Open Access Article
Open Access ArticleNanoparticle-induced chemoresistance: the emerging modulatory effects of engineered nanomaterials on human intestinal cancer cell redox metabolic adaptation†
Zhuoran Wu‡
a,
Magdiel Inggrid Setyawati‡a,
Hong Kit Lima,
Kee Woei Ng abc and
Chor Yong Tay
abc and
Chor Yong Tay
 *ab
*ab
aSchool of Materials Science and Engineering, Nanyang Technological University, 50 Nanyang Avenue, Singapore 639798, Singapore. E-mail: cytay@ntu.edu.sg
bEnvironmental Chemistry and Materials Centre, Nanyang Environment & Water Research Institute, 1 Cleantech Loop, CleanTech One, Singapore 637141, Singapore
cCenter for Nanotechnology and Nanotoxicology, Department of Environmental Health, Harvard T.H. Chan School of Public Health, Boston, MA 02115, USA
First published on 7th September 2022
Abstract
The widespread use of engineered nanomaterials (ENMs) in food products necessitates the understanding of their impact on the gastrointestinal tract (GIT). Herein, we screened several representative food-borne comparator ENMs (i.e. ZnO, SiO2 and TiO2 nanoparticles (NPs)) and report that human colon cancer cells can insidiously exploit ZnO NP-induced adaptive response to acquire resistance against several chemotherapeutic drugs. By employing a conditioning and challenge treatment regime, we demonstrate that repeated exposure to a non-toxic dose of ZnO NPs (20 μM) could dampen the efficacy of cisplatin, paclitaxel and doxorubicin by 10–50% in monolayer culture and 3D spheroids of human colon adenocarcinoma cells. Structure–activity relationship studies revealed a complex interplay between nanoparticle surface chemistry and cell type in determining the chemoresistance-inducing effect, with silica coated ZnO NPs having a negligible influence on the anticancer treatment. Mechanistically, we showed that the pro-survival paracrine signaling was potentiated and propagated by a subset of ZnO NP “stressed” (Zn2++/ROS+) cells to the surrounding “bystander” (Zn2++/ROS−) cells. Transcriptome profiling, bioinformatics analysis and siRNA gene knockdown experiments revealed the nuclear factor erythroid 2-related factor 2 (Nrf2) as the key modulator of the ZnO NP-induced drug resistance. Our findings suggest that a ROS-inducing ENM can emerge as a nano-stressor, capable of regulating the chemosensitivity of colon cancer cells.
1. Introduction
The rapid development of nanotechnology has led to a vast application of engineered nanomaterials (ENMs) in every walk of our life including the food and agriculture sectors.1 To date, the Center for Food Safety registry has identified titanium dioxide (TiO2 E171), silica dioxide (SiO2 E551), silver (Ag) and zinc dioxide (ZnO) as the most commonly utilized ENMs in nano-enabled food and food contact products.2 Typically, TiO2 E171 nanoparticles (NPs) are used as coloring agents, flavor enhancers and coating materials for candies,3 while SiO2 E551 NPs are used as anti-caking agents to avoid clumping.4 Leveraging on their anti-microbial properties, Ag and ZnO NPs are widely exploited as food packaging materials to prevent food contamination.4,5 In this regard, human exposure to ENMs via the gastrointestinal tract (GIT) route is inevitable. It was estimated that TiO2 and SiO2 food additives alone contribute to the daily ingestion of approximately 1012–1013 particles per person.6,7 In a more recent in vitro study, Moreno-Olivas et al.5 detected the presence of ZnO NPs in canned food products, presumably due to the leaching of NPs from can linings, resulting in an estimated dietary intake of 3.5 × 1015 NPs per serving of canned food alone. Approximately 55% of ZnO NPs was demonstrated to be present in the intestinal stage after a 120 min in vitro digestion process (NanoReg D5.03). In a recent study, it was also shown that regardless of the type of food matrix (i.e. starch, milk, and oil), ZnO NPs can be generated de novo in the intestinal juice.8 Thus, there appears to be several pathways that can contribute to ENM–intestinal interaction, be it via the direct intake route or the indirect biotransformation route. Although ingestion of these food-borne ENMs at environmental levels is deemed unlikely to cause overt toxicity due to their low absorption in the GIT, recent findings have demonstrated that food-borne ENMs can translocate through the Peyer's patch of the intestine.6,7,9,10 Moreover, their intake has been linked to several pathological conditions such as Crohn's disease, colitis, diabetes, metabolic syndrome, and cancer.11,12 Although, some progress has been made in the understanding of the effects of the exposure to these food borne ENMs on human health, the long-term impact arising from the interaction of ENMs with components of the GIT remain elusive to date.From the materials standpoint, the toxic potential of ENMs is largely governed by their physiochemical properties such as size, density, morphology, surface area, surface chemistry, and colloidal stability.13,14 The particular ENM-induced biological outcome can vary significantly from one cell type to another, and also with respect to the ENM.15,16 However, the predominant mode-of-action of nanotoxicity is the generation of reactive oxygen species (ROS) which could cause irreversible oxidative damage to proteins, membranes, organelles and DNA.17–19 Consequently, several studies have exploited the ROS inducing properties of ENMs for anti-cancer nanomedicine applications.20–26 However, the pathophysiological significance of ROS imbalance caused by ENM exposure remains elusive.
Previously, we showed that SiO2, TiO2 and ZnO NPs (<30 nm) could invoke considerable increase in intracellular ROS and inflammation in several human intestinal cell lines such as DLD-1, SW480 and NCM460.27 However, in recent years, emerging studies have shown that low levels of ENM-induced ROS can activate several evolutionary conserved stress response pathways to adapt to or resist oxidative stress. Such ENM invoked adaptive response (nano-adaptation) has been documented in a broad spectrum of biological systems, ranging from bacteria to complex eukaryotes and plants.28,29 Accordingly, our earlier studies provided solid evidence for activation of endogenous adaptive response to intermittent brief episodes of non-cytotoxic doses of ZnO NP exposure via nuclear factor erythroid 2-related factor 2 (Nrf2) mediated cytoprotective stress responses.30,31 The Nrf2 signaling pathway functions as an important molecular rheostat that would allow the cells to respond adaptively to xenobiotic and oxidative stress via the expression of antioxidants and phase II enzymes.32,33 Extended ZnO NP exposure resulted in a persistent increase in Nrf2 mediated proteolytic activity, which can lead to enhanced tolerance against subsequent toxic concentrations of ZnO NPs.30 Conversely, a defective Nrf2-dependent redox signaling in chronically inflamed cells increased the susceptibility to ENM-induced oxidative stress and cell death.31 Recently, in a proteome-wide assessment involving 11 different types or metal and metal oxide NPs, the expression levels of numerous ROS-sensitive and adaptive response proteins (i.e., HMOX1, HS71B, DNJB1, and SQSTM1) were found to be significantly altered in human THP-1 derived macrophages at cytotoxic doses.34 These studies highlight the need to consider ENM invoked defensive and homeostatic mechanisms to better appreciate the nanotoxicological outcomes.
However, pathways involving ROS triggered responses may also potentially confer collateral adaptive advantages to pathological states, such as cancer. Numerous studies have shown that cancer cells frequently exhibited an elevated endogenous ROS content and antioxidant production.35,36 It is therefore not surprising to learn that the constitutive expression of Nrf2 is significantly higher in many cancer cells compared to healthy cells. Such an arrangement would appear to be advantageous for cancer cells to leverage ROS as a signaling molecule to drive cancer biology, while subduing the ROS pool before it reaches a harmful level. These ROS-adaptive responses have been documented in numerous cancer cell types to promote cancer cell survival via metabolic rewiring and contribute to disease progression by promoting chemo- and radio-resistance.35–37 Besides chemotherapeutic drugs,38 chronic exposure to ROS-inducing environmental chemicals and pollutants such as particulate matter (PM2.5),39 benzo-α-pyrene,40 and pesticides41 has also been implicated in chemoresistance. Despite several lines of evidence suggesting that adaptive response to oxidative stress and drug resistance may share common mechanisms, the plausible role of ROS-inducing ENMs as a chemo-disruptive agent has never been studied before.
Considering the prevalence of nano-enabled food products, we therefore ask whether these ROS-inducing ENMs could contribute to chemoresistance in colorectal cancer. Herein, ZnO, SiO2 and TiO2 NPs (<200 nm) were chosen as representative food-borne ENMs to examine their ability to alter the responsiveness to three commercialized anticancer drugs, i.e., cisplatin (CDDP), paclitaxel (PTX), and doxorubicin (DOX) in NCM460, SW480 and Caco-2 human colorectal cells (Fig. 1). Following our recently developed conditioning and challenge (2Cs) methodology,30,31 colorectal cells were exposed to a nontoxic dose (20 μM) of ENMs on a daily basis, up to three days, to evaluate the acquired resistance to chemotherapeutics by the cells (if any). From our in vitro screening experiments, ZnO NPs were uncovered as a novel nanomodulator of drug sensitivity in the SW480 and Caco-2 colorectal cancer cells, but not in the healthy NCM460 colon cells. To gain further structure–activity relationship insights, 3 distinct types of ZnO NPs bearing different surface charges and solubility profiles, i.e., amine-functionalized ZnO NPs (NH2-ZnO), carboxylic acid-functionalized ZnO NPs (COOH-ZnO), and silica-coated ZnO NPs (Si-ZnO), were synthesized and subjected to 2Cs treatment. Mechanistic studies were performed to identify important post-exposure alterations (redox balance, proteins, and genes) in the cancer cells that are responsible for ZnO NP-induced chemoresistance. In addition, bioinformatics analysis was employed to identify the key molecular player that mediates the ZnO NP-induced chemoresistance acquisition process. The capability of the two colorectal cancer cell models to acquire chemoresistance to the panel of drugs was further examined using a three-dimensional (3D) tumor spheroid model to further validate the phenomenon in a more biomimetic scenario.
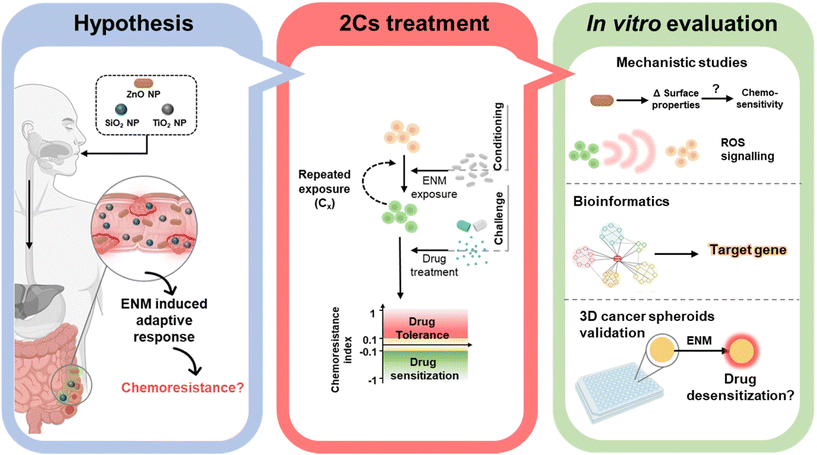 | ||
| Fig. 1 Schematic showing the central hypothesis, as well as the key experimental approaches and methods employed in this study. Illustrations created with BioRender.com. | ||
2. Experimental section
2.1. Engineered nanomaterials
ZnO NPs were purchased from Sigma Aldrich (USA), while TiO2 NPs (E171) were obtained from Zhengzhou Sino Chemical Co. Ltd (China). SiO2 NPs were synthesized by the Stöber method as per the procedure described in our previous study.42Amine-functionalized ZnO NPs (NH2-ZnO) were prepared following a published work.43 Briefly, 20 mg of ZnO NPs was first suspended in 40 mL of DI water and the pH of the NP suspension was adjusted to ∼6.5 with HCl (1M). The NP suspension was then equilibrated at room temperature with vigorous stirring at 400 rpm for 1 h. Thereafter, 15 μL of 3-aminopropyl-triethoxysilane (APTES, Sigma Aldrich) was added into the suspension and the reaction mixture was left for another 24 h. To obtain carboxylic acid-conjugated ZnO NPs (COOH-ZnO), 10 mg of ZnO NPs was mixed with 5 mg of sodium citrate in 5 mL of HEPES buffer (0.02 M; Gibco, USA). The NP suspension was equilibrated by vigorous stirring at 400 rpm for 1 h. Thereafter, the modified nanoparticles were collected by centrifugation at 8000 rpm and washed thoroughly with HEPES buffer.44 Finally, silica-coated ZnO NPs (Si-ZnO) were synthesized by suspending 25 mg of ZnO NPs in 20 mL of ethanol, followed by vigorous stirring at 400 pm for 1 h at room temperature. Thereafter, to the equilibrated NP suspension, 20 μL of tetraethyl orthosilicate (TEOS; Sigma Aldrich) and 1 mL of ammonia (28–30%; Sigma Aldrich) were subsequently added. The mixture was left to react in an ultrasonic water bath (Fisherbrand, 150 W, 50–60 Hz) for 1 h.45 Unless stated otherwise, the functionalized ZnO NPs were centrifuged at 8000 rpm, washed thoroughly with ethanol, dried at 60 °C overnight and stored in a desiccator until further use. Stock suspensions of ENMs were prepared in ultrapure water at a concentration of 1 mM and maintained at ambient temperature.
2.2. Transmission electron microscopy
The morphological shape and primary size of the ENMs were characterized with a transmission electron microscope (TEM, Carl Zeiss Libra 120 Plus). Briefly, 30 μL of ENMs (100 mg mL−1) suspended in absolute ethanol was pipetted onto a carbon coated TEM copper grid and the samples were allowed to be air dried at a temperature of 50 °C for 1 h. The samples were then imaged using TEM at a voltage of 120 kV and magnification ranging from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× to 70
000× to 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×. The primary particle size was measured with ImageJ freeware.46
000×. The primary particle size was measured with ImageJ freeware.46
2.3. Dynamic light scattering and zeta potential analysis
The hydrodynamic diameter (DH) and zeta potential (ζ) of the comparator ENMs were determined by dynamic light scattering (DLS) analysis using a Zetasizer Nano ZS (Malvern, UK). The ENMs were sonicated in an ultrasonic water bath (Fisherbrand, 150 W, 50–60 Hz) for 30 min to form a stable aqueous suspension. To better characterize the physiochemical properties of the NPs in the biological milieu, the DH and ζ of the nanoparticles in the complete cell culture medium dispersant were also analyzed. In this case, the suspension was prepared by adding the NPs into the cell culture medium followed by incubation in an incubator for 30 min at 37 °C and 5% CO2. Subsequently, the NPs were collected by ultracentrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and allowed to resuspend in DI water for further analysis.
000 rpm for 10 min and allowed to resuspend in DI water for further analysis.
2.4. Fourier-transform infrared spectroscopy (FTIR)
The successful surface modification of ZnO NPs was confirmed using an FTIR spectrometer (PerkinElmer Frontier, USA). 0.5 mg of ZnO NPs was mixed with KBr powder and compacted into a pellet for FTIR examination. FTIR spectra were recorded in the wave number range of 400–4000 cm−1.2.5. Energy dispersive X-Ray analysis (EDX)
EDX analysis was performed to validate the formation of a silica coat on the ZnO NP surface. 2 mg of ZnO or Si-ZnO NP dry powder was mounted on carbon tape and adhered to the sample holder. The sample-containing holder was then loaded into a scanning electron microscope (JEOL FE-SEM 7600F) and analyzed using an integrated EDX analysis system from Oxford instruments (UK).2.6. Cell culture and tumor spheroid formation
Colorectal cancer cells, SW480 (ATCC, USA) and Caco-2 (ATCC) and the normal colon mucosal cell line NCM460 (INCELL, USA) were grown in Dulbecco's Modified Eagle's Medium (DMEM; GE Hyclone, USA) supplemented with 10% (v/v) fetal bovine serum (FBS, GE Hyclone, USA) and 1% penicillin–streptomycin (GE Hyclone, USA) cocktail. Standard culture conditions (37 °C, 5% CO2) were used and the cells were passaged when they reached 80–90% confluence. For 2D cell culture studies, the cells were seeded one day prior to any treatment with initial seeding densities of 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 cells per cm2, 112
500 cells per cm2, 112![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 cells per cm2 and 60
500 cells per cm2 and 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 for SW480, Caco-2 and NCM460, respectively. These seeding densities were applied to the following experiments unless otherwise stated.
000 cells per cm2 for SW480, Caco-2 and NCM460, respectively. These seeding densities were applied to the following experiments unless otherwise stated.
3D tumor spheroids were formed with the help of non-adherent agarose micro-molds as previously described.47 Briefly, non-adherent micro-molds were formed by casting 2% agarose solution in a MicroTissues® 3D Petri Dish (array 5 × 7; Sigma-Aldrich). The formed micro-molds thereafter were placed in a 24-well plate, sterilized with UV irradiation for 30 min and soaked overnight with complete DMEM. Thereafter, Caco-2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells) or SW480 (500
000 cells) or SW480 (500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells) cells were added and allowed to incubate for 24 h to generate 3D tumor spheroids.
000 cells) cells were added and allowed to incubate for 24 h to generate 3D tumor spheroids.
2.7. Conditioning and challenging (2Cs) treatment of colorectal cells
The in vitro conditioning and challenging (2Cs) protocol was employed to examine the potential chemoresistance-inducing properties of ENMs in the human colorectal cell lines.30,31 Briefly, the cells were conditioned by repeatedly exposing them to the comparator ENMs at a non-toxic concentration of 20 μM. A typical conditioning exposure last for 24 h, in which the number of conditioning rounds is denoted by the subscript (i.e., Cn). After the completion of the conditioning step, the cells were then challenged by the panel of anticancer drugs at their respective EC50 (determined from the drug specific dose–response curves). Cell viability was thereafter examined using an alamarBlue assay (ThermoFisher, USA; see ESI†) and the chemoresistance index was determined with the following equation:Cells exhibiting increased resistance to the drugs will have a computed chemoresistance index of >0.1. A chemoresistance index between −0.1 and 0.1 denotes cells with unchanged drug tolerance, while a chemoresistance index of <−0.1 denotes an increased sensitivity to the drugs. The anti-cancer drugs included in this study are cisplatin (CCDP; European pharmacopoeia reference standard, Sigma Aldrich), doxorubicin hydrochloride (DOX; European pharmacopoeia reference standard, Sigma Aldrich), and paclitaxel (PTX; European pharmacopoeia reference standard, Sigma Aldrich). To examine the ZnO NP-induced chemoresistance in a more realistic scenario, SW480 and Caco-2 tumor spheroids were formed, and 2Cs experiments were repeated on the tumor spheroids except the preconditioning dosage was changed to 75 μM, as predetermined by dose response profiling as a nonlethal concentration to both types of tumor spheroids. Cell viability was evaluated as described earlier and the chemoresistance index was computed using the equation as shown earlier. To confirm the formation of chemo-resistant phenotype in the nanoparticle pre-exposed tumor spheroids, the viability of spheroids post-2Cs experiments was examined using SYTOX Green stain (see the ESI†).
2.8. Cytotoxicity assay
Colorectal cells were seeded in a 96-well plate 24 h prior treatment to the comparator ENMs at concentrations of 0–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 μM or anticancer drugs, i.e., CDDP, PTX, DOX, at concentrations of 0–100 μM for the indicated time of exposure. At the end of treatment, cell viability was ascertained by the alarmarBlue cell viability stain (ThermoFisher, USA) for an incubation period of 2 h. Thereafter, the supernatant was collected, and data were obtained from a high-throughput microplate reader (Molecular Devices SpectraMax M2) using a maxima excitation/emission wavelengths of 530/590 nm. The same cytotoxicity assay was employed to ascertain the chemoresistance following the 2Cs protocol.
500 μM or anticancer drugs, i.e., CDDP, PTX, DOX, at concentrations of 0–100 μM for the indicated time of exposure. At the end of treatment, cell viability was ascertained by the alarmarBlue cell viability stain (ThermoFisher, USA) for an incubation period of 2 h. Thereafter, the supernatant was collected, and data were obtained from a high-throughput microplate reader (Molecular Devices SpectraMax M2) using a maxima excitation/emission wavelengths of 530/590 nm. The same cytotoxicity assay was employed to ascertain the chemoresistance following the 2Cs protocol.
For tumor spheroids, cytotoxicity was ascertained with SYTOX Green (ThermoFisher, USA) staining. Briefly, at the end of the treatment, the cells were washed, collected and redispersed with trypsinization (0.25% Trypsin, 0.53 mM EDTA in PBS). The cells were then collected, redispersed in PBS and subjected to sequential staining with cell impermeant SYTOX Green (5 μM, 5 min) followed by cell permeant with Hoescht 33342 (1 μg mL−1, 5 min). Cell suspensions were introduced into a KOVA glasstic slide (Fisher Scientific, USA), and fluorescence images were taken with the help of an inverted fluorescence microscope (IX53, Olympus, Japan), after which the images were analyzed with the help of ImageJ software. The number of dead cells was determined by counting the SYTOX Green stained cells, while the total number of cell populations was determined with the help of Hoescht 33342 staining. The same cytotoxicity assay was employed to ascertain the tumor spheroid chemoresistance following the 2Cs approach.
2.9. Dissolution study
The dissolution properties of ZnO NPs were investigated using inductively coupled plasma mass spectrometry (ICP-MS). Briefly, to simulate the lysosomal pH, the pH of DI water was first adjusted to ∼4.7 by the addition of 1M HCl. ZnO NPs (1 mg ml−1) were added to the dissolution solution and allowed to mix by stirring (100 rpm) at 25 °C for 24 h. Subsequently, the solutions were decanted separately into a 15 mL centrifuge tube and subjected to centrifugation at 8000 rpm for 20 min to separate the particles remaining in the solution. The concentration of the Zn ion for each sample was determined by ICP-MS.2.10. Immunostaining
Cells were fixed with 4% paraformaldehyde for 15 min at room temperature and the cells were further permeabilized using PBS buffer containing 0.2% Triton X-100 for 10 min. Thereafter, the cells were washed with PBS and blocked with 2% bovine serum albumin (BSA) solution for 1 h. Rabbit anti Nrf-2 primary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Cell Signaling Technology, USA) in 2% BSA solution was added to the cells and further incubated at 4 °C overnight. Afterward, the samples were washed 3 times with PBS and counterstained with Hoechst 33342 (2 μg mL−1; ThermoFisher, USA) and chicken anti-rabbit IgG secondary antibody conjugated with Alexa Fluor 488 (1
200; Cell Signaling Technology, USA) in 2% BSA solution was added to the cells and further incubated at 4 °C overnight. Afterward, the samples were washed 3 times with PBS and counterstained with Hoechst 33342 (2 μg mL−1; ThermoFisher, USA) and chicken anti-rabbit IgG secondary antibody conjugated with Alexa Fluor 488 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; ThermoFisher, USA) in the dark at room temperature for 1 h. The samples were then imaged using a fluorescence microscope (Carl Zeiss AxioObserver Z1).
200; ThermoFisher, USA) in the dark at room temperature for 1 h. The samples were then imaged using a fluorescence microscope (Carl Zeiss AxioObserver Z1).
2.11. Immunoblotting
Immunoblotting was used to check the Nrf-2 expression level after siRNA transfection (as described in the previous section). Upon transfection, the transfection medium was changed with fresh cell culture medium, and the cells were further cultivated for up to 3 days. Each day, one group of cells was treated with 10 μM of MG132 (Sigma Aldrich) for 8 h before being harvested with trypsinization for immunoblotting analysis. Non-transfected cells were subjected to the same MG132 treatment and used as a control. Laemmli buffer (BioRad, USA) supplemented with HaltTM protease and phosphatase inhibitor cocktail (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; ThermoFisher, USA) was used to lyse the cells. The protein lysate was then separated by SDS-polyacrylamide gel electrophoresis (SDS PAGE; Mini Protean, Biorad, USA) on a gel casted with TGX and TGX Stain-Free FastCast Acrylamide Kits (Biorad, USA). The resolved proteins thereafter were transferred onto an Immun-Blot PVDF membrane (Biorad, USA), and the membrane was then blocked with 5% BSA solution for 1 h and incubated with primary antibodies (4 °C, overnight). Subsequently, the membrane was washed three times and incubated with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h. The membrane then was exposed to Clarity Western ECL Substrate (Biorad, USA) and the protein bands were visualized with a chemiluminescence imaging system, ChemiDoc (Biorad, USA). Tris buffered saline with Tween 20 (TBST; comprising of 137 mM NaCl, 20 mM Tris-HCl pH 7.6, and 0.05% Tween 20) was used for washing and preparing the blocking and antibody solutions. Rabbit-anti-Nrf-2 (1
100; ThermoFisher, USA) was used to lyse the cells. The protein lysate was then separated by SDS-polyacrylamide gel electrophoresis (SDS PAGE; Mini Protean, Biorad, USA) on a gel casted with TGX and TGX Stain-Free FastCast Acrylamide Kits (Biorad, USA). The resolved proteins thereafter were transferred onto an Immun-Blot PVDF membrane (Biorad, USA), and the membrane was then blocked with 5% BSA solution for 1 h and incubated with primary antibodies (4 °C, overnight). Subsequently, the membrane was washed three times and incubated with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h. The membrane then was exposed to Clarity Western ECL Substrate (Biorad, USA) and the protein bands were visualized with a chemiluminescence imaging system, ChemiDoc (Biorad, USA). Tris buffered saline with Tween 20 (TBST; comprising of 137 mM NaCl, 20 mM Tris-HCl pH 7.6, and 0.05% Tween 20) was used for washing and preparing the blocking and antibody solutions. Rabbit-anti-Nrf-2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Cell Signaling Technology, USA) and mouse-anti-α-tubulin (1
1000; Cell Signaling Technology, USA) and mouse-anti-α-tubulin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000; Sigma Aldrich, USA) primary antibodies were used in this study. Additionally, HRP-conjugated secondary antibodies were obtained from Abcam (USA) and used at a dilution of 1
5000; Sigma Aldrich, USA) primary antibodies were used in this study. Additionally, HRP-conjugated secondary antibodies were obtained from Abcam (USA) and used at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
2.12. Real-time PCR
Total RNA was isolated with the PureLink RNA Mini Kit (Life Technologies, USA). Reverse transcription of RNA samples was performed with the iScript cDNA synthesis kit (Biorad, USA) in accordance with the manufacturer's protocol. Quantitative real-time PCR was conducted using a CFX 96 real-time PCR detection system (Biorad, USA) and SYBR FAST qPCR Master Mix (2×) Universal Kit (KAPA Biosystem, Roche, USA) under the following thermal cycling conditions: enzyme activation at 95 °C for 3 min; followed by 40 cycles of denaturation at 95 °C for 3 s and annealing/extension/data acquisition at 60 °C for 20 s. Melt-curve analysis was also performed to assess the purity of the amplicon/product. Primer sequences (listed in Table 1) were obtained from the validated primer bank (https://pga.mgh.harvard.edu/primerbank/) and commercially synthesized (Sigma Aldrich, Singapore). Ingenuity pathway analysis (IPA) (Qiagen Inc., Germany), gene set enrichment analysis and network analysis were performed as reported.48| Gene | RT-PCR primer sequence | |
|---|---|---|
| Sense | Anti-sense | |
| B2M (housekeeping) | GAGGCTATCCAGCGTACTCCA | CGGCAGGCATACTCATCTTTT |
| ABCC1 | CTCTATCTCTCCCGACATGACC | AGCAGACGATCCACAGCAAAA |
| ABCC5 | GAACTCGACCGTTGGAATGC | TCATCCAGGATTCTGAGCTGAG |
| ABCC11 | GTGAATCGTGGCATCGACATA | GCTTGGGACGGAAGGGAATC |
| P53 | ACTTGTCGCTCTTGAAGCTAC | GATGCGGAGAATCTTTGGAACA |
| ABCG2 | CAGGTGGAGGCAAATCTTCGT | ACCCTGTTAATCCGTTCGTTTT |
| ABCB1 | TTGCTGCTTACATTCAGGTTTCA | AGCCTATCTCCTGTCGCATTA |
| HIF-1A | GAACGTCGAAAAGAAAAGTCTCG | CCTTATCAAGATGCGAACTCACA |
| NF-KB1 | AACAGAGAGGATTTCGTTTCCG | TTTGACCTGAGGGTAAGACTTCT |
| NF-KB2 | ATGGAGAGTTGCTACAACCCA | CTGTTCCACGATCACCAGGTA |
| PIK3/CA | CCACGACCATCATCAGGTGAA | CCTCACGGAGGCATTCTAAAGT |
| PIK3/CB | TATTTGGACTTTGCGACAAGACT | TCGAACGTACTGGTCTGGATAG |
| STAT3 | CAGCAGCTTGACACACGGTA | AAACACCAAAGTGGCATGTGA |
| EGFR | AGGCACGAGTAACAAGCTCAC | ATGAGGACATAACCAGCCACC |
| BAX | CCCGAGAGGTCTTTTTCCGAG | CCAGCCCATGATGGTTCTGAT |
| BCL-2 | ATGTGTGTGGAGAGCGTCAA | CAGGAGAAATCAAACAGAGGC |
| MCL-1 | TGCTTCGGAAACTGGACATCA | TAGCCACAAAGGCACCAAAAG |
| HMOX-1 | AAGACTGCGTTCCTGCTCAAC | AAAGCCCTACAGCAACTGTCG |
| NQO1 | GAAGAGCACTGATCGTACTGG | GGATACTGAAAGTTCGCAGGG |
| GSTM3 | TACCTCTTATGAGGAGAAACGGT | AGGAAAGTCCAGGTCTAGCTTG |
| UGT | CATGCTGGGAAGATACTGTTGAT | GCCCGAGACTAACAAAAGACTCT |
| GSS | GGGAGCCTCTTGCAGGATAAA | GAATGGGGCATAGCTCACCAC |
| GCLM | TGTCTTGGAATGCACTGTATCTC | CCCAGTAAGGCTGTAAATGCTC |
| CAT | TGGAGCTGGTAACCCAGTAGG | TGGTACCTTTGCCTTGGAGTATT |
| SOD | GCTCCGGTTTTGGGGTATCTG | GCGTTGATGTGAGGTTCCAG |
| P21 | GTCACTGTCTTGTACCCTTGTG | CGGCGTTTGGAGTGGTAGAAA |
| TOP3B | CTGTGCTCATGGTTGCTGAAA | CAGCCCTTTGTGTGAGGACA |
| DNMT1 | AGGCGGCTCAAAGATTTGGAA | GCAGAAATTCGTGCAAGAGATTC |
| TYMS | CTGCTGACAACCAAACGTGTG | GCATCCCAGATTTTCACTCCCTT |
| HSPA6 | CAAGGTGCGCGTATGCTAC | GCTCATTGATGATCCGCAACAC |
| HSPB2 | ACCGCCGAGTACGAATTTG | GAGGCCGGACATAGTAGCCA |
| MDM2 | GAATCATCGGACTCAGGTACATC | TCTGTCTCACTAATTGCTCTCCT |
| KRAS | CCTGCTGTGTCGAGAATATCCA | TTGACGATACAGCTAATTCAGAATCA |
| TGF-β1 | CTAATGGTGGAAACCCACAACG | TATCGCCAGGAATTGTTGCTG |
| TGF-α | AGGTCCGAAAACACTGTGAGT | AGCAAGCGGTTCTTCCCTTC |
| PSMB1 | CCTCTACAGCCATGTATTCGGC | CGTTGAAAACGTAGGGCGAAAAT |
| PSMB5 | AGGAACGCATCTCTGTAGCAG | AGGGCCTCTCTTATCCCAGC |
| PSMB7 | CAACTGAAGGGATGGTTGTTGC | GCACCAATGTAACCTTGATACCT |
| AKT | CCTCCACGACATCGCACTG | TCACAAAGAGCCCTCCATTATCA |
| TNF-α | GAGGCCAAGCCCTGGTATG | CGGGCCGATTGATCTCAGC |
| CYP1A1 | TCGGCCACGGAGTTTCTTC | GGTCAGCATGTGCCCAATCA |
| CYP2E1 | GTGATGCACGGCTACAAGG | GGGTGGTCAGGGAAAACCG |
| ALDH1A1 | GCACGCCAGACTTACCTGTC | CCTCCTCAGTTGCAGGATTAAAG |
| PRDX1 | CCACGGAGATCATTGCTTTCA | AGGTGTATTGACCCATGCTAGAT |
| TXNRD1 | ATATGGCAAGAAGGTGATGGTCC | GGGCTTGTCCTAACAAAGCTG |
| TXNDC5 | AGGTTCGTGGCTATCCCACT | TCCGTCGCTCCAGTCTCTG |
2.13. Intracellular reactive oxygen species (ROS) and Zn2+ expression detection
The intracellular ROS expression and Zn2+ content were profiled with CellROX deep red (ThermoFisher, USA) and Newport Green™ DCF diacetate (ThermoFisher, USA), respectively. After the completion of the NP conditioning (24 h) step, the cells were washed thrice with PBS to remove the membrane-bound NPs. Thereafter, the cells were harvested by trypsinization and centrifugation (1500 rpm for 3 min), followed by washing with PBS. The cells then were resuspended in culture medium and counterstained with CellROX deep red (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) and DCF green reagent (1
500) and DCF green reagent (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800). Pluronic® F-127 (0.25 mg mL−1) was also added into the staining solution to facilitate the permeation of DCF green reagent into the cell cytoplasm. The samples were left to stand for 30 min in the dark at room temperature. The samples were then mounted on a glass slide and imaged using a fluorescence microscope (Carl Zeiss AxioObserver Z1) at excitation/emission wavelengths of 505/535 (DCF green) and 640/665 (CellROX deep red). The raw images were captured and analyzed using ImageJ software.
800). Pluronic® F-127 (0.25 mg mL−1) was also added into the staining solution to facilitate the permeation of DCF green reagent into the cell cytoplasm. The samples were left to stand for 30 min in the dark at room temperature. The samples were then mounted on a glass slide and imaged using a fluorescence microscope (Carl Zeiss AxioObserver Z1) at excitation/emission wavelengths of 505/535 (DCF green) and 640/665 (CellROX deep red). The raw images were captured and analyzed using ImageJ software.
2.14. siRNA transfection
To develop Nrf2 depleted colorectal cancer cells, Nrf2 siRNA (Assay ID 107966, ThermoFisher) was transfected into the cancer cells as per the manufacturer's instruction. In brief, Nrf2 siRNA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and HiPerFect transfection reagent (7.5
1000) and HiPerFect transfection reagent (7.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Qiagen, USA) were pre-mixed in serum-free culture medium and allowed to equilibrate at room temperature for 30 min prior to use. The cells were transfected with the prepared transfection reagent mixture at 37 °C for 10 h. The knockdown efficiency of the cells (on days 1, 2, and 3) was ascertained by immunoblotting.
1000; Qiagen, USA) were pre-mixed in serum-free culture medium and allowed to equilibrate at room temperature for 30 min prior to use. The cells were transfected with the prepared transfection reagent mixture at 37 °C for 10 h. The knockdown efficiency of the cells (on days 1, 2, and 3) was ascertained by immunoblotting.
2.15. Statistical analysis
All experiments in this study were carried out in triplicate. Data are presented as the mean ± standard deviation (SD). Origin 9 (OriginLab) was used for statistical analysis. Statistical significance was ascertained with either one-way or two-way analysis of variance (ANOVA) Tukey's post-hoc HSD. Statistical differences are indicated with the probability value (p-value) in the associated text or figure caption.3. Results and discussion
3.1. ZnO NPs promote resistance in colorectal cancer cells to chemotherapeutics
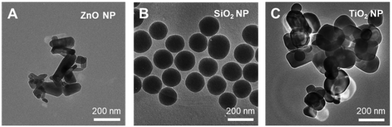
Transmission electron microscopy revealed that the ENMs employed in this study showed varied morphologies. The ZnO NPs are rod-shaped, while the SiO2 NPs and TiO2 NPs are predominately spherical (Fig. 2). On average, the ZnO, SiO2 and TiO2 NPs had an average primary particle size of 97 ± 45 nm, 155 ± 12 nm and 130 ± 50 nm, respectively.Due to their high surface energy to volume ratio, the ENMs agglomerated upon dispersing in ultrapure water to reduce their free surface energy, as evidenced from the hydrodynamic size (DH) measurements (Table 2). Notably, we observed a further increase (1.2–18 fold) in the DH of the ENMs in serum containing DMEM. The tested ENMs also showed negatively charged surfaces in the DMEM dispersant, which could be attributed to the formation of a protein corona enwrapping the NPs.13
| ENMs | ZnO | SiO2 | TiO2 | |
|---|---|---|---|---|
| a Data are presented as the mean ± S.D. b Determined from TEM. | ||||
| Primary sizeb/nm | 97 ± 45 | 155 ± 12 | 130 ± 50 | |
| Hydrodynamic size (DH)/nm | H2O | 174 ± 26 | 199 ± 3 | 298 ± 2 |
| DMEM | 210 ± 17 | 361 ± 16 | 448 ± 5 | |
| Zeta potential (ζ)/mV | H2O | 26.6 ± 2.1 | −24.9 ± 0.2 | −37.2 ± 2.1 |
| DMEM | −24.8 ± 6.9 | −22.4 ± 0.5 | −24.4 ± 0.2 | |
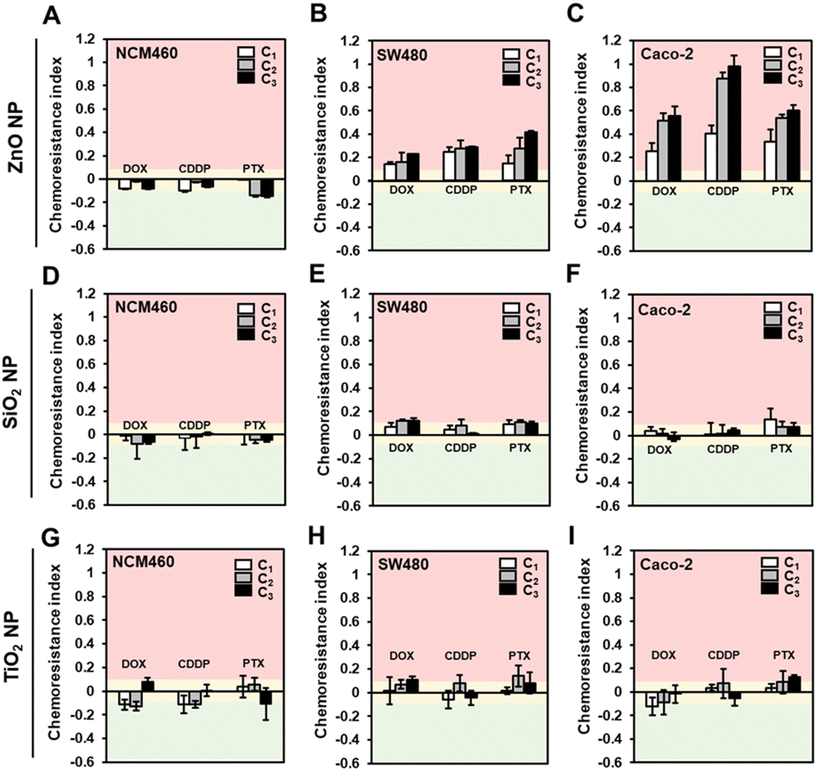
Next, to test the hypothesis that ENMs could induce chemoresistance in colorectal cells, we employed the conditioning and challenge (2Cs) approach30 in which the cells were conditioned through repeated exposure to the ENMs at a non-toxic dose prior to being challenged with chemotherapeutics at their respective EC50 (Fig. 1). In order to determine the conditioning dose, we first performed a cytotoxicity assessment of the comparator ENMs in a panel of human colorectal cells with different (epi)genetic profiles and disease progression statuses, i.e., colorectal adenocarcinoma cells, SW480 and Caco-2, and normal colorectal cells NCM460.49 In general, except for the SiO2 NPs, acute exposure of the ZnO and TiO2 NPs resulted in a dose-dependent cytotoxicity on all three colorectal cells, regardless of their disease status (Fig. S1†). Significant cytotoxicity (>20% cell death) was only observed at a high concentration of ZnO NPs, with the onset of toxicity ranging from 150 to 200 μM.
Therefore, it was deemed that a conditioning dose of 20 μM is apt for our study, since there is clearly no detectable toxic effect and is well within the range that could invoke the cells to mount an adaptive response.30,31 Normalized toxicity against unconditioned colorectal cells (i.e., cells that received no ENM treatment; C0) were used to compute the chemoresistance indices, allowing us to measure any change in the cells’ sensitivity to the chemotherapeutics. A chemotherapeutic index of >0.1 denotes a reduction in the effectiveness of the drugs, since ZnO NP exposure improved the cell tolerance towards the anti-cancer drugs. Conversely, a chemotherapeutic index of <−0.1 indicates increased sensitivity towards the anti-cancer drugs. A chemotherapeutic index range of ± 0.1 suggests that ZnO NPs has a marginal effect on modulating the cellular response to the chemotherapeutics. The EC50 values of CDDP, PTX and, DOX were found to be comparable to reported values and determined to be 750 μM, 300 μM, and 50 μM for NCM460, 50 μM, 9 μM and 1.5 μM for SW480 cells, and 85 μM, 1 μM and 1.5 μM for Caco-2 cells, respectively (Fig. S2†).
The chemotherapeutic indices of the various ENMs, cell types and drug combinations are shown in Fig. 3. In the case of ZnO NPs conditioned NCM460 cells, there was little evidence to suggest that the ZnO NP exposure could modulate the chemotherapeutic sensitivity (Fig. 3A). Specifically, the computed chemoresistance indices range from −0.01 to −0.14 even after three rounds of ZnO NP conditioning. In stark contrast, the increase in resistance against the tested panel of drugs following the 2Cs treatment was notable in the colorectal cancer cells. Both SW480 (Fig. 3B) and Caco-2 (Fig. 3C) cells exhibited increased tolerance to the chemotherapeutics as early as after 1 round of conditioning with ZnO NPs (C1) and this phenomenon became more prominent with increasing rounds of ZnO NP conditioning. (i.e., C2 and C3). Pre-exposing SW480 cells to ZnO NPs resulted in approximately 15–40% increased resistance to the anti-cancer drugs (Fig. 3B). On the other hand, the effect of ZnO NP treatment was more dramatic in Caco-2 cells with an overall increase in chemoresistance indices by 25–100% relative to the unconditioned cells (Fig. 3C). Conversely, the chemoresistance indices for cells treated with concentration-matched SiO2 (Fig. 3D and E) and TiO2 (Fig. 3G–I) NPs fall within the ± 10% range, which indicates that the drug responses were not significantly altered. This suggest that on a “pound-for-pound” basis, ZnO NPs served as a highly potent ENM to elicit an adaptive response in SW480 and Caco-2 cells. A plausible explanation could be due to the amphoteric nature of ZnO NPs, making them more prone to dissolution in the lysosomes than SiO2, and TiO2 NPs, thereby increasing the bioavailability of Zn2+ intracellularly to trigger ROS production.45 More importantly, our findings revealed for the first time that ZnO NPs could promote chemoresistance in human colorectal cancer cells.
3.2. ZnO NPs invoked chemo-disruptive effects governed by surface chemistries
Since ENM surface chemistries are known to profoundly influence the adversarial biological responses,13,14 we next examined if the ZnO NP-induced chemoresistance phenomenon could be modulated by varying the particle surface charge and coating. Therefore, size-matched amine (NH2), carboxylic acid (COOH) and silica (Si) functionalized ZnO NPs were prepared. The corresponding primary particle size, surface charge and hydrodynamic diameter of the ZnO NP variants are shown in Table 3. Fig. S3A† shows the FTIR spectra of the respective ZnO NPs. The characteristic peaks at 1383 and 1555 cm−1 can be attributed to the symmetric and asymmetric stretchings of COO–, which correspond to the presence of citrate in the COOH-ZnO NPs.50 Conversely, the peak detected at 1587 cm−1 is attributable to the amine (NH2) group on the NH2-ZnO NPs.42 Asymmetric stretching vibration of the Si–O–Si bond (1041 cm−1) was detected in the Si-ZnO NP sample (Fig. S3A†).42 Consistent with the FTIR results, both EDX analysis and TEM confirmed the formation of a silica layer (∼5.6 nm) encapsulating the ZnO NPs (Fig. S3B and C†). Next, the chemoresistance imparting properties of the ZnO NPs were assessed using the 2Cs approach in SW480 and Caco-2 cells. Similar to our earlier results, a conditioning dose of 20 μM was employed as we did not observe any significant cytotoxic outcomes (Fig. S1†). A ZnCl2 (20 μM) group was also added to compare the chemoresistance-inducing effect of ZnO NPs and that of free Zn2+ ions.| ENMs | COOH-ZnO | NH2-ZnO | Si-ZnO | |
|---|---|---|---|---|
| a Data are presented as the mean ± S.D. b Determined from TEM. | ||||
| Primary sizeb/nm | 106 ± 50 | 108 ± 67 | 94 ± 53 | |
| Hydrodynamic size (DH)/nm | H2O | 141 ± 13 | 185 ± 20 | 244± 8 |
| DMEM | 215 ± 4 | 198 ± 4 | 302 ± 6 | |
| Zeta potential (ζ)/mV | H2O | −36.6 ± 2.2 | 37.5 ± 0.4 | −38.7 ± 0.5 |
| DMEM | −27.3 ± 0.3 | −23.9 ± 9.1 | −23.7 ± 6.9 | |
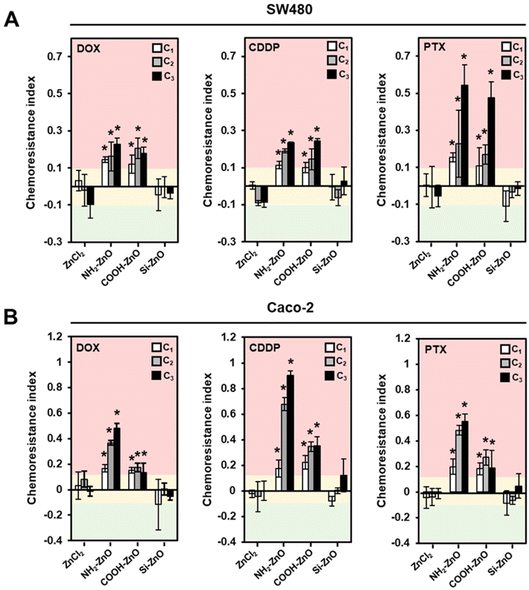
Preconditioning the colorectal cancer cells with ZnCl2 did not result in the induction of chemoresistance in the cells as we observed no perceivable change in the chemoresistance indices (± 10%) across the tested drugs (Fig. 4). This finding agrees with our earlier observation, which showed that ZnO must be presented as a nanoparticulate form before it could trigger an innate adaptive response in the cells that are subjected to a low level of oxidative stress.30 In comparison, cancer cells exposed to COOH-ZnO NPs and NH2-ZnO NPs showed significant enhancement in resistance to the chemotherapeutics drugs by 12–90% (Fig. 4A). Positively charged ENMs are known to favor cell–nanomaterial interactions and thus expected to exhibit greater modulatory effects in biological processes such as metabolism,51 immunoreactivity,52 and defense responses.53 Therefore, from the structure–activity relationship (SAR) standpoint, the chemoresistance-inducing properties are expected to be higher in the positively charged NH2-ZnO NPs compared to the negatively charged COOH-ZnO. Although this trend holds true for Caco-2 cells (Fig. 4B), we did not observe any significant drug-resistance enhancement effect of the NH2-ZnO NPs over the negatively charged COOH-ZnO NPs in the SW480 cells (Fig. 4A). Nonetheless, it is noteworthy that the chemoresistance-inducing effect of ZnO NPs could be circumvented with the aid of a silica coat, as in this case of Si-ZnO NPs. This observation is interesting, since silica coating has been documented as an effective strategy to limit the intracellular dissolution of ZnO NPs and thereby reducing their toxic potential.45 In this regard, we uncovered that this “safer-by-design” concept is amenable in the context of ZnO NP-induced chemoresistance. Collectively, our findings suggest that the SAR is less straightforward in the context of low dose ZnO NP exposure and chemoresistance, as multiple determinants such as ENM properties, drug-type, and genetic and phenotypic characteristics of the cancer cells are simultaneously at play.
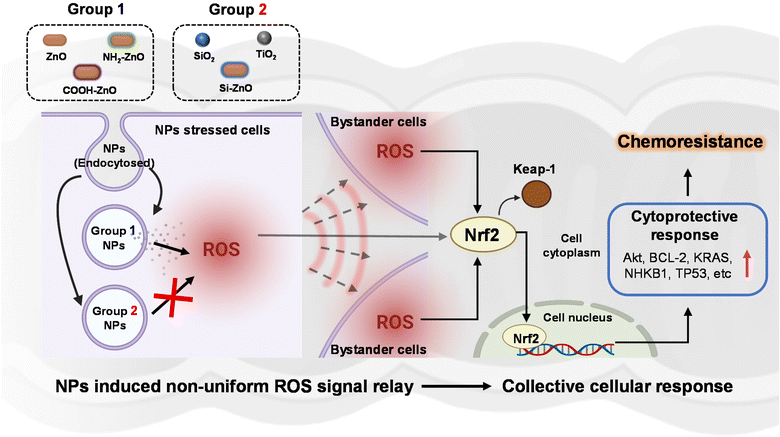
3.3. ZnO NP stressed cells stimulate ROS production in bystander cells
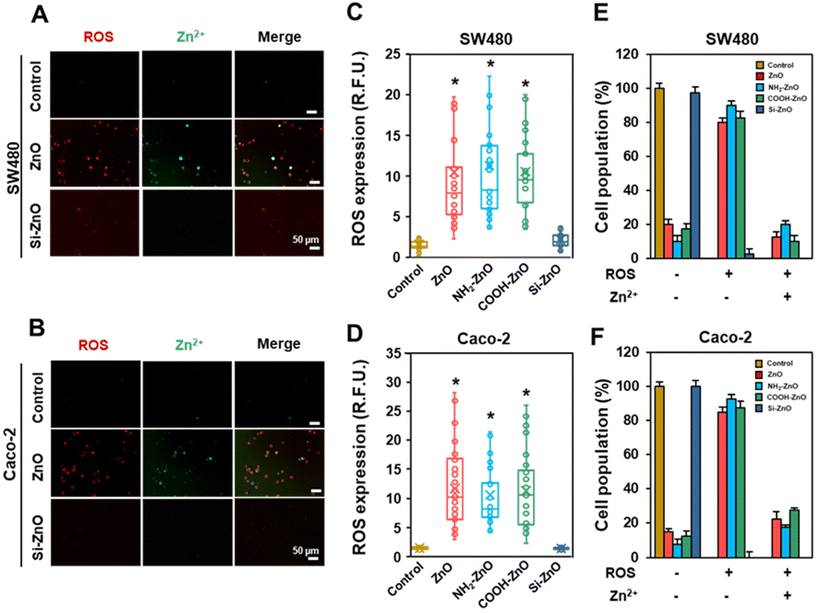
The mitigation of ZnO NP-induced chemoresistance effects with the silica coat strongly implies that the dissolution of ZnO NPs in the acidic lysosomal compartment and subsequent release of Zn2+ ions are important obligatory upstream events.45 Indeed, in vitro dissolution tests showed that the silica coating could significantly suppress Si-ZnO NPs dissolution at acidic pH (Fig. S3D†). Even so, it is unclear how would exposing the cells to a low dose (limiting) of ZnO NPs (20 μM) invoke a systematic acquired resistance to the anti-cancer drugs. Since low levels of ROS are well documented to offer cytoprotective effects through hormesis, we postulated that the ROS from the ZnO NPs stressed cells may be relayed to the “bystander” cells, which stimulate adaptation against the chemotherapeutic drugs. To investigate the involvement of oxidative stress, we first measured the percentage of ROS positive cells in each of the 4 h post-treatment. As expected, except for the Si-ZNO NP group, the intracellular levels of ROS were significantly elevated in the ZnO, NH2-ZnO and COOH-ZnO NP treated cells, averaging 5.8–7.5-fold higher relative to the untreated cells. (Fig. 5A–D). Experiments performed using concentration-matched SiO2 and TiO2 NPs similarly did not increase the intracellular ROS level in either cell types (Fig. S4†), corroborating the lack of ENM-induced adaptive stress responses (Fig. 3E, F H, and I). Interestingly, visual inspection of cells counter-stained with CellROX and DCF revealed that only a limited number of cells were both positively stained for ROS (red) and Zn2+ (green) (Fig. 5E and F). Specifically, ∼10–27.5% of the cell population was found to express elevated levels of ROS and Zn2+ regardless of the cell type. In stark contrast, majority (>70%) of the cells were ROS (+) but Zn2+ (−). Taken together, our results suggest that ROS can be relayed between the ZnO NP sensing cells and the rest of the bystander cells to coordinate a systemic ZnO NP-induced adaptive response.3.4. ZnO NPs activate Nrf2 related stress response pathways to confer chemoresistance in colorectal cancer cells
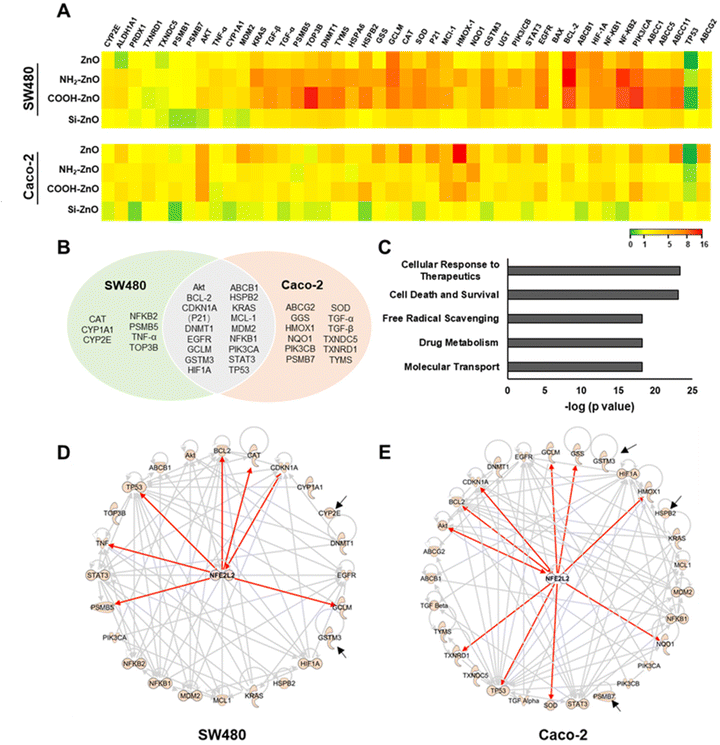
Acquisition of chemoresistance in cancer cells is a complex process in which multiple functional pathways to work are orchestrated in a concerted manner to ensure the survival of these cells against the onslaught of cancer drugs.54 Many of these pathways are modulated by cellular master transcriptional regulators in response to ROS homeostasis perturbance caused by chemotherapeutics.54,55 Thus, to gain a mechanistic understanding of ZnO NP-induced chemoresistance, we employed a real-time PCR array to screen a panel of genes that are associated with oxidative stress response and/or chemoresistance (Fig. 6A).56–59 Compared to the untreated control, Si-ZnO NP exposure has negligible effects on colorectal cancer cell gene expression. Cells treated with ZnO, NH2-ZnO and COOH-ZnO NPs, however, resulted in 24 and 29 differentially expressed (DE) genes (with an absolute fold change of >1.5) in SW480 and Caco-2 cells, respectively, with 17 genes found to be differentially expressed in both cell types (Fig. 6B). We further analyzed these DE genes in correlation to their known canonical pathways based on the ingenuity knowledge base (IKB),60 and identified the top 5 associated canonical pathways (p < 10−15) which were enriched following ZnO NP exposure, namely cellular response to therapeutics, drug metabolism, molecular transport, free radical scavenging and cell death and survival.Next, we identified pathways associated with cellular response to therapeutics (p = 4.6 × 10−24), drug metabolism (p = 6.5 × 10−19), and molecular transport (p = 6.5 × 10−19) to be significantly enriched (Fig. 6C). Alteration of genes within these pathways have been implicated to augment chemotherapeutics resistance in many cancer cells. For example, repeated exposure of cultured tumor cells or tumors in vivo with DOX has been reported to increase the expression of ABCB1 genes, resulting in the overexpression of P-glycoprotein (P-gp), a crucial drug-efflux transporter, and subsequent development of multidrug resistance (MDR) cancer.61,62 In another study, HIF-1 activation was demonstrated to promote therapeutic resistance through inhibition of apoptosis and senescence and the activation of drug efflux and cellular metabolism.63 Interestingly, majority of these enriched pathways converge with ROS, a known modulator.56,59,63,64 For instance, ROS overproduction via NADPH oxidase 4 activation was demonstrated to be an important upstream signaling event needed to upregulate the expression levels of HIF-1 and P-gp in the build-up of resistance against DOX.65 An enhanced ROS level also triggers a multitude of signal transductions involved in the maintenance of redox balance through various enzymatic scavengers66,67 and the repression of apoptotic death.33,35,37 Consistent with these findings, we noted pathways associated with free radical scavenging (p = 6.0 × 10−19) and cell death and survival (p = 8.7 × 10−24) processes were significantly enriched (Fig. 6C), thereby further supporting the important role of ENM-induced ROS (non-toxic level) production in the colorectal cancer cells in the context of chemoresistance (Fig. 5).
The data thus far suggested that ZnO NP-induced chemoresistance is a multifaceted process, necessitating the involvement of a ROS-sensitive cytoprotective molecular regulator. The Nrf2 protein is a well-established master regulator of antioxidative responses and cellular defense mechanisms, and recently linked to the chemoresistance acquisition in colon cancer.14,19 Under basal conditions, Nrf2 is maintained at a low level through the formation of the Nrf2-Keap1 complex to facilitate its proteasomal degradation by the 26S proteasome. In response to exogenous stressors, Nrf2 would dissociate from Keap1 and translocate to the nucleus to form a transcriptionally active complex with proteins such as Mafs, thereby driving the expression of more than 500 cytoprotective genes encoding for antioxidants, detoxification or metabolic enzymes and multi drug resistance-associated protein transporters.33,67,68 Therefore, we postulate that Nrf2 may be implicated in ZnO-induced chemoresistance. Indeed, functional network analysis of ZnO NP treated SW480 revealed that 7 genes (i.e., BCL-2, CAT, TP53, CDKN1A, GCLM, PSMB5, and TNF-α) were directly mediated by Nrf2, with additional 15 genes that are indirectly regulated by Nrf2 (Fig. 6D). In the case of Caco-2 cells, we identified that 10 genes (i.e., AKT, BCL-2, CDKN1A(P21), GCLM, GSS, HMOX1, NQO1, SOD, TP53, and TXNRD1) were under direct control by Nrf2 while another 16 genes were mediated indirectly by Nrf2 (Fig. 6E). These functional network analyses support the postulation that Nrf2 serves as a key transcriptional factor mediating ZnO NP-induced adaptive cellular response in colorectal cancers.
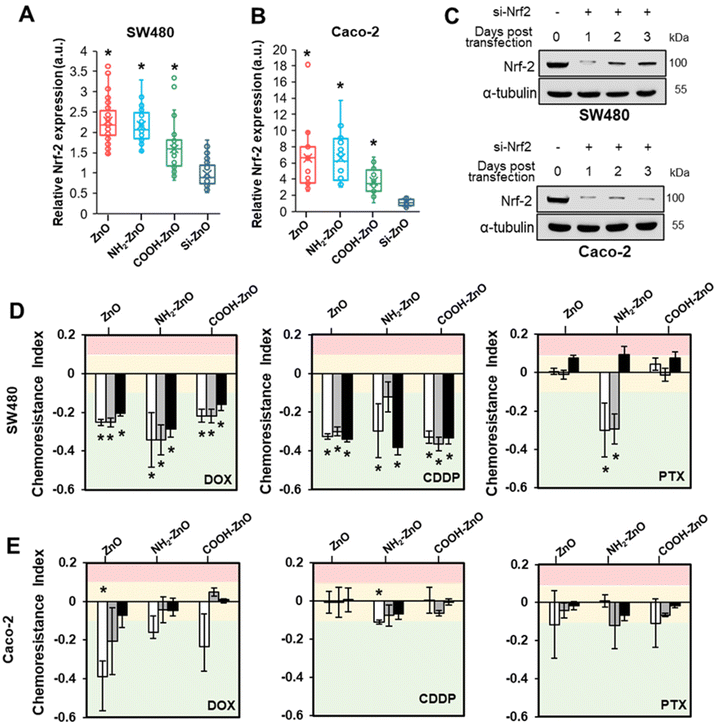
3.5. Activation of Nrf2 is required for ZnO NP-induced chemoresistance in colorectal cancers
To ascertain the precise chemoresistance regulatory role of Nrf2, immunofluorescence staining was performed on the cancer cells to visualize the expression level and subcellular distribution of Nrf2 (Fig. S5†). In the untreated control and Si-ZnO NPs treated cells, Nrf2 was present at low levels and was detected in a diffused pattern. ZnO, NH2-ZnO and COOH-ZnO NP treatment caused a significant Nrf2 signal intensity increase in the cell nucleus. The enhancement in Nrf2 staining is consistent with the PCR data and suggest that ZnO NPs can trigger the Nrf2 stress response pathway, presumably under the influence of oxidative stress (Fig. 3). Quantitative analysis showed that the nucleus bound Nrf2 expressions are 1.6- to 2.3-fold (SW480) and 3.7- to 6.5-fold (Caco-2) higher in the ZnO, NH2-ZnO and COOH-ZnO NP treated cells compared to the control (Fig. 7A and B).So far, our findings showed that the development of chemoresistance due to ZnO NP treatment is intimately associated with the constitutive upregulation of Nrf2 protein expression. Therefore, we asked if inhibiting Nrf2 function may disrupt the chemoresistance-inducing effect of ZnO NPs. To examine this possibility, siRNA Nrf2 was stably transfected into SW480 and Caco-2 cells. According to the immuno-blotting results, the Nrf2 protein levels in both cell lines decreased significantly over the course of 3 days in the siRNA Nrf2 transfected cells, suggesting that the Nrf2 expression was successfully knocked down (Fig. 7C). To determine the effect of Nrf2 knockdown, we subjected the siRNA Nrf2 transfected cells to the 2Cs treatment. As shown in Fig. 7D and E, we observed Nrf2 knockdown abrogated ZnO NP-induced multidrug resistance in both types of cancer cells. Additionally, ZnO NP conditioning increased the sensitivity of the Nrf2 deficient cells to DOX and CDDP, resulting in increased cell death compared to the non-conditioned control. The chemoresistance indices of the ZnO, NH2-ZnO NP and COOH-ZnO NP conditioned SW480 cells range between 0.2 and 0.4. However, only the NH2-ZnO NP exposed cells showed an increased susceptibility to PTX. The chemotherapeutic sensitization effect is less apparent in Nrf2 knockdown Caco-2. These results suggest that the chemoresistance-inducing of ZnO NPs is contingent on an intact Nrf2 stress response mechanism in SW480 and Caco-2 cells.
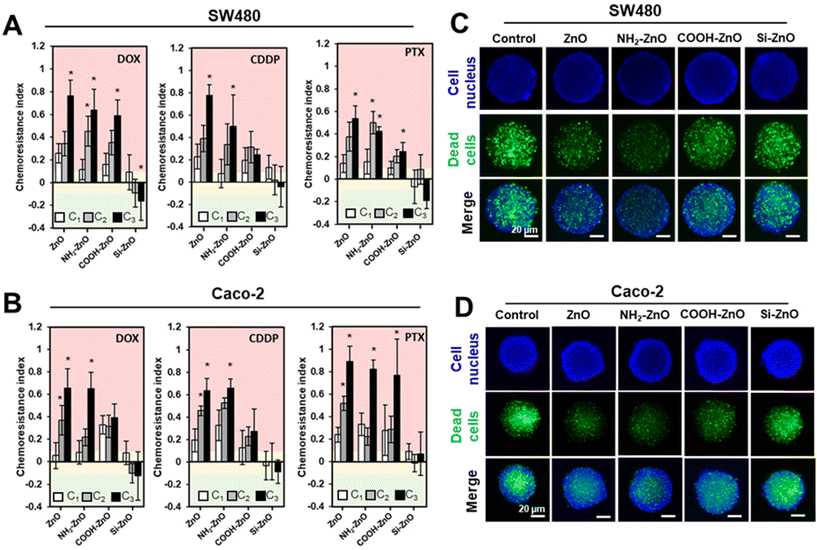
3.6. ZnO nanoparticle-induced chemoresistance in colorectal cancer spheroids
The clinical relevance of our findings was next evaluated using biomimicking 3D cancer spheroids models. Compared to conventional 2D culture, 3D cancer spheroids could better recapitulate the in vivo tumor microenvironment, matrix organization, oxygen distribution, and signaling, resulting in a model with better clinical presentation and response to treatment of the tumor.42,47,69–71 To re-ascertain the non-cytotoxic ZnO NP level and the EC50 of the panel of drugs to be used in the 2Cs experiments for the colorectal tumor spheroids, we determined the cell viability by double staining The cells were co-stained with SYTOX Green (green) and Hoescht 33342 (blue), which labels the dead cells and cell nucleus, respectively (Fig. S6†). After 2 rounds of conditioning the colorectal tumor spheroids with ZnO, NH2-ZnO and COOH-ZnO NPs (75 μM), chemoresistance indices were significantly increased against the tested anticancer drugs (i.e., CDDP, PTX, and DOX) in SW480 (Fig. 8A) and Caco-2 (Fig. 8B) spheroids. Consistent with the 2D data, Si-ZnO NP exposure, however, could influence the chemosensitivity of the tumor spheroids. These results were further supported by the representative fluorescence images of the PTX treated cancer spheroids, following 3 rounds (i.e., the C3 group) of ZnO, NH2-ZnO and COOH-ZnO NP exposure (Fig. 8C and D). Compared to the unconditioned control, there is an apparent decrease in the number of dead cells (green+) for the ZnO NP conditioned spheroids, except for the Si-ZnO NP group.4. Conclusions
In the present study, we examined the potential association between ENM exposure and chemoresistance in colorectal cancer cells, as an indicative public health implication in disease aggravation. Although there is currently no direct evidence of ingested ENM interaction with intestinal cancer in humans, this possibility becomes evident when one considers the organization of colorectal tumor in the colonic lumen. Since the pathological presentation of in situ colon carcinomas typically takes the form of protruding features in the luminal surface of the colon, it can therefore be argued that the likelihood of the ingested ENMs interfacing with the cancerous colon epithelial cells to be similar as normal colonic epithelial cells. Using the developed 2Cs approach, we experimentally provided the earliest demonstration of ZnO NP (∼100 nm)-induced resistance to CDDP, DOX, and PTX in SW480 and Caco-2 human colon cancer cell lines, but not normal NCM460 colon cells. However, the ZnO NP-induced chemotherapeutics resistance was also found to vary considerably according to particle surface chemistry, with the pristine, NH2- and COOH- functionalized ZnO NPs showing the highest potency. In contrast, Si-ZnO NP conditioning has negligible bearing on the efficacy of the anti-cancer drugs, suggesting that the “safe-by-design” strategy45,72,73 can be extended to remit ENM-induced chemoresistance. Detailed cellular profiling showed that ZnO NPs activated the intrinsic adaptive mechanism in response to the ENM-triggered oxidative stress could be relayed at the systemic level through ROS overproduction. To gain further mechanistic insights, we employed the transcriptomics approach to monitor changes in stress response associated gene expression following ZnO NP exposure. In combination with IPA bioinformatics analysis and siRNA knockdown experiments, Nrf2 was revealed to be a key transcription factor coordinating the ZnO NP mediated cytoprotective effects against the chemotherapeutics in colorectal cancer cells. Therefore, the non-toxic dose of ZnO NPs could concomitantly induce Nrf2-dependent oxidative stress and redox metabolic re-wiring to impart resistance against chemotherapeutic agents. These salient findings are succinctly summarized in Fig. 9. Our findings may point to the real possibility that certain subpopulations are likely to present higher susceptibility to ZnO NP exposure through ingestion. Based on our findings we recommend that future studies should be directed to (i) cross-validate the biological significance of ENM-induced chemoresistance using relevant in vivo colon carcinogenesis models, (ii) examine the possible role of ENM–drug interaction in mediating the chemo-disruptive process, and (iii) further investigate the link between specific ENM sources/ physicochemical properties and their chemoresistance-inducing effects. This would enable a better discernment on the disproportionate adversarial effects of ENM exposure in the context of cancer development and drug resistance.Author contributions
Conceptualization: TCY. Methodology: TCY, ZW, MIS, and HKL. Investigation: ZW, MIS, and HKL. Data analysis: TCY, ZW, MIS, and HKL. Funding acquisition: TCY. Supervision: TCY and NKW. Writing – original draft: TCY, ZW, and MIS. Writing – review and editing: TCY, MIS, ZW, NKW, and HKL.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge support from the Nanyang Technological University (NTU)-Harvard School of Public Health (HSPH) Initiative for Sustainable Nanotechnology Program (NTU-HSPH 18002). The authors would also like to acknowledge the Facility for Analysis, Characterization, Testing and Simulation (FACTS), Nanyang Technological University, Singapore, for the use of their electron microscopy facilities.References
- M. I. Setyawati, Z. Zhao and K. W. Ng, Small, 2020, 16, 2001246 CrossRef CAS PubMed.
- Nanotechnology in Food, https://www.centerforfoodsafety.org/nanotechnology-in-food, (accessed April 2022).
- A. Weir, P. Westerhoff, L. Fabricius, K. Hristovski and N. von Goetz, Environ. Sci. Technol., 2012, 46, 2242–2250 CrossRef CAS PubMed.
- Q. Chaudhry, M. Scotter, J. Blackburn, B. Ross, A. Boxall, L. Castle, R. Aitken and R. Watkins, Food Addit. Contam., Part A: Chem., Anal., Control, Exposure Risk Assess., 2008, 25, 241–258 CrossRef CAS PubMed.
- F. Moreno-Olivas, E. Tako and G. J. Mahler, Food Chem. Toxicol., 2019, 124, 112–127 CrossRef CAS.
- A. B. da Silva, M. Miniter, W. Thom, R. E. Hewitt, J. Wills, R. Jugdaohsingh and J. J. Powell, Curr. Opin. Toxicol., 2020, 19, 112–120 CrossRef PubMed.
- M. C. Lomer, R. P. Thompson and J. J. Powell, Proc. Nutr. Soc., 2002, 61, 123–130 CrossRef PubMed.
- L. Voss, P. E. J. Saloga, V. Stock, L. Böhmert, A. Braeuning, A. F. Thünemann, A. Lampen and H. Sieg, ACS Appl. Nano Mater., 2020, 3, 724–733 CrossRef CAS.
- K. Nakagawa, in Nano– and Microencapsulation for Foods, ed. H. S. Kwak, John Wiley & Sons, Ltd, UK, 2014, ch. 10, pp. 249–267 Search PubMed.
- L. Kish, N. Hotte, G. G. Kaplan, R. Vincent, R. Tso, M. Ganzle, K. P. Rioux, A. Thiesen, H. W. Barkema, E. Wine and K. L. Madsen, PLoS One, 2013, 8, e62220 CrossRef CAS.
- K. Williams, J. Milner, M. D. Boudreau, K. Gokulan, C. E. Cerniglia and S. Khare, Nanotoxicology, 2015, 9, 279–289 CrossRef CAS PubMed.
- A. Pietroiusti, E. Bergamaschi, M. Campagna, L. Campagnolo, G. De Palma, S. Iavicoli, V. Leso, A. Magrini, M. Miragoli, P. Pedata, L. Palombi and I. Iavicoli, Part. Fibre Toxicol., 2017, 14, 47 CrossRef PubMed.
- C. Y. Tay, M. I. Setyawati, J. Xie, W. J. Parak and D. T. Leong, Adv. Funct. Mater., 2014, 24, 5936–5955 CrossRef CAS.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- K. Ariga, Nanoscale Horiz., 2021, 6, 364–378 RSC.
- K. Ariga, Mater. Chem. Front., 2017, 1, 208–211 RSC.
- K. W. Ng, S. P. K. Khoo, B. C. Heng, M. I. Setyawati, E. C. Tan, X. Zhao, S. Xiong, W. Fang, D. T. Leong and J. S. C. Loo, Biomaterials, 2011, 32, 8218–8225 CrossRef CAS PubMed.
- M. I. Setyawati, C. Y. Tay and D. T. Leong, Biomaterials, 2013, 34, 10133–10142 CrossRef CAS.
- S. M. Toprani, D. Bitounis, Q. Huang, N. Oliveira, K. W. Ng, C. Y. Tay, Z. D. Nagel and P. Demokritou, ACS Nano, 2021, 15, 4728–4746 CrossRef CAS PubMed.
- S. Azimee, M. Rahmati, H. Fahimi and M. A. Moosavi, Life Sci., 2020, 248, 117466 CrossRef CAS PubMed.
- O. Margalit, A. J. Simon, E. Yakubov, R. Puca, A. Yosepovich, C. Avivi, J. Jacob-Hirsch, I. Gelernter, A. Harmelin, I. Barshack, G. Rechavi, G. D'Orazi, D. Givol and N. Amariglio, Int. J. Cancer, 2012, 131, E562–E568 CrossRef CAS PubMed.
- M. Xu, Y. Hu, W. Ding, F. Li, J. Lin, M. Wu, J. Wu, L. P. Wen, B. Qiu, P. F. Wei and P. Li, Biomaterials, 2020, 258, 120308 CrossRef CAS PubMed.
- X. Cai, Y. Luo, W. Zhang, D. Du and Y. Lin, ACS Appl. Mater. Interfaces, 2016, 8, 22442–22450 CrossRef CAS PubMed.
- Y. H. Miao, L. P. Mao, X. J. Cai, X. Y. Mo, Q. Q. Zhu, F. T. Yang and M. H. Wang, World J. Gastroenterol., 2021, 27, 3851–3862 CrossRef CAS PubMed.
- Y. Hu, H. R. Zhang, L. Dong, M. R. Xu, L. Zhang, W. P. Ding, J. Q. Zhang, J. Lin, Y. J. Zhang, B. S. Qiu, P. F. Wei and L. P. Wen, Nanoscale, 2019, 11, 11789–11807 RSC.
- N. Tripathy, R. Ahmad, H. A. Ko, G. Khang and Y. B. Hahn, Nanoscale, 2015, 7, 4088–4096 RSC.
- M. I. Setyawati, C. Y. Tay and D. T. Leong, Small, 2015, 11, 3458–3468 CrossRef CAS PubMed.
- A. Pinsino, N. G. Bastús, M. Busquets-Fité, L. Canesi, P. Cesaroni, D. Drobne, A. Duschl, M.-A. Ewart, I. Gispert, J. Horejs-Hoeck, P. Italiani, B. Kemmerling, P. Kille, P. Procházková, V. F. Puntes, D. J. Spurgeon, C. Svendsen, C. J. Wilde and D. Boraschi, Environ. Sci.: Nano, 2020, 7, 3216–3232 RSC.
- C. Zhang, R. Sun and T. Xia, Nano Today, 2020, 34, 100909 CrossRef CAS.
- Z. Wu, H. Yang, G. Archana, M. Rakshit, K. W. Ng and C. Y. Tay, Nanotoxicology, 2018, 12, 1215–1229 CrossRef CAS PubMed.
- Z. Wu, P. Shi, H. K. Lim, Y. Ma, M. I. Setyawati, D. Bitounis, P. Demokritou, K. W. Ng and C. Y. Tay, Small, 2020, 16, e2000963 CrossRef PubMed.
- T. Nguyen, P. J. Sherratt and C. B. Pickett, Annu. Rev. Pharmacol. Toxicol., 2003, 43, 233–260 CrossRef CAS PubMed.
- E. Panieri, P. Telkoparan-Akillilar, S. Suzen and L. Saso, Biomolecules, 2020, 10, 791 CrossRef CAS PubMed.
- T. Zhang, M. J. Gaffrey, D. G. Thomas, T. J. Weber, B. M. Hess, K. K. Weitz, P. D. Piehowski, V. A. Petyuk, R. J. Moore, W.-J. Qian and B. D. Thrall, NanoImpact, 2020, 17, 100194 CrossRef.
- S. Wu, H. Lu and Y. Bai, Cancer Med., 2019, 8, 2252–2267 CrossRef PubMed.
- G. Barrera, M. A. Cucci, M. Grattarola, C. Dianzani, G. Muzio and S. Pizzimenti, Antioxidants, 2021, 10, 510 CrossRef CAS PubMed.
- M. R. de la Vega, E. Chapman and D. D. Zhang, Cancer Cell, 2018, 34, 21–43 CrossRef PubMed.
- W. A. Hammond, A. Swaika and K. Mody, Ther. Adv. Med. Oncol., 2016, 8, 57–84 CrossRef CAS PubMed.
- R. Merk, K. Heßelbach, A. Osipova, D. Popadić, W. Schmidt-Heck, G. J. Kim, S. Günther, A. G. Piñeres, I. Merfort and M. Humar, Int. J. Environ. Res. Public Health, 2020, 17, 8193 CrossRef CAS PubMed.
- K. Dzobo, N. Hassen, D. A. Senthebane, N. E. Thomford, A. Rowe, H. Shipanga, A. Wonkam, M. I. Parker, S. Mowla and C. Dandara, Molecules, 2018, 23, 930 CrossRef.
- O. Doğanlar, Z. B. Doğanlar, A. K. Kurtdere, T. Chasan and E. S. Ok, Ecotoxicol. Environ. Saf., 2020, 202, 110940 CrossRef PubMed.
- Z. Wu, H. K. Lim, S. J. Tan, A. Gautam, H. W. Hou, K. W. Ng, N. S. Tan and C. Y. Tay, Small, 2020, e2003757, DOI:10.1002/smll.202003757.
- F. Grasset, N. Saito, D. Li, D. Park, I. Sakaguchi, N. Ohashi, H. Haneda, T. Roisnel, S. Mornet and E. Duguet, J. Alloys Compd., 2003, 360, 298–311 CrossRef CAS.
- K. M. Kim, M. H. Choi, J. K. Lee, J. Jeong, Y. R. Kim, M. K. Kim, S. M. Paek and J. M. Oh, Int. J. Nanomed., 2014, 9(Suppl 2), 41–56 Search PubMed.
- S. L. Chia and D. T. Leong, Heliyon, 2016, 2, e00177 CrossRef.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- S. L. Chia, C. Y. Tay, M. I. Setyawati and D. T. Leong, Small, 2015, 11, 702–712 CrossRef CAS PubMed.
- H. S. Cheng, C. Marvalim, P. Zhu, C. L. D. Law, Z. Y. J. Low, Y. K. Chong, B. T. Ang, C. Tang and N. S. Tan, Theranostics, 2021, 11, 5127–5142 CrossRef CAS.
- J. M. Biazik, K. A. Jahn, Y. Su, Y. N. Wu and F. Braet, World J. Gastroenterol., 2010, 16, 2743–2753 CrossRef PubMed.
- M. Nouri-Khezrabad, A. P. Luz, V. R. Salvini, F. Golestani-Fard, H. R. Rezaie and V. C. Pandolfelli, Ceram. Int., 2015, 41, 3051–3057 CrossRef CAS.
- C. Y. Tay, Y. Yu, M. I. Setyawati, J. Xie and D. T. Leong, Nano Res., 2014, 7, 805–815 CrossRef CAS.
- S. Singha, K. Shao, K. K. Ellestad, Y. Yang and P. Santamaria, ACS Nano, 2018, 12, 10621–10635 CrossRef CAS PubMed.
- A. Nagy, A. Steinbruck, J. Gao, N. Doggett, J. A. Hollingsworth and R. Iyer, ACS Nano, 2012, 6, 4748–4762 CrossRef CAS PubMed.
- H. C. Zheng, Oncotarget, 2017, 8, 59950–59964 CrossRef PubMed.
- X. J. Wang, Z. Sun, N. F. Villeneuve, S. Zhang, F. Zhao, Y. Li, W. Chen, X. Yi, W. Zheng, G. T. Wondrak, P. K. Wong and D. D. Zhang, Carcinogenesis, 2008, 29, 1235–1243 CrossRef CAS PubMed.
- E. K. Kim, M. Jang, M. J. Song, D. Kim, Y. Kim and H. H. Jang, Antioxidants, 2019, 8, 471 CrossRef CAS PubMed.
- B. Mansoori, A. Mohammadi, S. Davudian, S. Shirjang and B. Baradaran, Adv. Pharm. Bull., 2017, 7, 339–348 CrossRef CAS PubMed.
- A. D. Steg, M. R. Burke, H. M. Amm, A. A. Katre, Z. C. Dobbin, D. H. Jeong and C. N. Landen, Oncotarget, 2014, 5, 7065–7080 CrossRef PubMed.
- P. E. Lonning and S. Knappskog, Oncogene, 2013, 32, 5315–5330 CrossRef CAS PubMed.
- A. Kramer, J. Green, J. Pollard Jr. and S. Tugendreich, Bioinformatics, 2014, 30, 523–530 CrossRef.
- I. Genovese, A. Ilari, Y. G. Assaraf, F. Fazi and G. Colotti, Drug Resistance Updates, 2017, 32, 23–46 CrossRef PubMed.
- R. W. Robey, K. M. Pluchino, M. D. Hall, A. T. Fojo, S. E. Bates and M. M. Gottesman, Nat. Rev. Cancer, 2018, 18, 452–464 CrossRef CAS PubMed.
- N. A. Warfel and W. S. El-Deiry, Curr. Med. Chem., 2014, 21, 3021–3028 CrossRef CAS PubMed.
- K. A. Conklin, Integr. Cancer Ther., 2004, 3, 294–300 CrossRef CAS PubMed.
- N. A. Seebacher, D. R. Richardson and P. J. Jansson, Br. J. Pharmacol., 2015, 172, 2557–2572 CrossRef CAS PubMed.
- J. K. Kwee, BioMed Res. Int., 2014, 2014, 209845–209845 Search PubMed.
- C. Gonzalez-Donquiles, J. Alonso-Molero, T. Fernandez-Villa, L. Vilorio-Marques, A. J. Molina and V. Martin, PLoS One, 2017, 12, e0177549 CrossRef CAS PubMed.
- B. M. Hybertson, B. Gao, S. K. Bose and J. M. McCord, Mol. Aspects Med., 2011, 32, 234–246 CrossRef CAS PubMed.
- H. Cao, H. S. Cheng, J. K. Wang, N. S. Tan and C. Y. Tay, Acta Biomater., 2021, 132, 448–460 CrossRef CAS.
- C. Y. Tay, M. S. Muthu, S. L. Chia, K. T. Nguyen, S.-S. Feng and D. T. Leong, Adv. Funct. Mater., 2016, 26, 4046–4065 CrossRef CAS.
- D. T. Leong and K. W. Ng, Adv. Drug Delivery Rev., 2014, 79–80, 95–106 CrossRef CAS PubMed.
- R. Li, Z. Ji, C. H. Chang, D. R. Dunphy, X. Cai, H. Meng, H. Zhang, B. Sun, X. Wang, J. Dong, S. Lin, M. Wang, Y.-P. Liao, C. J. Brinker, A. Nel and T. Xia, ACS Nano, 2014, 8, 1771–1783 CrossRef CAS PubMed.
- G. A. Sotiriou, C. Watson, K. M. Murdaugh, T. H. Darrah, G. Pyrgiotakis, A. Elder, J. D. Brain and P. Demokritou, Environ. Sci.: Nano, 2014, 1, 144–153 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr03893e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |