Open Access Article
Open Access ArticleA near-infrared AIE probe for super-resolution imaging and nuclear lipid droplet dynamic study†
Ming-Yu
Wu‡
 ab,
Jong-Kai
Leung‡
ab,
Jong-Kai
Leung‡
 b,
Chuen
Kam
b,
Chuen
Kam
 b,
Tsu Yu
Chou
b,
Tsu Yu
Chou
 b,
Dong
Wang
b,
Dong
Wang
 c,
Shun
Feng
a and
Sijie
Chen
c,
Shun
Feng
a and
Sijie
Chen
 *b
*b
aSchool of Life Science and Engineering, Southwest Jiaotong University, Chengdu 610031, China
bMing Wai Lau Centre for Reparative Medicine, Karolinska Institutet, Hong Kong. E-mail: sijie.chen@ki.se
cCenter for AIE Research, College of Materials Science and Engineering, Shenzhen University, Shenzhen 518060, China
First published on 19th February 2021
Abstract
In contrast to the lipid droplets (LDs) in the cytoplasm, nuclear lipid droplets (nLDs), generated from the metabolically active inner nuclear membrane, contribute to the nuclear lipid homeostasis, locally regulate the availability of signaling lipids, and exchange the proteins between LDs and nuclei. Since the currently available fluorescence tools for imaging and tracking nLDs are limited, exploring a new nLD probe would be highly beneficial for studying the biogenesis and functional significance of nLDs. Herein, we developed a near-infrared probe, namely DTZ-TPA-DCN, which possessed the aggregation-induced emission (AIE) property with good biocompatibility for cytoplasm LD and nLD imaging. DTZ-TPA-DCN could be used for the super-resolution imaging of LDs with the structured illumination microscopy. By labelling nLDs, we could monitor the nLD formation under an ER stress, and for the first time, we observed that the signaling lipid, namely diacylglycerol (DAG), enhanced the formation of the nLDs. Together, DTZ-TPA-DCN provided a promising imaging tool for nLD studies, which contributed to a better understanding of the physiological significance of nLDs.
Introduction
Lipid droplets (LDs) are indispensable in lipid metabolism and energy storage.1 Although they are primarily responsible for the lipid storage, LDs are also crucial in numerous biological processes, including the regulation of the protein degradation,2 the homeostasis of the plasma membrane,3 and cell signaling.4 In addition, LDs play a protective role against lipotoxicity and endoplasmic reticulum (ER) stress.5 Moreover, researches have also revealed that LDs are implicated in the progression of numerous diseases, such as cancer,6 diabetes,7 fatty liver disease,8 atherosclerosis,9 hyperlipidemia,10 and neurodegeneration.11In general, LDs are thought to be originated from ER and further translocated to cytoplasm.12 However, a sub-population of LDs are found inside the nucleus of hepatocytes.13 These nuclear lipid droplets (nLDs) are derived from apolipoprotein B-free lumenal LDs, which highly accumulate inside the inner nuclear membrane (INM) upon an ER stress.14 In yeasts, on the other hand, nLDs are generated in the metabolically active territory of INM, which contributes to the nuclear lipid homeostasis.15 However, due to the limitation of research tools, the origin and physiological significance of nLDs are still poorly understood.
Fluorescence imaging has been widely used for visualizing biological molecules and structures, tracking physiological and pathological processes, and diagnosing diseases in biological systems.16 Conventional techniques, such as immunofluorescence and ectopically expressed genes fused with fluorescent protein, are commonly used to monitor the LDs in nucleus, which involve time-consuming experimental procedures.17 Although a considerable number of fluorescent probes have been developed recently for imaging cytoplasmic LDs, BODIPY 493/503 is the only one used for imaging nLDs.18 Nevertheless, most of the conventional dyes suffer from the inherent aggregation-caused quenching (ACQ) property of being easily self-quenched when the luminophores are aggregated. Probes with aggregation-induced emission (AIE) properties have received wide attention since the first reporting of the phenomenon in 2001.19 Compared with the ACQ fluorophores, AIE luminophores show weak or undetectable emissions in dilute solutions but emit strong fluorescence in the aggregated or solid states. These characteristics of AIE luminophores allow the fluorescence imaging with low background, high signal-to-noise ratio, and good photostability, making them ideal tools for the fluorescence imaging of biological targets and even the ultrastructural characterization of biological targets via super-resolution imaging.20
In this contribution, a near-infrared (NIR) fluorescent probe, namely DTZ-TPA-DCN, for nLD imaging was developed based on a D–π–A framework, in which non-planar triphenylamine (TPA) was the donor to evade the ACQ effect and the lipophilic cyano groups were the acceptors. Furthermore, the bithiophene and carbon–carbon double bonds were conjugation groups that could extend the absorption or emission wavelength.19e
DTZ-TPA-DCN exhibited AIE properties and could be used for imaging LDs in both live and fixed cells with high selectivity. By using DTZ-TPA-DCN, we could clearly observe the LDs in the nucleus of cells in two-dimensional (2D) and three-dimensional (3D) images. Thanks to its good photostability, DTZ-TPA-DCN could also be used for structured-illumination microscopy (SIM)-based super-resolution imaging of LDs. Furthermore, the dynamics of LDs inside the nucleus was also highlighted and investigated using DTZ-TPA-DCN.
Results and discussion
Synthesis and photophysical properties
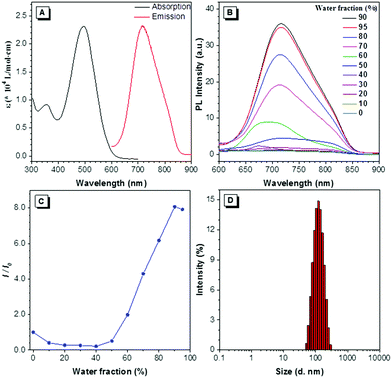
DTZ-TPA-DCN was synthesized via a simple Knoevenagel condensation reaction of aldehyde and malononitrile (Scheme 1) with 87.5% yield. It was characterized by 1H NMR, 13C NMR and high-resolution mass spectrometry (ESI†). The optical properties of DTZ-TPA-DCN are illustrated in Fig. 1. From photoluminescence spectrometry results, it was evident that DTZ-TPA-DCN showed its maximum absorption peak at 495 nm and near-infrared emission peak at 715 nm with a large Stokes shift of 220 nm (Fig. 1A). The emission spectra displayed a strong solvent effect from toluene (605 nm) to acetonitrile (700 nm), representing a 105 nm bathochromic shift (Fig. S1, ESI†). To further investigate the AIE features of DTZ-TPA-DCN, acetonitrile/water (MeCN/H2O) solvent mixture with different H2O fractions was utilized as the solvent system. As depicted in Fig. 1B and C, DTZ-TPA-DCN showed a weak fluorescence emission at 700 nm with a 0.04% quantum yield (QY) in a pure MeCN solution. When adding H2O to the MeCN solution, the maximum emission gradually red-shifted from 700 nm to 715 nm, and the fluorescence intensity slowly decreased at first as the water fractions were increased from 10% to 40%. This result could be attributed to the twisted intramolecular charge transfer (TICT) effect. When the ratio of water in the mixed solvent was further increased from 40% to 90%, the fluorescence intensity of DTZ-TPA-DCN rebounded dramatically due to the formation of the probe aggregates, and the fluorescence intensity of DTZ-TPA-DCN in the 90% H2O solution (QY: 0.28%) was 8.1-fold of that observed in pure MeCN solution. The aggregate formed with an average size of approximately 113.5 nm in diameter was confirmed via the dynamic light scattering technique (Fig. 1D). QY in the solid state is 2.3%, which is much higher than that observed in the aggregated and solution states. As a result, these data affirmatively revealed the AIE characteristics of DTZ-TPA-DCN.Cytotoxicity and co-localization experiment
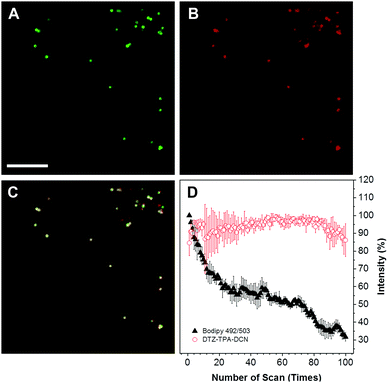
The cytotoxicity of DTZ-TPA-DCN in live cells was evaluated using a standard Cell Counting Kit-8 (CCK-8) assay, and HepG2 cells were treated with different concentrations of DTZ-TPA-DCN (from 2.5 to 20 μM). As illustrated in Fig. S2 (ESI†), the cells were still highly viable even after incubated with 20 μM DTZ-TPA-DCN for 24 h, which demonstrated an excellent biocompatibility of DTZ-TPA-DCN. We further examined the cellular uptake and the subcellular pattern of DTZ-TPA-DCN in HepG2 cells. After incubating the cells with 10 μM DTZ-TPA-DCN for 60 min, a number of punctate structures in the cytoplasm were observed under the confocal laser scanning microscopy. These puncta highly colocalized with the commercial LD probe BODIPY 493/503 (Fig. 2A–C) with a Pearson's correlation coefficient (Rr) of 0.937 (Fig. S3, ESI†), which indicated the high labelling specificity of DTZ-TPA-DCN to LDs. Besides live-cell imaging, DTZ-TPA-DCN also demonstrated a good performance in fixed cell imaging (Fig. S4, ESI†). Moreover, as depicted in Fig. 2, 85% of the fluorescence intensity of DTZ-TPA-DCN-stained LDs remained unchanged after 100 repetitive scanning under confocal microscopy, while the fluorescence intensity of BODIPY 493/503-stained LDs dropped to only 30% of its original value when using the same scanning parameter (Fig. 2D). Compared with BODIPY 493/503, DTZ-TPA-DCN showed a superior photostability, and this prompted further investigation of its performance under super-resolution fluorescence imaging, which could only be achieved using the fluorophores with an outstanding photostability.21Super-resolution imaging of LDs with SIM
There have been few reports using AIE probes for SIM imaging till now.22 To test the feasibility of using DTZ-TPA-DCN for super-resolution imaging, SIM was used in the imaging of DTZ-TPA-DCN-stained LDs in HepG2 cells pre-treated with oleic acid (OA). OA is a fatty acid that exists naturally in numerous animal fats and vegetable oils, which can stimulate LD formation in both nucleus and cytoplasm by activating the long-chain fatty acid receptor.23 After treating HepG2 cells with 0.4 mM OA for 24 h, not only the quantity but also the diameters of LDs in HepG2 cells were increased (Fig. S5, ESI†). As shown in Fig. 3, the LD image acquired by the SIM exhibited an enhanced spatial resolution and an improved image quality compared with the images obtained from the confocal laser scanning microscopes (Nikon and ZEISS LSM 880, Fig. 3A, C and Fig. S6, ESI†). With respect to SIM, LDs with sharper signals were observed, making them easier to be distinguished from each other in clusters, while the confocal images showed fuzzy LD signals. The line-scan plot of individual LD also demonstrated an enhanced spatial resolution (Fig. 3B and D), where the lateral full-width at half maximum (FWHM) of LDs in the SIM imaging was narrower than those observed in confocal imaging (Fig. 3B). These results suggested that DTZ-TPA-DCN was applicable for imaging LDs by SIM.Fluorescence imaging of nLDs with DTZ-TPA-DCN
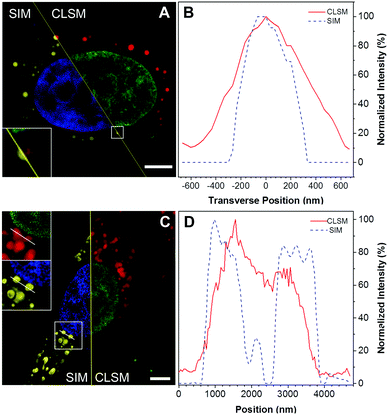
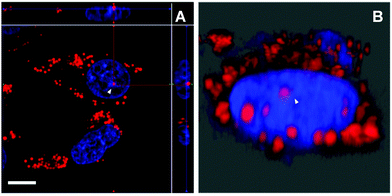
Aside from the cytoplasmic LDs that originated from ER, the nLDs generated in the INM have drawn considerable attention because of its non-canonical role in the nuclear lipid homeostasis.14,15 However, the physiological functions, cellular regulatory machinery, and membrane contact site of nLDs remain to be explored. Taking advantage of DTZ-TPA-DCN possessing near-infrared emission, good LD selectivity, and practical application to super-resolution imaging, we applied this fluorophore to the study of nLDs. After treating HepG2 cells with OA, the cells were counterstained with 10 μM DTZ-TPA-DCN and 20 μM Hoechst 33342. As shown in Fig. 4A, most near-infrared DTZ-TPA-DCN puncta localized around the Hoechst-labelled nuclei. However, there were several DTZ-TPA-DCN-labelled LDs localized inside the nuclei, which were possibly the nLDs (Fig. 4A, white arrowheads). Through the 3D reconstructed images obtained by the Z-stack acquisition, we further verified that these red puncta were localized inside the nuclei instead of in close proximity to the nuclear surface (Fig. 4B and Fig. S7, Video S1, ESI†). As indicated by the white arrowheads in Fig. 4, the red signal is suspended in the cell nuclei. These results demonstrated the ability of DTZ-TPA-DCN to label nLDs and prompted us to further track the dynamics of nLDs by DTZ-TPA-DCN.Monitor the nLD dynamics
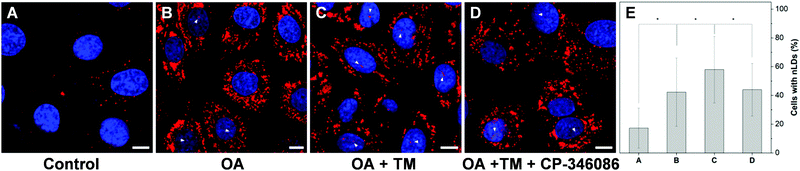
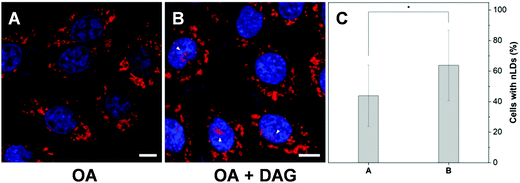
To further examine the capability of DTZ-TPA-DCN in the monitoring of nLD dynamics, drugs which induced or suppressed the formation of nLDs were applied to the OA-treated HepG2 cells. It was reported that ER stress led to the formation of the nLDs,14,24 and tunicamycin (TM) induced the ER stress, which in turn enhanced the nLD formation in OA-treated hepatic cell lines.25 In our experiment, after treating the HepG2 cells with 0.4 mM OA and 5 μgmL−1 TM for 12 h, the cells were stained with DTZ-TPA-DCN, followed by imaging. We then counted and analyzed the percentage of cells enriched with nLDs in each population with different treatments. For the statistical analysis, around 100 cells were sampled for each of the population in triplicate. As shown in Fig. 5 and Fig. S8 (ESI†), the OA-treated group shows a drastic increase in the nLDs compared with that of the control group. When TM was further added into the OA-treated group, the percentage of cells with nLDs also increased from 42.2% to 57.9% (Fig. 5A–C and E). On the other hand, CP-346086, a microsomal triglyceride transfer protein (MTP) activity inhibitor, was used to suppress the nLD formation.26 When the OA/TM-treated cells were further treated with CP-346086, the percentage of cells with nLDs dropped and restored to a comparable level as compared to the group with the OA treatment only (Fig. 5D and E). Moreover, DTZ-TPA-DCN staining also revealed that CP-346086 could suppress the nLD formation with the OA treatment (Fig. S9 and S10, ESI†). These results showed that DTZ-TPA-DCN could be used for the monitoring of the nLD formation. By using DTZ-TPA-DCN, the nLD dynamics in the hepatic cell lines was successfully tracked.In addition, researches have shown that the active lipid metabolism and the distinct lipid composition of the INM affect the ability of the cells to generate nLDs.15 For instance, triacylglycerols (TAGs), which are the major neutral lipids stored in LDs, are essential factors in the biogenesis of LDs.1b,27 To promote the formation of LDs, the TAG in cells needs to reach a critical concentration.28 Moreover, the synthesis of TAGs from DAGs was catalysed by diacylglycerol acyltransferase (DGAT). Interestingly, DAGs were also highly related to the formation of nLDs.1b,27,29 Therefore, we would like to investigate whether the addition of DAG could promote the hepatocellular nLD formation. As shown in Fig. 6 and Fig. S11 (ESI†), by using DTZ-TPA-DCN staining, we can observe that the nLDs were unevenly dispersed in cells with 0.4 mM OA treatment. Interestingly, the percentage of cells containing nLDs increased from around 44.5% to more than 61.4% when the cells were treated with 0.4 mM OA and DAG (10 μM), suggesting that DAG enhanced the formation of nLDs in the hepatic cells (Fig. 6C). Some researchers pointed out that the seipin-deficient yeasts might form more nLDs when compared to the wild-type yeasts, and DAG is one of the factors involved in this phenomenon. The lack of seipin is believed to cause an increase in phosphatidic acid (PA), a precursor of DAG, in yeasts. When the free PA level inside the cells increased, excess PA would diffuse into the INM. These PAs would then be converted to DAG by PA phosphatases, which further upregulated the TAG synthesis and eventually contributed to more nLD formation.15,27 Despite this pathway was demonstrated using yeasts as a model organism only, it was postulated that the connection of lipid metabolism and nLD biogenesis were evolutionarily conserved in higher eukaryotes. This experiment suggested that DTZ-TPA-DCN could be a useful tool to explore the unravelled mechanism of nLD biogenesis and regulation.
Conclusions
In summary, a lipophilic near-infrared fluorescent probe, namely DTZ-TPA-DCN, was developed based on a common D–π–A framework via a simple Knoevenagel condensation reaction. DTZ-TPA-DCN exhibited AIE properties with large Stokes shift and a distinct solvatochromic effect with high sensitivity towards environmental polarity. It was highly biocompatible and was able to label LDs specifically in both live and fixed cells. Owing to its excellent photostability, DTZ-TPA-DCN was successfully used in the SIM imaging of LDs. Furthermore, the nLDs were clearly observed in 2D or 3D imaging using DTZ-TPA-DCN in the labelling. The favourable optical properties of DTZ-TPA-DCN enabled further monitoring and tracking of the nLD dynamics under ER stress or by a lipid metabolism of the INM. It was shown for the first time that the important signaling lipid, DAG, induced nLD formation in mammalian cells. Conclusively, this study provided a versatile and reliable tool, DTZ-TPA-DCN, in the studies of nLDs, and its great potential for exploring the functional significance of nLDs was envisioned.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
M.-Y. W. thanks the National Natural Science Foundation of China (21708030) and the Applied Basic Research of Sichuan Province (2021YJ0397). S. C. acknowledges the start-up funding from Ming Wai Lau Centre for Reparative Medicine, Karolinska Institutet.Notes and references
- (a) T. C. Walther and R. V. Farese Jr, Lipid droplets and cellular lipid metabolism, Annu. Rev. Biochem., 2012, 81, 687–714 CrossRef CAS PubMed; (b) A. R. Thiam, R. V. Farese Jr and T. C. Walther, The biophysics and cell biology of lipid droplets, Nat. Rev. Mol. Cell Biol., 2013, 14, 775–786 CrossRef CAS PubMed.
- J. A. Olzmann, C. M. Richter and R. R. Kopito, Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 1345–1350 CrossRef CAS PubMed.
- J. K. Zehmer, Y. Huang, G. Peng, J. Pu, R. G. W. Anderson and P. Liu, A role for lipid droplets in inter-membrane lipid traffic, Proteomics, 2009, 9, 914–921 CrossRef CAS PubMed.
- S. Martin and R. G. Parton, Lipid Droplets: A unified view of a dynamic organelle, Nat. Rev. Mol. Cell Biol., 2006, 7, 373–378 CrossRef CAS PubMed.
- (a) J. A. Olzmann and P. Carvalho, Dynamics and functions of lipid droplets, Nat. Rev. Mol. Cell Biol., 2019, 20, 137–155 CrossRef CAS PubMed; (b) J. Boren and K. M. Brindle, Apoptosis-induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation, Cell Death Differ., 2012, 19, 1561–1570 CrossRef CAS PubMed.
- (a) L. Tirinato, F. Pagliari, T. Limongi, M. Marini, A. Falqui, J. Seco, P. Candeloro, C. Liberale and E. Di Fabrizio, An overview of lipid droplets in cancer and cancer stem cells, Stem Cells Int., 2017, 2017, 1656053 CAS; (b) Q. Liu, Q. Luo, A. Halim and G. Song, Targeting lipid metabolism of cancer cells: a promising therapeutic strategy for cancer, Cancer Lett., 2017, 401, 39–45 CrossRef CAS PubMed.
- N. Krahmer, R. V. Farese and T. C. Walther, Balancing the fat: lipid droplets and human disease, EMBO Mol. Med., 2013, 5, 973–983 CrossRef PubMed.
- G. Onal, O. Kutlu, D. Gozuacik and S. D. Emre, Lipid droplets in health and disease, Lipids Health Dis., 2017, 16, 128 CrossRef PubMed.
- A. Paul, B. H.-J. Chang, L. Li, V. K. Yechoor and L. Chan, Deficiency of adipose differentiation-related protein impairs foam cell formation and protects against atherosclerosis, Circ. Res., 2008, 102, 1492–1501 CrossRef CAS PubMed.
- E. Kiss, B. Kranzlin, K. Wagenbalaβ, M. Bonrouhi, J. Thiery, E. Grone, V. Nordstrom, D. Teupser, N. Gretz, E. Malle and H.-J. Grone, Lipid droplet accumulation is associated with an increase in hyperglycemia-induced renal damage: prevention by liver X receptors, Am. J. Pathol., 2013, 182, 727–741 CrossRef CAS PubMed.
- N. Roitenberg and E. Cohen, Lipid assemblies at the crossroads of aging, proteostasis, and neurodegeneration, Trends Cell Biol., 2019, 29, 954–963 CrossRef CAS PubMed.
- (a) V. Choudhary, G. Golani, A. S. Joshi, S. Cottier, R. Schneiter, W. A. Prinz and M. M. Kozlov, Architecture of lipid droplets in endoplasmic reticulum is determined by phospholipid intrinsic curature, Curr. Biol., 2018, 19(28), 915–926 CrossRef PubMed; (b) T. C. Walther and R. V. Farese Jr, Lipid droplets and cellular lipid metabolism, Annu. Rev. Biochem., 2012, 81, 687–714 CrossRef CAS PubMed.
- (a) J. P. Layerenza, P. González, M. M. García de Bravo, M. P. Polo, M. S. Sisti and A. Ves-Losada, Nuclear lipid droplets: a novel nuclear domain, Biochim. Biophys. Acta, 2013, 1831, 327–340 CrossRef CAS PubMed; (b) R. Uzbekov and P. Roingeard, Nuclear lipid droplets identified by electron microscopy of serial sections, BMC Res. Notes, 2013, 6, 386 CrossRef PubMed; (c) L. Wang, Y. Wang, Y. Liang, J. Li, Y. Liu, J. Zhang, A. Zhang, J. Fu and G. Jiang, Physical key-protected one-time pad, Sci. Rep., 2013, 3, 217 Search PubMed.
- K. Sołtysik, Y. Ohsaki, T. Tatematsu, J. Cheng and T. Fujimoto, Nuclear lipid droplets derive from a lipoprotein precursor and regulate phosphatidylcholine synthesis, Nat. Commun., 2019, 10, 473 CrossRef PubMed.
- A. Romanauska and A. Köhler, The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets, Cell, 2018, 174, 700–715 CrossRef PubMed.
- (a) R. Yuste, Fluorescence microscopy today, Nat. Methods, 2005, 2, 902–904 CrossRef CAS PubMed; (b) J. W. Lichtman and J.-A. Conchello, Fluorescence microscopy, Nat. Methods, 2005, 2, 910–919 CrossRef CAS PubMed.
- (a) A. Malhas, C. Goulbourne and D. J. Vaux, The nucleoplasmic reticulum: form and function, Trends Cell Biol., 2011, 21, 362–373 CrossRef CAS PubMed; (b) R. Bernardi and P. P. Pandolfi, Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies, Nat. Rev. Mol. Cell Biol., 2007, 8, 1006–1016 CrossRef CAS PubMed; (c) Y. Ohsaki, T. Kawai, Y. Yoshikawa, J. Cheng, E. Jokitalo and T. Fujimoto, PML isoform II plays a critical role in nuclear lipid droplet formation, J. Cell Biol., 2016, 212, 29–38 CrossRef CAS PubMed.
- (a) M. Collot, T. K. Fam, P. Ashokkumar, O. Faklaris, T. Galli, L. Danglot and A. S. Klymchenko, Ultrabright and fluorogenic probes for multicolor imaging and tracking of lipid droplets in cells and tissues, J. Am. Chem. Soc., 2018, 140, 5401–5411 CrossRef CAS PubMed; (b) H. Xu, H. Zhang, G. Liu, L. Kong, X. Zhu, X. Tian, Z. Zhang, R. Zhang, Z. Wu, Y. Tian and H. Zhou, Coumarin-based fluorescent probes for super-resolution and dynamic tracking of lipid droplets, Anal. Chem., 2019, 91, 977–982 CrossRef CAS PubMed; (c) M. Collot, S. Bou, T. K. Fam, L. Richert, Y. Mely, L. Danglot and A. S. Klymchenko, Probing polarity and heterogeneity of lipid droplets in live cells using a push-pull fluorophore, Anal. Chem., 2019, 91, 1928–1935 CrossRef CAS PubMed; (d) D. Dang, H. Liu, J. Wang, M. Chen, Y. Liu, H. H.-Y. Sung, I. D. Williams, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Highly emissive AIEgens with multiple functions: facile synthesis, chromism, specific lipid droplet imaging, apoptosis monitoring, and in vivo imaging, Chem. Mater., 2018, 30, 7892–7901 CrossRef CAS; (e) T. K. Fam, A. S. Klymechenko and M. Collot, Recent advances in fluorescent probes for lipid droplets, Materials, 2018, 11, 1768 CrossRef PubMed; (f) D. Wang, H. Su, R. T. K. Kowk, G. Shan, A. C. S. Leung, M. M. S. Lee, H. H. Y. Sung, I. D. Williams, J. W. Y. Lam and B. Z. Tang, Facile synthesis of red/NIR AIE luminogens with simple structures, bright emissions, and high photostabilities, and their applications for specific imaging of lipid droplets and image-guided photodynamic therapy, Adv. Funct. Mater., 2017, 27, 1704039 CrossRef; (g) L. Guo, M. Tian, R. Feng, G. Zhang, R. Zhang, X. Li, Z. Liu, X. He, J. Z. Sun and X. Yu, Interface-targeting strategy enables two-photon fluorescent lipid droplet probes for high-fidelity imaging of turbid tissues and detecting fatty liver, ACS Appl. Mater. Interfaces, 2018, 10, 10706–10717 CrossRef CAS PubMed; (h) L. Shi, K. Li, L.-L. Li, S.-Y. Chen, M.-Y. Li, Q. Zhou, N. Wang and X.-Q. Yu, Novel easily available purine-based AIEgens with colour tunability and applications in lipid droplet imaging, Chem. Sci., 2018, 9, 8969–8974 RSC; (i) J.-S. Ni, H. Liu, J. Liu, M. Jiang, Z. Zhao, Y. Chen, R. T. K. Kwok, J. W. Y. Lam, Q. Peng and B. Z. Tang, The unusual aggregation-induced emission of coplanar organoboron isomers and their lipid droplet-specific applications, Mater. Chem. Front., 2018, 2, 1498–1507 RSC; (j) N. Zhao, Y. Li, W. Yang, J. Zhuang, Y. Li and N. Li, Multifunctional pyrazoline based AIEgens: real-time tracking and specific protein “fishing” of lipid droplets, Chem. Sci., 2019, 10, 9009–9016 RSC; (k) W. Xu, M. M. S. Lee, Z. Zhang, H. H. Y. Sung, I. D. Willams, R. T. K. Kwok, J. W. Y. Lam, D. Wang and B. Z. Wang, Facile synthesis of AIEgens with wide color tunability for cellular imaging and therapy, Chem. Sci., 2019, 10, 3494–3501 RSC; (l) H. Tian Jr, A. C. Sedgwick, H.-H. Han, S. Sen, G.-R. Chen, Y. Zang, J. L. Sessler, T. D. James, J. Li and X.-P. He, Fluorescent probes for the imaging of lipid droplets in live cells, Coord. Chem. Rev., 2021, 427, 213577 CrossRef.
- (a) J. Luo, Z. Xie, J. W. Y. Lam, L. Cheng, H. Chen, C. Qiu, H. S. Kowk, X. Zhan, Y. Liu, D. Zhu and B. Z. Tang, Aggregation-induced emission of 1-methyl-1,2,3,4,5-pentaphenylsilole, Chem. Commun., 2001, 1740–1741 RSC; (b) J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Aggregation-induced emission: together we shine, united we soar!, Chem. Rev., 2015, 115(21), 11718–11940 CrossRef CAS PubMed; (c) Y. Hong, J. W. Y. Lam and B. Z. Tang, Aggregation-induced emission, Chem. Soc. Rev., 2011, 40, 5361–5388 RSC; (d) X. Cai and B. Liu, Aggregation-induced emission: recent advances in materials and biomedical applications, Angew. Chem., Int. Ed., 2020, 59, 9868–9886 CrossRef CAS PubMed; (e) S. Xu, Y. Duan and B. Liu, Precise molecular design for high-performance luminogens with aggregation-induced emission, Adv. Mater., 2019, 32, 1903530 CrossRef PubMed; (f) Z. Guo, C. Yan and W.-H. Zhu, High-performance quinoline-malononitrile core as a building block for the diversity-oriented synthesis of AIEgens, Angew. Chem., Int. Ed., 2020, 59, 9812–9825 CrossRef CAS PubMed; (g) J. Mei, Y. Huang and H. Tian, Progress and trends in AIE-based bioprobes: a brief overview, ACS Appl. Mater. Interfaces, 2018, 10, 12217–12261 CrossRef CAS PubMed.
- (a) M.-Y. Wu, J.-K. Leung, L. Liu, C. Kam, K. Y. K. Chan, R. A. Li, S. Feng and S. Chen, A small-molecule AIE chromosome periphery probe for cytogenetic studies, Angew. Chem., Int. Ed., 2020, 59, 10327–10331 CrossRef CAS PubMed; (b) L. Shi, Y.-H. Liu, K. Li, A. Sharam, K.-K. Yu, M. S. Ji, L.-L. Li, Q. Zhou, H. Zhang, J. S. Kim and X.-Q. Yu, An AIE-based probe for rapid and ultrasensitive imaging of plasma membranes in biosystems, Angew. Chem., Int. Ed., 2020, 59, 9962–9966 CrossRef CAS PubMed.
- L. Wang, M. S. Frei, A. Salim and K. Johnsson, Small-molecule fluorescent probes for live-cell super-resolution microscopy, J. Am. Chem. Soc., 2019, 141, 2770–2781 CrossRef CAS PubMed.
- X. Ge, M. Gao, B. Situ, W. Feng, B. He, X. He, S. Li, Z. Ou, Y. Zhong, Y. Lin, X. Ye, X. Hu, B. Z. Tang and L. Zheng, One-step, rapid fluorescence sensing of fungal viability based on a bioprobe with aggregation-induced emission characteristics, Mater. Chem. Front., 2020, 4, 957–964 RSC.
- A. Rohwedder, Q. Zhang, S. A. Rudge and M. J. O. Wakelam, Lipid droplet formation in response to oleic acid in Huh-7 cells is mediated by the fatty acid receptor FFAR4, J. Cell Sci., 2014, 127, 3104–3115 CAS.
- L. Ozcan and I. Tabas, Role of endoplasmic reticulum stress in metabolic disease and other disorders, Annu. Rev. Med., 2012, 63, 317–328 CrossRef CAS PubMed.
- J.-S. Lee, R. Mendez, H. H. Heng, Z.-Q. Yang and K. Zhang, Pharmacological ER stress promotes hepatic lipogenesis and lipid droplet formation, Am. J. Transl. Res., 2012, 4, 102–113 CAS.
- G. F. Gibbons, K. Islam and R. J. Pease, Mobilisation of triacylglycerol stores, Biochim. Biophys. Acta, 2000, 1483, 37–57 CrossRef CAS.
- H. F. Hashemi and J. M. Goodman, The life cycle of lipid droplets, Curr. Opin. Cell Biol., 2015, 33, 119–124 CrossRef CAS PubMed.
- A. R. Thiam and M. Beller, The why, when and how of lipid droplet diversity, J. Cell Sci., 2017, 130, 315–324 CAS.
- (a) A. D. Barbosa, K. Lim, M. Mari, J. R. Edgar, L. Gal, P. Sterk, B. J. Jenkins, A. Koulman, D. B. Savage, M. Schuldiner, F. Reggirori, P. A. Wigge and S. Siniossoglou, Compartmentalized synthesis of triacylglycerol at the inner nuclear membrane regulates nuclear organization, Dev. Cell, 2019, 50, 755–766 CrossRef CAS PubMed; (b) M. Gao, X. Huang, L. Bao and H. Yang, The biogenesis of lipid droplets: Lipids take center stage, Prog. Lipid Res., 2019, 75, 100989 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis processes and characterization, photophysical properties, cell viability, and cell imaging results. See DOI: 10.1039/d0qm00914h |
| ‡ These authors contributed equally to this work. |
| This journal is © the Partner Organisations 2021 |