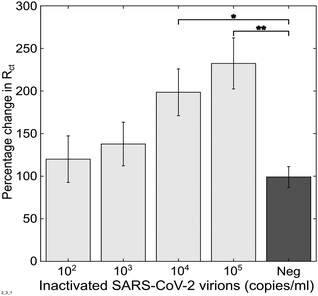
Next, assay performance in more complex samples was explored. Due to biosafety restrictions, the sensor was tested with inactivated virus samples at different concentrations alongside a negative sample from a commercial kit. All samples were provided in viral transport medium which contains proteins and dead eukaryotic cells used to culture the virus and was an appropriate proxy for a complex human tissue such as saliva. Furthermore, these kits are used to validate diagnostic tests in clinical biomedical laboratories because of their close resemblance to clinical samples.9 When a viral dose response curve was constructed, it was possible to detect the inactivated virus with increasing concentration (Fig. 2). Curve fitting and comparison to blank solution yielded an LoD of 38.6 copies mL−1. At 104 and 105 copies mL−1, the impedance change was significantly different to the negative control (P = 0.004 and P = 6.765E-4 respectively). The background signal increase observed in the negative control is most likely due to residual non-specific surface interactions from the complex viral sample medium (proteins, cellular debris etc.). To provide context, RNA levels for SARS-CoV-2 in saliva samples have been estimated in the range of 104–1011 copies per mL.10,11 Therefore, these findings show it was possible to detect inactivated SARS-CoV-2 from a complex sample in 30 minutes at clinically relevant detection levels. Coupling this to the sensor being designed for manufacture in mass production environments, it raises the possibility of developing a Covid-19 assay using established sensors and instrumentation from the blood glucose industry.
To assess performance of the sensor more comprehensively, two human saliva samples were tested in bio secure conditions. One SARS-CoV-2 positive human saliva sample, confirmed through PCR based clinical diagnostics (Ct = 26 cycles, indicating high viral load), was transferred into inactivation solution called VPSS containing a proprietary cell culture medium and Triton-X (to denature the viral envelope whilst leaving proteins largely intact). This was compared against a negative human saliva sample (confirmed via PCR) which had undergone the same handling processes as the positive sample. To carry out this experiment an improved assay protocol was used. Firstly, the positive and negative samples were premixed with ACE2 and then allowed to physisorb onto the PFDT modified sensor surface with a measurement taken immediately after sample addition. This increased the practicality of the assay by accelerating time to result and has the potential to make storage, transport easier and shelf life longer because sensor strips can be shipped without the ACE2 enzyme being preprinted. Secondly, the clinical samples were tested in VPSS medium. Once inactivated in VPSS, suspected SARS-CoV-2 positive samples can be manipulated at Biosafety Level 2 in contrast to untreated patient samples, or samples in Viral Transport Medium (VTM), which require Biosafety Level 3 facilities. The ability to detect SARS-CoV-2 in VPSS using the biosensor reported here means that as well as for self testing, the assay is compatible with more centralised testing facilities where it is important to protect an operator running multiple samples such as hospital wards, GP surgeries, care homes, airports and workplaces. Adopting this revised approach, the RCT changes in the positive and negative samples were found to be significantly different (Fig. 3, P = 1.2 × 10−7). Changes to the assay approach with clinical samples resulted in an apparent drop in RCT. This is attributed to a baseline artefact caused by Triton-X and sensor surface interactions in VPSS. Further investigation is being performed to fully understand and quantify these effects.
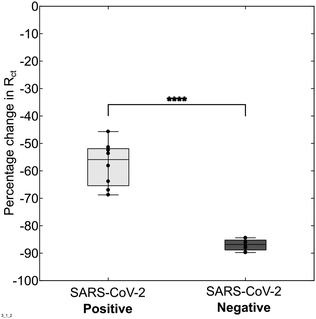 | ||
| Fig. 3 Assay signal response following incubation with one SARS-CoV-2 positive patient sample (Ct = 26) and a negative sample. N = 8, horizontal line = mean. | ||
Numerous SARS-CoV-2 biosensor approaches have been developed and reported recently. Magnetic beads, carbon black electrodes and multiple antibodies were employed to achieve a detection limit of 6.5 plaque forming units mL−1 in whole virus.12 An ultrasensitive super sandwich-type assay has been presented to detect viral RNA from SARS-CoV-2 with an LoD of 200 copies mL−1,13 and a FET biosensor capable of detecting spike protein with a LoD of 242 copies mL−1 in transport medium has also been reported.9 Plasmonic photothermal biosensors have been shown to detect down to 0.22 pM of viral RNA, which was estimated to be around 2.26 × 104 copies.14 Other electrochemical sensors have also shown good sensitivity, including molecular imprinted polymers (15 fM)15 and graphene and gold nanostars (1.68 × 10−22 μg mL−1).16 The sensor developed here has comparable and clinically relevant sensitivity (LoD for inactivated virus of 38.6 copies mL−1), with a clear route to manufacture. Importantly, the design of the sensor reported here is resistant to mutations in the spike protein. In terms of the chosen surface chemistry and the use of PFDT in a SAM layer, there are examples in the literature of other sensor systems with good anti-biofouling properties and these have been well reviewed.17 Demonstrations of anti-fouling properties in ternary SAMs have recently been made18,19 and there are advantages to these approaches for more complex sensors. However, in our experiments we could not reproduce the ACE2 insertion behaviour with standard hydrocarbon SAMs (Fig. S2, ESI†) and because of the emphasis on eventual sensor manufacture, PFDT was found to be well suited to the end application where monolayer coverage was sufficient to both retain the enzyme via amphiphobic interactions and provide anti-fouling properties.
Specificity of this sensor has been demonstrated against IL6, streptavidin, virial transport medium and one negative clinical saliva sample. This suggests that biofouling and non-specific binding of other commonly encountered biological proteins does not compromise assay sensitivity or specificity. Another consideration is other viral pathogens that bind to ACE2. Of the three viruses that are known to bind to ACE2, SARS-CoV-2 has the greatest affinity. SARS-CoV-1 is not present in the community and is therefore unlikely to impact the performance of this sensor. HCoV-NL63 primarily affects paediatric patient groups, which could limit the sensor to adult diagnostics if signal stratification is not possible. The current time to result based on the presented results is 30 min, which is competitive with existing SARS-CoV-2 assays but significant scope exists to reduce our assay time to faster sample turnaround times.
Further work will include sensor verification with a large set of clinical samples to determine, (1) whether low, medium and high viral load SASR-CoV-2 samples can be stratified alongside HCoV-NL63 samples or whether positive vs negative is the only feasible assay output, (2) fully understanding the measurement artefact effects of surfactants and detergents from VPSS and VTM, (3) testing shorter incubation times to reduce the overall assay time to result below the current 30 minutes and (4) testing simpler electrochemical measurements (e.g. amperometry) to determine if more straight forward measurement circuits (and therefore a lower cost reader can be employed) than those required for impedance.
These results present the basis of a novel, sensitive, selective and scalable biosensor assay which is based upon the principle of immobilising ACE2 into a layer with amphiphobic character. This sensor assay has sensitivity (1.68 ng mL−1) with recombinant spike protein and specificity against two proteins (IL-6 and streptavidin). When evaluated with a SARS-CoV-2 analytical grade laboratory inactivated virus kit, an LoD of 38.6 copies mL−1 was achieved. Finally, testing with high viral load clinical sample (Ct = 26) showed strong discrimination against a negative sample following inactivation in VPSS medium. By taking advantage of glucose biosensor production approaches (per sensor cost >£0.2) it is possible to demonstrate a clear path to translation through established manufacturing techniques, utilisation of existing sensor substrates and read out devices with prior CE marking. These findings together point towards a useful diagnostic tool, which can be rapidly deployed at low cost to screen for SARS-CoV-2 in a range of settings.
Conflicts of interest
There are no conflicts to declare.Notes and references
- P. S. Masters, Adv. Virus Res., 2006, 65, 193–292 CrossRef.
- J. Lan, J. Ge, J. Yu, S. Shan, H. Zhou, S. Fan, Q. Zhang, X. Shi, Q. Wang, L. Zhang and X. Wang, Nature, 2020, 581, 215–220 CrossRef CAS PubMed.
- A. C. Mathewson, A. Bishop, Y. Yao, F. Kemp, J. Ren, H. Chen, X. Xu, B. Berkhout, L. van der Hoek and I. M. Jones, J. Gen. Virol., 2008, 89, 2741–2745 CrossRef CAS PubMed.
- F. J. Warner, A. I. Smith, N. M. Hooper and A. J. Turner, Cell. Mol. Life Sci., 2004, 61, 2704–2713 CrossRef CAS PubMed.
- M. Cametti, B. Crousse, P. Metrangolo, R. Milani and G. Resnati, Chem. Soc. Rev., 2012, 41, 31–42 RSC.
- Q. J. Cai, M. B. Chan-Park, Q. Zhou, Z. S. Lu, C. M. Li and B. S. Ong, Org. Electron., 2008, 9, 936–943 CrossRef CAS.
- P. Towler, B. Staker, S. G. Prasad, S. Menon, J. Tang, T. Parsons, D. Ryan, M. Fisher, D. Williams, N. A. Dales, M. A. Patane and M. W. Pantoliano, J. Biol. Chem., 2004, 279, 17996–18007 CrossRef CAS PubMed.
- National Institute for Health and Care Excellence, Blood glucose testing strips, https://bnf.nice.org.uk/medical-device-type/blood-glucose-testing-strips-2.html, 2021.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. I. Kim, Correction to Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor, ACS Nano, 2020 DOI:10.1021/acsnano.0c02823.
- Y. M. Bar-On, A. Flamholz, R. Phillips and R. Milo, SARS-CoV-2 (COVID-19) by the numbers, eLife, 2020 DOI:10.7554/eLife.57309.
- R. Wölfel, V. M. Corman, W. Guggemos, M. Seilmaier, S. Zange, M. A. Müller, D. Niemeyer, T. C. Jones, P. Vollmar, C. Rothe, M. Hoelscher, T. Bleicker, S. Brünink, J. Schneider, R. Ehmann, K. Zwirglmaier, C. Drosten and C. Wendtner, Nature, 2020, 581, 465–469 CrossRef PubMed.
- L. Fabiani, M. Saroglia, G. Galatà, R. De Santis, S. Fillo, V. Luca, G. Faggioni, N. D’Amore, E. Regalbuto, P. Salvatori, G. Terova, D. Moscone, F. Lista and F. Arduini, Biosens. Bioelectron., 2021, 171, 112686 CrossRef CAS PubMed.
- H. Zhao, F. Liu, W. Xie, T. C. Zhou, J. OuYang, L. Jin, H. Li, C. Y. Zhao, L. Zhang, J. Wei, Y. P. Zhang and C. P. Li, Sens. Actuators, B, 2021, 327, 128899 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS.
- S. A. Hashemi, N. G. Golab Behbahan, S. Bahrani, S. M. Mousavi, A. Gholami, S. Ramakrishna, M. Firoozsani, M. Moghadami, K. B. Lankarani and N. Omidifar, Biosens. Bioelectron., 2021, 171, 112731 CrossRef CAS PubMed.
- A. Raziq, A. Kidakova, R. Boroznjak, J. Reut, A. Öpik and V. Syritski, Biosens. Bioelectron., 2021, 178, 113029 CrossRef CAS PubMed.
- A. Hasan and L. M. Pandey, Polym.-Plast. Technol. Eng., 2015, 54, 1358–1378 CrossRef CAS.
- A. Miodek, E. M. Regan, N. Bhalla, N. A. E. Hopkins, S. A. Goodchild and P. Estrela, Sensors, 2015, 15, 25015–25032 CrossRef PubMed.
- S. Campuzano, F. Kuralay, M. J. Lobo-Castañón, M. Bartošík, K. Vyavahare, E. Paleček, D. A. Haake and J. Wang, Biosens. Bioelectron., 2011, 26, 3577–3583 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc00936b |
| This journal is © The Royal Society of Chemistry 2021 |