Emerging SERS biosensors for the analysis of cells and extracellular vesicles
Mohammad
Tavakkoli Yaraki
 ,
Anastasiia
Tukova
,
Anastasiia
Tukova
 and
Yuling
Wang
and
Yuling
Wang
 *
*
School of Natural Sciences, Faculty of Science and Engineering, Macquarie University, Sydney, NSW 2109, Australia. E-mail: yuling.wang@mq.edu.au
First published on 19th September 2022
Abstract
Cells and their derived extracellular vesicles (EVs) or exosomes contain unique molecular signatures that could be used as biomarkers for the detection of severe diseases such as cancer, as well as monitoring the treatment response. Revealing these molecular signatures requires developing non-invasive ultrasensitive tools to enable single molecule/cell-level detection using a small volume of sample with low signal-to-noise ratio background and multiplex capability. Surface-enhanced Raman scattering (SERS) can address the current limitations in studying cells and EVs through two main mechanisms: plasmon-enhanced electric field (the so-called electromagnetic mechanism (EM)), and chemical mechanism (CM). In this review, we first highlight these two SERS mechanisms and then discuss the nanomaterials that have been used to develop SERS biosensors based on each of the aforementioned mechanisms as well as the combination of these two mechanisms in order to take advantage of the synergic effect between electromagnetic enhancement and chemical enhancement. Then, we review the recent advances in designing label-aided and label-free SERS biosensors in both colloidal and planar systems to investigate the surface biomarkers on cancer cells and their derived EVs. Finally, we discuss perspectives of emerging SERS biosensors in future biomedical applications. We believe this review article will thus appeal to researchers in the field of nanobiotechnology including material sciences, biosensors, and biomedical fields.
1. Introduction
Raman spectroscopy is a spectroscopic technique that is based on the inelastic scattering of photons that cause vibration of chemical bonds in the molecules, first predicted theoretically in 1923 by Smekal and co-workers1 and discovered by C.V. Raman in 1928, which led to a Nobel Prize in physics for Raman in 1930.2 The rich vibrational information on changes in composition, structure, or phase transitions in the sample provided by Raman spectroscopy has made it a unique approach in analytical chemistry. However, Raman signals for bulk (solid) or concentrated samples are not strong enough for further analysis. Therefore, development of new techniques to enhance the Raman signal for a sample at low concentration is essential for further applications, especially in the biomedical field.3–5Surface-enhanced Raman scattering (SERS), which was first discovered by Fleischmann et al.6 in 1974, is a technique in which the Raman signal of a molecule can be enhanced dramatically when placed in the vicinity (<10 nm) of some nanomaterials including plasmonic noble nanostructures, 2D nanomaterials, and semiconductors. In general, there are two main mechanisms for SERS: (i) the plasmon-enhancement mechanism, which is the dominant mechanism in noble metal-based SERS systems, and (ii) the chemical enhancement mechanism, which is the main mechanism in non-metal-based SERS systems and also occurs between molecules and the substrate. In the last decade, colloidal and planar SERS systems have found their place as strong analytical tools in biomedical field7–11 owing to their high sensitivity, which can be even at the single molecule level.12
Accurate analysis is imperative to inform health care providers in predicting the progression of a disease and implementing appropriate treatment. Biosensors can be used to observe and predict a patient's state of health by monitoring the presence of biomarkers in biological fluids such as blood, saliva and urine, tears, and cerebrospinal and synovial fluids.13–19 Extensive research has been done on the use of analytical methods to monitor biomarkers such as circulating tumour cells (CTC), specific proteins, circulating RNA and DNA, and small extracellular vehicles (EVs)20–26 to create fast, effective, and reliable assays and techniques to determine their activity and measure their concentration in patients’ samples.27–29 Among them, SERS-based systems have received tremendous attention owing to their high sensitivity and low signal-to-noise ratio background, as well as their non-invasive nature and requirement for a very low amount of sample.10,30–32 Additionally, SERS biosensors could enable multiplex detection of various analytes in the sample by one single measurement, owing to the unique distinct Raman peaks of Raman reporter molecules in the design of SERS nanotags. Plenty of excellent reviews have been recently published on this topic.33–35 However, it seems the chemical mechanism in SERS enhancement mechanism has been neglected, and no review specifically covers the use of SERS to analyse the surface biomarkers on cells and their EVs.
In this review, we first present an overview of SERS mechanisms with the focus on the chemical mechanism, which has been overlooked by many review articles. Then, we highlight different nanomaterials that have been used to design highly sensitive colloidal and planar SERS systems based on electromagnetic enhancement mechanism and chemical enhancement mechanism. Next, we discuss label-free and labelled SERS systems as two main approaches for the analysis of cancer cells and EVs, where various SERS systems based on plasmon enhancement and/or chemical enhancement have been designed to provide deeper insights into the molecular signatures of cells and EVs, and therefore for early detection or monitoring of the cancer treatment process. At the end, we propose our perspectives and future work using SERS as a sensitive tool for sensing cells and their EVs.
2. Overview of the SERS mechanism
The weak Raman scattering cross-section of molecules (10−28–10−30 cm2 per molecule) results in an inefficient Raman scattering, which has limited its application in real practice.36,37 SERS has solved this issue by enhancing the Raman signal of molecules placed in the vicinity of SERS nanomaterials (e.g., plasmonic nanostructures, semiconductors, etc.). Although the understanding of SERS enhancement has been one of the main topics in SERS studies for decades, debate about the mechanisms of SERS remains. Despite the many proposed models for SERS enhancement, there is still a need for a new SERS enhancement model to explain some complex systems. Fig. 1 shows the two common mechanisms that could be used to explain SERS enhancement in most systems based on different nanomaterials: (i) electromagnetic enhancement (Fig. 1a, i.e., plasmon enhancement); and (ii) chemical enhancement (Fig. 1b with the two concepts of Macroscopic, where the charge transfer between nanomaterial (metal) and molecule occurs, and Microscopic, where nanomaterial and molecule form a complex with a new electronic energy state). In this section, these two main mechanisms are explained and the effective parameters of these mechanisms are reviewed.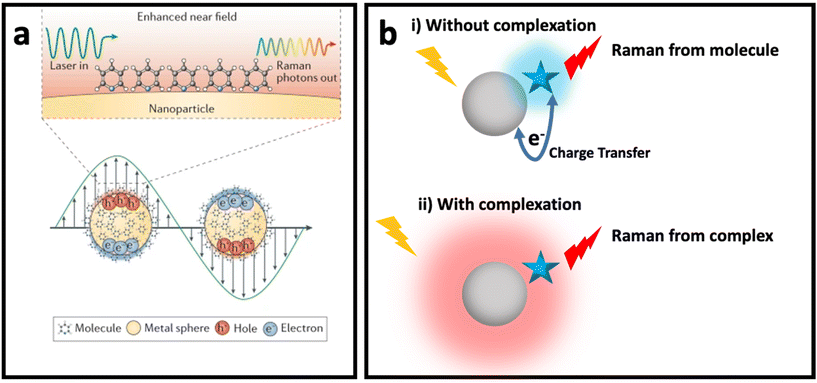 | ||
| Fig. 1 (a) Enhanced near field in a plasmonic nanomaterial due to localized surface plasmon resonance (LSPR) when it interacts with incident light with wavelength larger than its size (bottom) and SERS of a molecule placed on surface of the plasmonic nanomaterials due to electromagnetic enhancement mechanism (top). Adapted from ref. 38 with permission from Nature-Springer Group, copyright 2016. (b) Chemical enhancement mechanism with macroscopic (without complexation) and microscopic (with complexation and formation of new electronic state) point of view when a molecule (blue star) is adsorbed on surface of a nanomaterial (grey sphere). | ||
2.1. Electromagnetic enhancement mechanism
Electromagnetic enhancement is the result of localized surface plasmon resonance (LSPR). When light (i.e., electromagnetic wave) interacts with a metal or semiconductor nanostructure, oscillation of electrons at the surface of the metal or semiconductor nanostructure results in an enhanced electric field in the space surrounding the nanostructure, which can modulate the optical properties of molecules placed in the vicinity of the surface of the nanostructure, including Raman scattering.7Noble metal (i.e., Au, Ag, Pd, Pt, Cu) nanostructures have free electrons at their surface which lead to a considerably enhanced electric field in the surrounding environment, while the LSPR, and consequently the enhanced electric field under visible range excitation in semiconductors, is weak, since it depends on the electron density of the conduction band (CB) and a large electron density (1022–1024 cm−3) of the valence band (VB).39 Therefore, the electromagnetic enhancement mechanism usually refers to noble metal-based SERS systems. Mie theory explains that when incident light with a certain wavelength is much greater than the size of nano-object, incident light interacts with a metal nanosphere, and the extinction cross-section in the nanosphere can be explained by the following equation:
 | (1) |
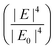 . The enhanced electric field around metal nanostructures possesses short decay (10–30 nm in sphere and <10 nm in anisotropic nanoparticles with sharp edges and corners) (Fig. 2a), but when a dramatically enhanced electric field occurs in the space between the particles, this is called a “hot-spot” (Fig. 2b).43–45,46 Therefore, the SERS enhancement factor is roughly proportional to d−12, where d is the distance between the molecule and the surface of the metal nanostructure.5 The enhanced electric field is size and shape dependent, and the material of the nanoparticle strongly affects the enhanced electric field. Therefore, appropriately matching the shape, size, and material of the nanoparticle with the excitation wavelength of the Raman scattering is of great importance for SERS applications. Fig. 2c shows how tuning the size of silver nanocubes affects the average enhanced electric field on the surface of these nanostructures under a common laser with the excitation wavelength. Although it was mentioned that semiconductors do not have strong LSPR, and therefore they do not have a strong enhanced electric field in the visible region, some attempts have been made to shift the LSPR of semiconductor-based SERS systems to the visible range by optimizing the size and geometry configurations as well as defect engineering in the semiconductor nanomaterials. It should be taken into account that the LSPR peak is a function of the morphology (size and shape) and the component of the nanoparticle. There is an optimal size of metallic nanoparticles to achieve the strongest SERS enhancement. Small nanoparticles (<10 nm) have an extreme surface damping, while large nanoparticles have a great radiation damping (>160 nm).47 It is important to note that for a different excitation wavelength the choice of optimal nanoparticle size for maximum SERS enhancement will vary. Surface plasmons are also strongly influenced by the shape of nanoparticles. For example, gold nanorods have two resonance modes at different wavelengths corresponding to the longitudinal and transverse direction of plasmon oscillation.48 Adjusting the aspect ratio of these nanoparticles would result in shifting of the LSPR peak, and as a result in different SERS enhancement. LSPR and SERS enhancement also depend on the surrounding environment. For example, Li et al. have investigated the optimal size of gold nanoparticles for SERS under different conditions.49 They found that under the same concentration and total surface area of gold nanoparticles the optimal size for the maximum SERS enhancement among the particles from 17 to 80 nm is 50 nm. SERS also depends on the component of NPs. Although Ag exhibits high plasmonic quality in the visible to near-infrared range and enhances strong Raman scattering, chemical stability and toxicity are two major concerns for its biomedical applications. Au exhibits SERS enhancement comparable to Ag in the long wavelength range (usually above 600 nm), but gold is widely used due to its high chemical stability and biocompatibility.
. The enhanced electric field around metal nanostructures possesses short decay (10–30 nm in sphere and <10 nm in anisotropic nanoparticles with sharp edges and corners) (Fig. 2a), but when a dramatically enhanced electric field occurs in the space between the particles, this is called a “hot-spot” (Fig. 2b).43–45,46 Therefore, the SERS enhancement factor is roughly proportional to d−12, where d is the distance between the molecule and the surface of the metal nanostructure.5 The enhanced electric field is size and shape dependent, and the material of the nanoparticle strongly affects the enhanced electric field. Therefore, appropriately matching the shape, size, and material of the nanoparticle with the excitation wavelength of the Raman scattering is of great importance for SERS applications. Fig. 2c shows how tuning the size of silver nanocubes affects the average enhanced electric field on the surface of these nanostructures under a common laser with the excitation wavelength. Although it was mentioned that semiconductors do not have strong LSPR, and therefore they do not have a strong enhanced electric field in the visible region, some attempts have been made to shift the LSPR of semiconductor-based SERS systems to the visible range by optimizing the size and geometry configurations as well as defect engineering in the semiconductor nanomaterials. It should be taken into account that the LSPR peak is a function of the morphology (size and shape) and the component of the nanoparticle. There is an optimal size of metallic nanoparticles to achieve the strongest SERS enhancement. Small nanoparticles (<10 nm) have an extreme surface damping, while large nanoparticles have a great radiation damping (>160 nm).47 It is important to note that for a different excitation wavelength the choice of optimal nanoparticle size for maximum SERS enhancement will vary. Surface plasmons are also strongly influenced by the shape of nanoparticles. For example, gold nanorods have two resonance modes at different wavelengths corresponding to the longitudinal and transverse direction of plasmon oscillation.48 Adjusting the aspect ratio of these nanoparticles would result in shifting of the LSPR peak, and as a result in different SERS enhancement. LSPR and SERS enhancement also depend on the surrounding environment. For example, Li et al. have investigated the optimal size of gold nanoparticles for SERS under different conditions.49 They found that under the same concentration and total surface area of gold nanoparticles the optimal size for the maximum SERS enhancement among the particles from 17 to 80 nm is 50 nm. SERS also depends on the component of NPs. Although Ag exhibits high plasmonic quality in the visible to near-infrared range and enhances strong Raman scattering, chemical stability and toxicity are two major concerns for its biomedical applications. Au exhibits SERS enhancement comparable to Ag in the long wavelength range (usually above 600 nm), but gold is widely used due to its high chemical stability and biocompatibility.
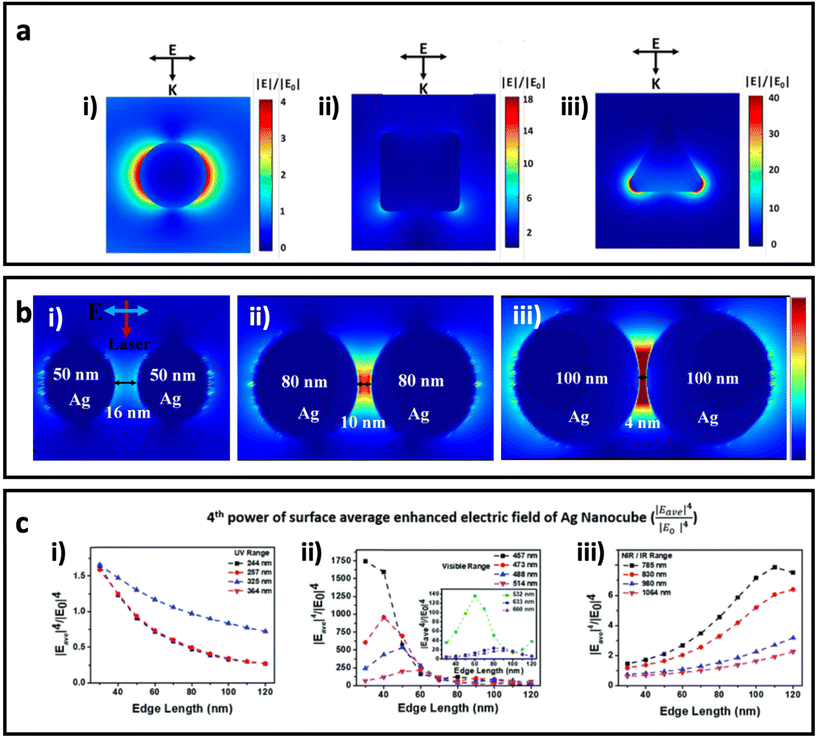 | ||
| Fig. 2 (a) Enhanced electric field distribution around a 130 nm Ag sphere (i), 84 nm Ag nanocube (ii) and 33 nm Ag triangle nanoplate with 7 nm thickness (iii) under excitation wavelength of 550 nm, which is well matched with the maximum LSPR of each nanoparticle. Adapted from ref. 50 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2020. (b) The enhanced electric field between AgNPs with different sizes and gaps under 633 nm incident light, adapted from ref. 46 with permission from Nature-Springer Group, copyright 2019. (c) The fourth power of surface average enhanced electric field (|Eave|4/|E0|4 on Ag nanocubes with different sizes (based on edge length) under a common Raman laser with the excitation wavelength in the (i) UV region (200–400 nm), (ii) visible region (400–700 nm), and (iii) NIR/IR region (700–1100 nm). Adapted from ref. 45 with permission from the Royal Society of Chemistry, copyright 2020. K is the wave vector, and E is the incident polarization. | ||
2.2. Chemical enhancement mechanism
Although electromagnetic enhancement usually occurs in metal nanostructure-based SERS systems and is the dominant mechanism, chemical enhancement is the dominant mechanism when a molecule locates on the surface of a semiconductor (metal oxides) or non-metal nanostructure (carbon nanomaterials).9,51 In chemical enhancement, the charge transfer process occurs between the metal or non-metal substrate and adsorbed molecule (Fig. 3a). This charge transfer process can happen in two different ways depending on the electronic states of the nanomaterial (SERS substrate) and the adsorbed molecule.52–54 First, the substrate and molecule may form a complex that has a new energy state, where the resonance of such a complex could enhance the Raman scattering under excitation by incident light. Second, there is no complex state, but the Fermi level of the nanomaterial substrate could match the highest occupied molecular orbital (HOMO) or the lowest unoccupied molecular orbital (LUMO) of the molecule. In this case, the electron can be transferred from the HOMO of the molecule to the unoccupied state above the Fermi level of the nanomaterial substrate, or from the LUMO of the molecule to the occupied state below the Fermi level of the nanomaterial substrate. A third possible pathway is excitation resonance, which is different from charge transfer system and happens when excitons (i.e., electron–hole pairs) are being generated as a result of electron transition from valence band to conductive band. When the size of nanoscale semiconductor is smaller than the distance between hole and pair (i.e., Bohr radius), the bandgap increases. If the wavelength of incident light is matched with the bandgap energy, the SERS performance of semiconductor nanoparticles through the chemical enhancement effect (i.e., exciton resonance) can be improved.55,56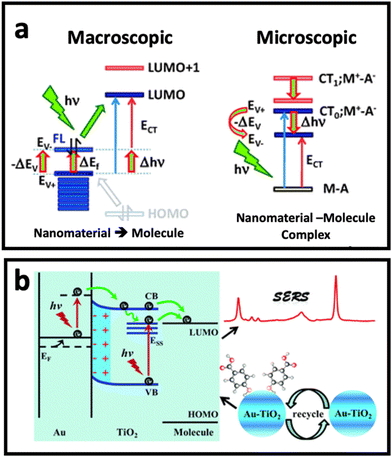 | ||
| Fig. 3 (a) Chemical enhancement mechanism based on resonant charge transfer model with macroscopic and microscopic point of view when a molecule is adsorbed (A: adsorbate) on surface of nanomaterial (in this case M: metal). Adapted from ref. 68 with permission from the Royal Society of Chemistry, copyright 2011. (b) Schematic diagram of the SERS enhancement mechanism of 4-mercaptobenzoic acid (4-MBA) molecules adsorbed on Au–TiO2 nanocomposite. Adapted from ref. 65 with permission from the Royal Society of Chemistry, copyright 2017. | ||
2.3. Synergic effect between electromagnetic enhancement and chemical enhancement mechanisms
The combination of the two mechanisms, taking advantage of the synergic effect between the two well-known mechanisms, in developing SERS substrates has been one of the hot topics in this research area. While surface defect-engineered semiconductors have been used as potential SERS substrates for some biomedical applications,57–59 it seems that the application of semiconductor nanomaterials in bioanalysis needs further investigation to solve the practical challenges that researchers face in biomedical applications, in particular in the analysis of cells and extracellular vesicles. Therefore, the combination of noble metal nanostructures and semiconductor nanomaterials has received tremendous attention in the design of SERS substrates with stronger SERS effect compared with noble metal-based substrates or semiconductor-based substrates alone.60–64 In such a composite, the presence of a noble metal results in stronger light absorption in the visible to near-infrared (NIR) region, depending on the size/shape/material of the noble metal nanostructure. This can modify the light absorption of the composite and lead to stronger excitation under Raman laser irradiation. On the other hand, since semiconductors have a generally higher Fermi level than noble metal nanostructures, the hole–electron separation effect in a semiconductor can be promoted by the presence of the metal nanostructure, and therefore the charge transfer efficiency of the semiconductor can be improved. Fig. 3b depicts how the combination of Au and TiO2 nanostructures could enhance the Raman signal. In this work, Jiang et al.65 deposited Au ions on TiO2 nanoparticles by a photocatalytic reduction approach, where it was found that a synergic effect between deposited Au and TiO2 nanoparticles plays an important role in the enhanced charge transfer from semiconductor to the adsorbed 4-mercaptobenzoic acid molecules, and in turn, the SERS effect of the nanocomposite. The synergistic effect between electromagnetic mechanism and chemical enhancement is not limited to metal–semiconductor substrates. Recently, MXenes made of Ti3C2, Nb2C, and Ta2C have shown up to 106-fold SERS enhancement factor, which was due to a combination of both enhanced electric field around the MXene structures and charge transfer between MXene and Raman molecule.66,67 We believe these nanomaterials have great potential for the analysis of surface biomarkers on cells or extracellular vesicles in the near future.To date, the potential of a wide range of nanomaterials for SERS enhancement has been investigated. Plasmonic noble nanostructures have shown promising SERS enhancement effects in both colloidal and planar systems through the electromagnetic enhancement (i.e., plasmon enhancement) mechanism. While chemical enhancement has also been found in metals, it is suppressed by the electromagnetic enhancement mechanism. However, in non-plasmonic nanomaterials such as semiconductors and 2D carbon nanostructures, the chemical enhancement effect is the dominant mechanism, and mainly occurs through the charge transfer process. Combination of these nanomaterials provides an opportunity to take advantage of the synergic effect between metal and non-metal nanomaterials, and to obtain dramatic SERS enhancement. In the following section, we discuss the nanomaterials that contribute to SERS enhancement via these mechanisms.
3. Nanomaterials for SERS
3.1. Nanomaterials for the plasmon enhancement effect
Noble metal nanostructures are the key nanomaterials that have been used for designing SERS systems in both colloidal and planar systems owing to their LSPR effect, and in turn, the dramatically enhanced electric field in their surrounding environment. As discussed earlier, the Raman signal of the molecule can be modulated when it is placed in this enhanced electric field area (i.e., typically the molecule is adsorbed on surface of metal nanostructure), which is proportional to the fourth power of the enhanced electric field. Among various noble metals, gold and silver have received more attention compared with other plasmonic metals (i.e., Cu, Pd, Pt), owing to their well-known size and shape-tunable chemical synthesis routes (i.e., bottom-up approaches), as well as the fewer technical challenges in the fabrication of nanostructures via top-down approaches (e.g., electrodeposition). In this section, we highlight nanomaterials that have been used for designing colloidal and planar SERS systems based on the plasmon enhancement mechanism.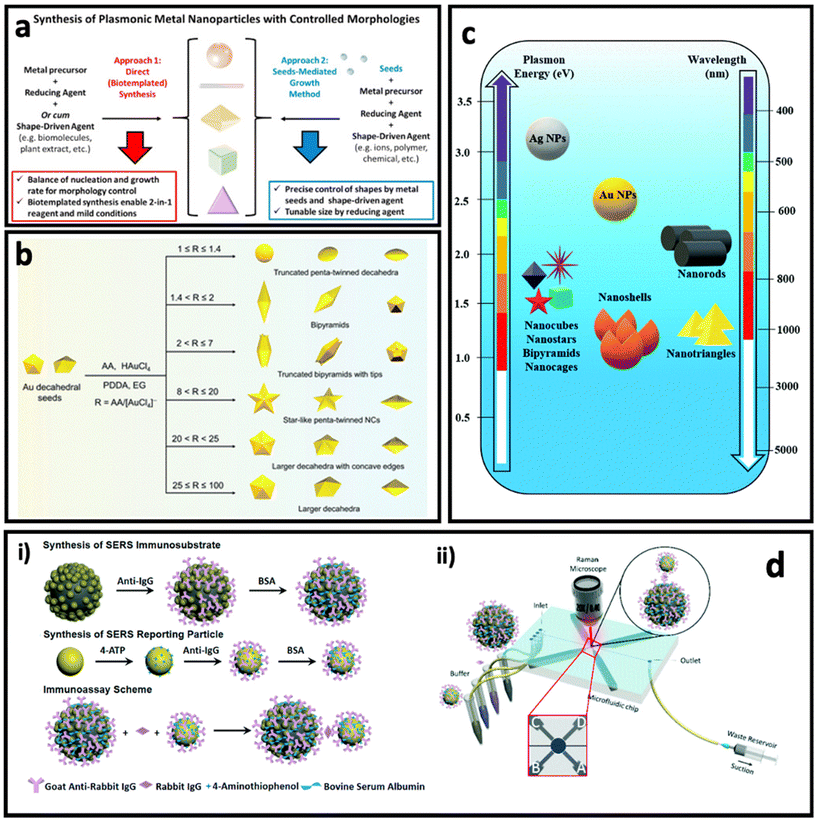 | ||
| Fig. 4 (a) Controlling the morphologies of metal nanoparticles via two main approaches: direct (seedless) synthesis (left) and seed-mediated growth method (right). Adapted from ref. 74 with permission from Elsevier, copyright 2020. (b) Controlled synthesis of penta-twinned Au nanocrystals by tuning the ascorbic acid (AA)/HAuCl4 ratio. Adapted from ref. 75 with permission from the Royal Society of Chemistry, copyright 2021. (c) Plasmon resonance and maximum LSPR peak of differently shaped gold (Au) and silver (Ag) nanoparticles (NPs). Adapted from ref. 76 with permission from the Royal Society of Chemistry, copyright 2021. (d) Synthesis of Fe3O4@AuNP@Ag as magnetic SERS nanotag (i) and its application in a microfluidics device for detection and quantification of immunoglobulin G. Adapted from ref. 77 with permission from the Royal Society of Chemistry, copyright 2017. | ||
In direct synthesis (Fig. 4a, left), a combination of mild reducing agent and shape-driving agent can lead to the formation of anisotropic nanoparticles, where bio-templated/biomolecules-based methods can satisfy these requirements. Biomolecules such as peptides, proteins and nucleic acids, proteins and peptides as well as plant extracts containing bioactive compounds could be used for the synthesis of anisotropic nanoparticles, due to the simultaneous presence of various sequences (i.e., nucleotides or amino acids) or chemical structures.78–80 For example, Tan et al.81,82 have reported the synthesis of triangular Au nanoplates using polypeptide as simultaneous reducing agent, shape-directing agent and capping agent under mild synthesis conditions such as neutral pH, room temperature and in aqueous solution. The as-synthesized nanostructures through these routes are fully covered with functional groups that enable further surface functionalization easily and provide high biocompatibility, which are essential for biomedical applications.83–85 In another study, Wang's group reported the seedless synthesis of spiky Au nanostars using ascorbic acid as reducing agent and capping agent in the presence of a trace amount of Ag+ ions as shape-directing agent, which showed a great SERS enhancement.86 In another work, using the same precursors, and just by changing the ratio between chemicals and synthesis conditions, Au–Ag alloy nanoboxes with a relatively smooth surface were synthesized.87
On the other hand, the seed-mediated growth approach (Fig. 4a, right) allows the manipulation of the shape and size of the anisotropic nanoparticles by introducing different amounts of seeds (i.e., usually small-sized nanoparticles with a size between 5 and 20 nm) as well as a shape-directing agent such as polyvinylpyrrolidone (PVP),88 poly(diallyldimethylammonium) chloride (PDDA),75 cetyltrimethylammonium bromide (CTAB),89 and a trace amount of metal ions (Ag+, Fe2+, etc.).50,90 The shape-directing agents usually have specific binding affinity for certain facets (e.g., {100}, {111}) in the crystal structure of the seeds, leading to reducing of metal ions and growth of the nanoparticle in specific directions. A careful control of the ratio between the reagents in the synthesis can result in anisotropic nanoparticles with well-defined shapes. For example, Ag bipyramids have been synthesized in the presence of CTAB, ascorbic acid and Cu2+ ions, where CTAB was specific to {100} facets of Ag seeds, and the combination of ascorbic acid and Cu2+ ions produced H2O2 that could etch the {110} and {111} facets on Ag seeds.50 Recently, Zhang et al.75 reported the synthesis of penta-twinned Au nanocrystals using Au decahedral seeds (Fig. 4b). Au decahedral seeds were first synthesized using ethylene glycol as solvent, ascorbic acid as reducing agent and PDDA as shape-directing agent at 220 °C. Then, the seeds were used for the synthesis of various penta-twinned nanocrystals (truncated penta-twinned decahedra, truncated bipyramids, bipyramids, truncated bipyramids with tips, star-like penta-twinned nanocrystals, decahedra with concave edges, and decahedra) just by tuning the ascorbic acid/HAuCl4 ratio in the presence of Ag+ ions. As can be seen in Fig. 4c, different shapes of gold and silver nanoparticles have different LSPR peaks, and therefore the SERS enhancement factor is maximum when the laser wavelength is well matched with the LSPR peak of the nanoparticle.
One of the interesting approaches to improve the SERS intensity in colloidal SERS nanotags is aggregation-induced SERS.91,92 In this approach, the SERS intensity increases dramatically due to formation of hot-spots between the SERS nanotags upon addition of the analyte.93 For example, Liu et al. have used porous Au@ZnS multi-yolk–shell structures derived from Au@ZIF-8 as SERS nanotags, where introducing the electrolyte resulted in a dramatic enhancement of the SERS intensity of these nanoparticles due to aggregation of them.94 This approach has also been used as the basis of a lot of magnetic bead–SERS nanotags biosensors. In this approach, both metal SERS nanotags and magnetic beads are functionalised by recognition elements that can interact with analyte molecules, resulting in the formation of magnetic bead/metal nanoparticles in the form of core/satellite shape. The metal nanoparticles (as satellites) in this structure become close enough to each other that many hot-spots are formed between them. In another method with a similar concept, magnetic SERS nanotags made of a magnetic core and grown metal nanoparticles on the surface can be designed.95 For example, Yap et al.77 designed Fe3O4@AuNP@Ag as magnetic SERS nanotags that were used to quantify immunoglobulin G protein using a microfluidic device (Fig. 4d). The external magnetic field causes the aggregation of the SERS nanotags, leading to a dramatic enhancement in SERS intensity.
The assembly of metal nanoparticles onto the substrate can be performed via electrostatic adsorption, a simple casting/drying process, solvent-mediated, sputtering, or even chemical binding.10 All these approaches usually result in a layer of metal nanoparticles on the surface of the substrate, where hot spots can be formed in the space between the metal nanoparticles.100 Another approach to make SERS substrate using metal nanoparticles is by combining metal nanoparticles with polymers and casting the polymer solution as a thin film. The performance of the metal nanoparticle-based SERS substrate strongly depends on the surface coverage and shape of the metal nanoparticles. Solís et al.96 have investigated the effect of surface coverage of spherical AuNPs, Au nanorods, and spiky Au nanostars on a glass substrate. As can be seen in Fig. 5a, the SERS enhancement factor is more sensitive to surface coverage in spherical nanoparticles than anisotropic nanoparticles, where the spiky Au nanostars showed less sensitivity. Additionally, the Au nanostars led to at least 1 order of magnitude higher SERS enhancement factor compared with spherical Au nanoparticles, which could be due to formation of stronger “hot-spots” between the sharp tips of the Au nanostars. In general, deposition of metal nanoparticles on substrates is an easy and cheap process; however, reproducibility of the quality of these SERS substrates should be considered in real biomedical applications.
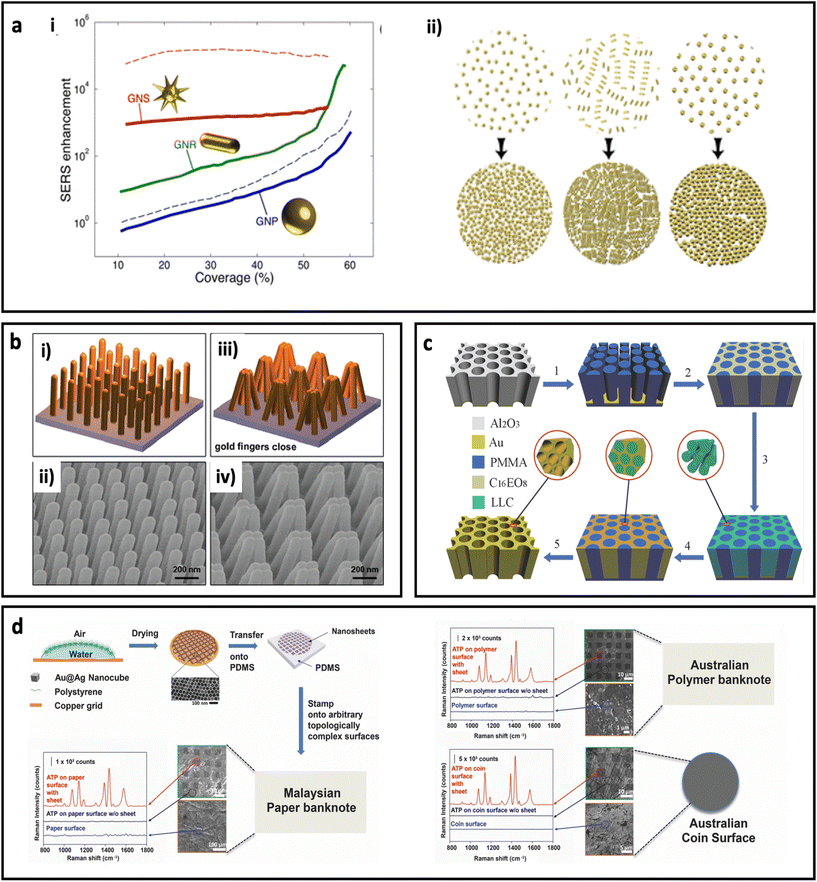 | ||
| Fig. 5 (a) Effect of surface coverage on SERS enhancement in a metal nanoparticle-based substrate (i) and schematic illustration of change in density of nanoparticles with increasing the deposition time. Au nanoparticles (GNPs, 51 nm diameter), Au nanorods (GNRs, 65 nm length, 21 nm diameter), and Au nanostars (GNSs, core with 20 nm diameter and 10 branches 15.5 nm long with tip apexes of 1 nm), solid curves for 785 nm excitation laser wavelength and dash curves for 633 nm excitation laser wavelength. Adapted from ref. 96 with permission from the American Chemical Society, copyright 2017. (b) Schematic illustration of plasmonic nanofinger and its SEM image before addition of analyte (i and ii) and after drying of analyte (iii, and iv). Adapted from ref. 97 with permission from the American Chemical Society, copyright 2010. (c) Schematic illustration of different steps for preparation of 3D hierarchically porous gold nanostructure. Adapted from ref. 98 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2015. (d) Schematic illustration of fabrication procedure for adhesive plasmonic SERS substrate using Au@Ag nanocube and polydimethylsiloxane (PDMS) (top left) and application of the fabricated SERS substrates on Malaysian paper banknote (bottom left), Australian polymer banknote and Australian coin surface (right). 4-Aminothiophenol (ATP) was used as Raman reporter molecule. Adapted from ref. 99 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2015. | ||
Deposition of metal atoms on the surface of the substrate using atomic electrodeposition equipment through a lithographic approach or using a template is another technique that enables researchers to investigate the performances of SERS substrates with unique, well-defined structures for both in-plane and out-of-plane vertically stacked plasmonic nanogaps. For example, Hu et al.97 have fabricated a flexible polymeric nanofinger coated with a thin layer of Au. As shown in Fig. 5b, the presence of the analyte molecule can bring the plasmonic nanofingers close to each other as result of capillary forces during the drying of the sample. This results in a dramatically enhanced electric field between the tips and, therefore, high SERS sensitivity. In another work, a dual-templating technique has been used to fabricate a 3D hierarchically porous gold nanostructure, by Zhang et al.98 In this approach (Fig. 5c), porous poly(methyl methacrylate) (PMMA) and octaethylene glycol monohexadecyl ether (C16EO8) were used as hard template and lyotropic liquid crystal (LLC), respectively. A porous anodic aluminium oxide (AAO) template was used to prepare a negative PMMA template, followed by formation of LLC and, finally, electrodeposition of gold. The synthesized nano-holes could harvest the incident light and therefore generate an electric field enhancement close to the edges of the nano-holes. The fabricated substrate showed considerable reproducibility and sensitivity for detection of benzenethiol at ultralow concentrations (0.1 pM). Although this approach provides unique SERS substrates at laboratory scale, the cost and time for the preparation of this type of SERS-active substrate should be taken into account when the scaling up of this type of SERS substrate is required. In another study, an ultrathin plasmene nanosheet was fabricated by stamping Au@Ag nanocubes on different substrates including paper banknote, polymer banknote and coin (Fig. 5d). The stamped nanostructures have shown dramatic SERS enhancement for 4-ATP molecules (as model Raman molecule). Careful tuning of the morphology of the nanoparticles in this approach could be used as a method to engineer the SERS performance.99 The self-assembly of nanoparticles on a flexible planar substrate has been reported in many studies.101,102 Huang's group has reported a new fabrication method based on tailoring the contact angle of nanoparticles to assemble the nanoparticles instantly (within 5 s) on silicon wafer.103 In another study, the same group has reported controlling the SERS intensity of a planar system (containing a monolayer of gold nanoparticles) by adjusting the shrinkage of the substrate through light. In this design, the substrate was made by photopolymerization of acrylamide, which undergoes the photopolymerization process under irradiation of UV light.101 This polymerization led to shrinkage of the substrate, and therefore the formation of hot-spots between nanoparticles that resulted in higher SERS intensity.104
3.2. Nanomaterials for the chemical enhancement effect
Although nanomaterials that have been used for developing SERS systems with the chemical enhancement effect are mostly based on inorganic semiconductors and carbon-based nanostructures (Table 1), metal nanoparticles have also shown the chemical enhancement effect when the adsorbed molecules experience the charge transfer to the metal surface. Since the plasmon effect is the dominant mechanism in metal nanoparticles, in this section, we only discuss the non-metallic nanostructures that have been used for designing SERS systems with the chemical enhancement effect. The first observation of SERS on TiO2 and NiO by Yamada et al.105,106 can be traced back to 1982, when they discovered the enhancement in Raman spectra of chemisorbed pyridine on the smooth surface of NiO-coated glass substrate. Since then, the research on using semiconductor nanomaterials as Raman enhancers has witnessed much progress, and various types of nanomaterials including metal oxides, single-element semiconductors, transition metal dichalcogenides, and organic semiconductors have been found as potential SERS substrates. In the following sections, we briefly review these nanomaterials and their applications as SERS substrates through the chemical enhancement mechanism.| Nanomaterial | Raman molecule | EF | Ref. |
|---|---|---|---|
| TiN | R6G | 1.0 × 105 | 117 |
| VO2 | R6G | 6.7 × 107 | 118 |
| d-MoN | R6G | 8.16 × 106 | 119 |
| TiO2nanoparticles | 4-MBA | 1 × 102 | 113 |
| MoO2 | R6G | 3.75 × 106 | 120 |
| h-MoO3 | MB | 6.99 × 105 | 121 |
| ZnO nanocrystal | 4-MPy | 103 | 122 |
| CuO nanocrystal | 4-MPy | 102 | 123 |
| Amorphous Rh3S6 | 4-NBT | 3 × 104 | 124 |
| Amorphous ZnO nanocages | 4-MBA | 6.62 × 105 | 125 |
| Amorphous TiO2nanosheets | 4-MBA | 1.86 × 106 | 126 |
| CdS nanoparticles | 4-MPy | 102 | 127 |
| N-Doped graphite | CV | 3.2 × 107 | 128 |
| Ozone-treated graphene | RhB | 2.5 × 104 | 129 |
| Black TiO2 | R6G | 1.2 × 106 | 112 |
| MoS2xOx | R6G | 1.4 × 105 | 62 |
| WO3 | 4MBA | 10 | 130 |
| ZnO1−x | R6G | 1.1 × 104 | 131 |
| WO3−x | R6G | 1.1 × 104 | 131 |
| Ga-doped ZnO nanoparticles | 4-MPy | 6.66 × 104 | 132 |
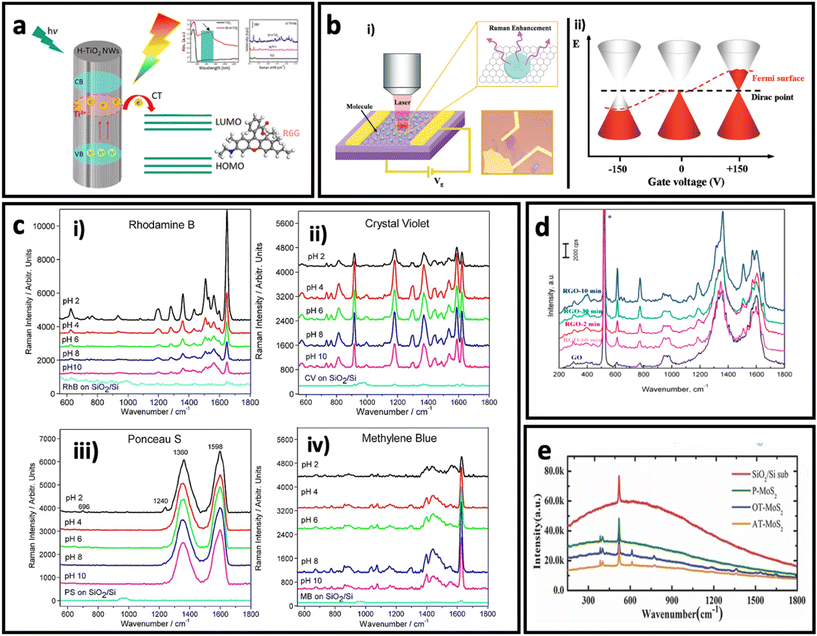 | ||
| Fig. 6 (a) Schematic illustration of charge transfer process in hydrogenated TiO2 nanowires to rhodamine 6G and its effect on SERS intensity of rhodamine 6G. Adapted from ref. 112 with permission from the American Chemical Society, copyright 2018. (b). Optical image of graphene and schematic illustration the setup (i), and effect of electrical field on Fermi level in a monolayer graphene (ii). Adapted from ref. 147 with permission from the American Chemical Society copyright 2011. (c) Effect of pH on SERS intensity of rhodamine B, 20 μM (i), crystal violet, 10 μM (ii), Ponceau S, 20 μM (iii), and methylene blue, 10 μM (iv). Adapted from ref. 161 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2013. (d) Effect of reduction time on SERS effect of reduced graphene oxide. Adapted from ref. 150 with permission from Elsevier, copyright 2016. (e) Raman spectra of R6G dye on different substrates: argon-plasma treated MoS2 (AT-MoS2), oxygen-plasma treated MoS2 (OT-MoS2), pristine MoS2 (P-MoS2), and SiO2/Si, adapted from ref. 160 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2014. | ||
Since the SERS performance is usually weak (enhancement factor between 102–105) in pure semiconductors,113,122,123,127 there are different strategies to engineer the bandgap of semiconductors, and in turn, improve their SERS effect through the chemical enhancement mechanism. Doping the structure of a semiconductor using a trace amount of another metal/non-metal is an effective strategy to engineer the bandgap of semiconductors. For example, Yang et al.59 have reported the synthesis of Mo-doped Ta2O5 nanowires that were used to make a SERS substrate. The fabricated SERS substrate exhibited a remarkable SERS sensitivity with an enhancement factor of 2.2 × 107 using methyl violet (MV). This value is 1–2 orders of magnitude larger than the enhancement factor reported for other metal oxides.133,134 Another approach is defect engineering by producing oxygen vacancies in the structure of semiconductors.135–137 Various methods such as lithium metal grinding reduction, irradiation, and high-temperature annealing under a reducing gas atmosphere have been employed to generate oxygen vacancies in the structure of semiconductors.138–141 For example, Cao et al.140 have successfully synthesized MoO2−x with a polycrystalline surface by treating MoO2 with 6 wt% Li. The as-synthesized MoO2−x showed better SERS performance compared with MoO2, which could be attributed to the produced oxygen vacancies on the surface of the semiconductor, confirmed by X-ray photoelectron spectroscopy.
Besides the graphene, the derivatives of graphene such as graphene oxide (GO) and reduced-graphene oxide (rGO) have also shown the SERS effect via the chemical enhancement mechanism. GO and rGO have high affinity to adsorb molecules with aromatic rings, good water dispersibility, and fluorescence-quenching capability, making them interesting candidates for SERS study. Since the pH determines the adsorption of organic molecules on the surface of GO via electrostatic interactions, a pH-dependent SERS effect has been observed for various molecules on GO (Fig. 6c). In rGO, Yin et al.150 found that the degree of reduction can determine the SERS effect. Fig. 6d shows that rGO has a better SERS effect compared with graphene, and 10 min reduction time results in maximum SERS effect when R6G molecules are adsorbed on surface of rGO.
3.3. Nanomaterials for plasmon/chemical enhancement synergic effect
In order to take advantage of both plasmon-enhancement effect and chemical enhancement effect, plasmonic nanomaterials are being combined with non-plasmonic nanomaterials (i.e., semiconductors, 2D nanomaterials, etc.) and are being used to prepare SERS substrates. This combination not only provides higher SERS enhancement through coupling the plasmon-enhancement and chemical enhancement mechanisms, but also protects the metallic part (in particular silver) from oxidation. For example, graphene and other 2D nanomaterials have extensively been used to develop AgNP/graphene-based SERS substrates to obtain high-performance SERS systems.162–165 The coupling between these two mechanisms can enhance the SERS effect through different pathways:60 (i) the metal nanostructure and semiconductor each plays its own role in the enhancement of the Raman signal through the plasmon-enhancement mechanism and chemical enhancement mechanism, respectively; (ii) the charge transfer rate increases due to the enhanced excitation rate in the semiconductor nanomaterial as a result of the enhanced electric field in the surrounding environment; (iii) the charge transfer rate in the semiconductor increases due to pumping electrons from noble metal to semiconductor (Fig. 7a); and (iv) distribution of the enhanced electric field can be changed and affect the Raman intensity of the adsorbed molecules. In particular, it has been reported that the enhanced electric field in between the metal islands which is usually maximum between the metal islands will shift to the surface of the metal islands in the presence of monolayered graphene. Additionally, the enhanced electric field region can penetrate into the graphene layer.166 In addition to the better SERS performance, the metal–semiconductor-based substrates possess reproducibility owing to the self-cleaning property that comes from the improved photoinduced catalytic properties of hybrid nanostructures. All of these properties suggest that the hybrid nanostructure is favourable for designing SERS substrates. Fig. 7b represents how metal, semiconductor and molecule could be in contact with each other in different SERS system designs.60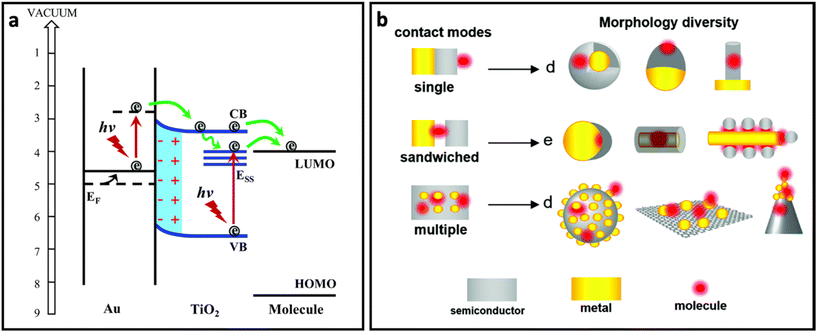 | ||
| Fig. 7 (a) The role of noble metal (Au) in pumping electrons to semiconductor (TiO2) and enhanced charge transfer from semiconductor to molecule, adapted from ref. 65 with permission from the Royal Society of Chemistry, copyright 2017. (b) Different contact modes for heterostructures containing metal (yellow), molecule (red) and semiconductor (grey) and morphology diversity for each contact mode, adapted from ref. 60 with permission from the Royal Society of Chemistry, copyright 2021. | ||
4. SERS analysis of biomarkers on cells and EVs
Traditional cancer diagnostics methods involve tissue biopsy, histopathological studies and imaging (with endoscope, MRI and others). Early detection of cancer is problematic due to the limited availability of clinical assessments for the general public.167 For most types of cancer, the above traditional diagnostics techniques are invasive and require the involvement of qualified professionals and expensive machinery, which makes cancer screening less available for patients. The limited availability of cancer screening leads to late diagnosis.168,169 As well as availability, in most cases these techniques are able to identify potentially dangerous tumours only when the number of tumour cells is large enough to cause an extensive release of biomarkers or metastasis has started.170As an analytical technique, SERS is a powerful tool for ultrasensitive analysis of small amounts of analyte, making it stand out among other techniques that require a larger amount of sample for biomarker detection. From this perspective, SERS can detect a single molecule at the designed condition.171 SERS-based techniques have been extensively used for the detection and discrimination of biomarkers.172–178 As discussed in sections 2 and 3 about the SERS enhancement mechanisms and substrates (nanomaterials), SERS is capable of providing a unique spectral profile for molecules on the SERS substrate – so-called “molecular fingerprints”.31,179 Additionally, the detection of cancer biomarkers with SERS is not invasive, which lowers the risk for patients and eases the analysis performance.180–182
Using the as-discussed SERS nanomaterials, the SERS-based analysis and detection of biomarkers on cancer cells and small extracellular vehicles (EVs) or exosomes can be classified into two main approaches: label-aided and label-free.76
Label-aided SERS analysis is an indirect sensing of the analyte based on the use of SERS tags, which consist of SERS-active nanoparticles functionalized with Raman-active or chromophore-based dyes that produce high-intensity and unique spectral bands/peaks. Most common label-aided detection methods utilise SERS tags that are bio-conjugated with targeting ligands, such as antibodies, to specifically target the analyte. This approach selectively identifies the target by monitoring the Raman spectrum from the SERS tag.183 Due to the high sensitivity of this method, label-aided SERS platforms have been widely utilised in the detection of biomarkers on cancer cells and EVs.15,16,184–187
Label-free SERS detects the intrinsic Raman spectrum of the analyte. This method allows for the identification of molecular fingerprint signals that originate from the biomolecules in close proximity to the SERS platform (i.e. from the surface of the cells or EVs). This approach is simple and easy to operate, and can provide information about analytes that are difficult to differentiate by targeting a specific biomarker.188,189 However, due to the complexity of biological systems, label-free SERS normally generates signals from all the molecules (known or unknown biomarkers) around the SERS substrate, as well as other molecules in the environment.188 Thus, multi-variant statistical analysis methods are usually required to enable the identification of target biomarkers on cells and EVs.
In this section we discuss recent advances in SERS platform designs with plasmon and chemical enhancement effects for the analysis of cells and EVs based on two major approaches: label-aided and label-free SERS.
4.1. Label-aided SERS
There are a large variety of SERS tags with different chemical and structural properties reported in the literature.183,189 Typically, the SERS tags’ design would include nanoparticles either metallic or non-metallic NPs as the core, coated with the Raman reporter molecule (which serves as label) and targeting molecules (Fig. 8).183,189–191 The rest of the design aims for high signal ratios to enable lower detection limits and specific targeting for reliable analysis and reproducible results. Development of label-SERS systems requires the conjugation of Raman reporter molecules to the surface of metal nanoparticles. Fig. 9 represents the chemical structure and corresponding Raman spectra of the molecules that are usually used as label or reporter in designing SERS nanotags. These molecules can be chemically attached to the surface of metal nanoparticles through strong metal–S bonds (in those that have the SH group). Metal NP@molecule (SERS nanotag) can be simply achieved by incubation of the metal nanoparticles with a solution of molecules. In order to enhance the selectivity of SERS nanotags, they can be further functionalized with various recognition elements (i.e., antibody, aptamer, DNA, lectins, etc.) for selective targeting of the analyte. One of the most common approaches for the functionalization of metal nanoparticles is using linkers that can bind to metal nanoparticles from one head and provide functional groups such as COOH and NH3 on their other head. The COOH or NH3 groups can be further linked to recognition elements through N-hydroxysuccinimide (NHS)/N′-ethylcarbodiimide hydrochloride (EDC) chemistry.192 Another approach is using a linker that can directly be attached to a recognition element. For example, we have reported the functionalization of AuNPs with lectins (i.e., WGA, cholera toxin B (CTB), Phaseolus vulgaris (PHA-L) and Aleuria aurantia lectin (AAL)) using (3,3′-dithiobis (sulfosuccinimidyl propionate)) (DTSSP) as linker. In this approach, lectin-DTSSPs were first incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mass ratio and the obtained complexes were incubated with AuNPs.193 Besides covalent functionalization, electrostatic interaction between the recognition element and metal nanoparticles covered by molecules is another approach. However, since this electrostatic interaction is loose, there is the need to consider the stability of the binding for biomedical applications. While the SERS nanotags are of great importance for in vitro and in vivo applications, their stability in the biological environment is one of the main concerns. To address this drawback, there are two main approaches: (i) formation of a silica shell to protect the metal core from oxidation (i.e., in the case of silver) and increase the colloidal stability; and (ii) coating with macromolecules (i.e., polymers, proteins, etc.) (Fig. 8). In a comparative study, we found that a layer of bovine serum albumin (BSA) on the SERS nanotag could result in more stability in biological media (i.e., 1X PBS, 1 M Tris-HCl, FPBS and RF-10) than the formation of a thin layer of silica. However, it was found that the stability of the antibody-functionalized BSA-coated SERS nanotags strongly depends on the type of antibody and also the thickness of this extra layer. Therefore, the stability of SERS nanotags should be investigated in each case. As discussed in previous sections, the nanoparticles that serve as the core for the SERS tag can provide plasmonic or non-plasmonic Raman signal enhancement as well as enabling hybrid SERS. The nature of the phenomenon depends on the chemistry of the core. In case of plasmonic label-aided SERS, nanoparticles are manufactured from noble metals like gold or silver.194,195 For the non-plasmonic label-aided SERS, the core of SERS tag can be any material that provides chemical Raman signal enhancement, such as semiconductors, carbon nanostructures and transition metals.178,196 In this section we discuss recent advances in label-aided SERS platforms that incorporate described SERS tags designs based on the plasmonic and chemical/hybrid enhancement of Raman signal.
1 mass ratio and the obtained complexes were incubated with AuNPs.193 Besides covalent functionalization, electrostatic interaction between the recognition element and metal nanoparticles covered by molecules is another approach. However, since this electrostatic interaction is loose, there is the need to consider the stability of the binding for biomedical applications. While the SERS nanotags are of great importance for in vitro and in vivo applications, their stability in the biological environment is one of the main concerns. To address this drawback, there are two main approaches: (i) formation of a silica shell to protect the metal core from oxidation (i.e., in the case of silver) and increase the colloidal stability; and (ii) coating with macromolecules (i.e., polymers, proteins, etc.) (Fig. 8). In a comparative study, we found that a layer of bovine serum albumin (BSA) on the SERS nanotag could result in more stability in biological media (i.e., 1X PBS, 1 M Tris-HCl, FPBS and RF-10) than the formation of a thin layer of silica. However, it was found that the stability of the antibody-functionalized BSA-coated SERS nanotags strongly depends on the type of antibody and also the thickness of this extra layer. Therefore, the stability of SERS nanotags should be investigated in each case. As discussed in previous sections, the nanoparticles that serve as the core for the SERS tag can provide plasmonic or non-plasmonic Raman signal enhancement as well as enabling hybrid SERS. The nature of the phenomenon depends on the chemistry of the core. In case of plasmonic label-aided SERS, nanoparticles are manufactured from noble metals like gold or silver.194,195 For the non-plasmonic label-aided SERS, the core of SERS tag can be any material that provides chemical Raman signal enhancement, such as semiconductors, carbon nanostructures and transition metals.178,196 In this section we discuss recent advances in label-aided SERS platforms that incorporate described SERS tags designs based on the plasmonic and chemical/hybrid enhancement of Raman signal.
 | ||
| Fig. 8 Different strategies for improving the stability of SERS nanotags in biological media. Adapted from ref.193 and 197 with permission from Elsevier, copyright 2019, and the Royal Society of Chemistry, copyright 2020, respectively. | ||
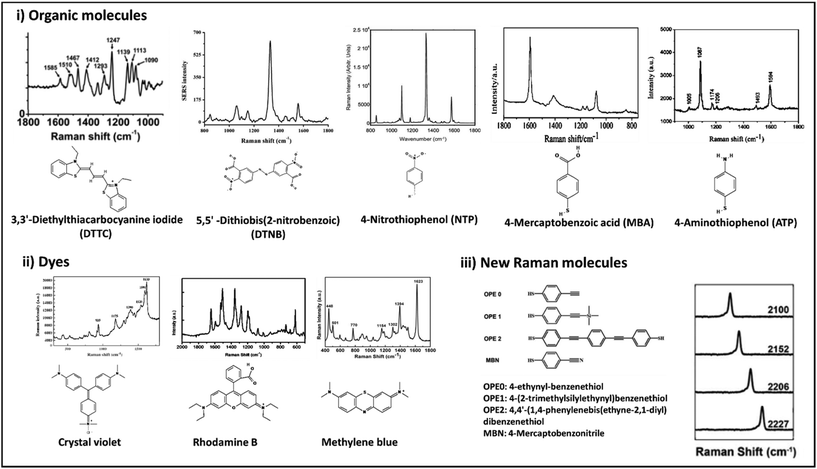 | ||
| Fig. 9 Chemical structures and Raman spectra of different Raman molecules including small organic molecules (i), dyes (ii), and newly designed Raman molecules (iii). DTTC: adapted from ref. 198 with permission from the American Chemical Society, copyright 2021. DTNB: adapted from ref. 199 with permission from Nature-Springer Group, copyright 2018. NTP: adapted from ref. 200 with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2020. MBA: adapted from ref. 201 with permission from the American Chemical Society, copyright 2016. ATP: adapted from ref. 202 with permission from the Royal Society of Chemistry, copyright 2012. Crystal violet: adapted from ref. 203 with permission from Elsevier, copyright 2012. Rhodamine B: adapted from ref. 204 with permission from Nature-Springer Group, copyright 2015. Methylene blue: adapted from ref. 205 with permission from Elsevier, copyright 2016. New Raman molecules: adapted from ref. 206 with permission from the American Chemical Society, copyright 2018. | ||
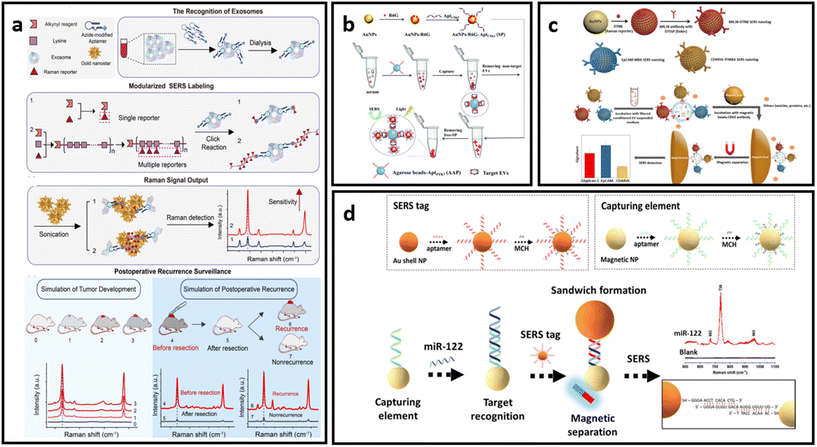 | ||
| Fig. 10 (a) Protocol of ultrasensitive exosome detection via SERS biosensor based on Au nanostars as substrate and Raman reporter coupled with lysine as SERS tags and targeting molecule, respectively; adapted from ref. 211 with permission from the American Chemical Society, copyright 2021; (b) sandwich-type SERS sensor based on dual aptamers and Au-enhanced Raman signal probes for the detection of tumour EVs; adapted from ref. 212 with permission from the Royal Society of Chemistry, copyright 2020; (c) detection of multiple cancer-specific surface proteins on EVs (molecular phenotype profiling) of CD63-positive EVs using SERS nanotags and CD63 antibody-functionalized magnetic beads; adapted from ref. 184 with permission from the American Chemical Society, copyright 2020. (d) Aptamer functionalized SERS-sandwich assay for ultrasensitive detection of miR-122 from exosomes, adapted from ref. 213 with permission from the American Chemical Society, copyright 2021. | ||
An interesting approach was developed to increase the detection sensitivity of EVs in which they are labelled with SERS tags conjugated with capturing substrates, enabling separation and accumulation of the analyte into a small area for ultrasensitive detection.184,212,213 This approach also eliminates the need for complex analyte isolation procedures from biological media. As indicated in Fig. 10b, Hou et al. developed a dual aptamer-based sandwich-type SERS sensor for quantitative detection of tumour-derived EVs.212 The EVs were immobilized on the surface of agarose beads functionalized with aptamers targeting PTK7 (tyrosine kinase-7) proteins, which are highly expressed in tumour-derived exosomes. Captured EVs were then labelled with SERS tags that consist of AuNPs decorated with R6G and aptamers targeting CD63 proteins which are pervasively expressed on targeted EVs. The concentrated precipitate could then be analysed via SERS. Another example of this approach was implemented by Zhang et al., where CD63-modified magnetic beads were used as capturing substrates.184 EVs immobilised on the magnetic beads were labelled with three different SERS tags – AuNP decorated with Raman reporter molecules (DTNB, MBA or TFMBA) and conjugated with monoclonal antibodies for specific binding (CD44V6, EpCAM and MIL38) (Fig. 10c). This approach enabled the simultaneous detection of multiple cancer-specific surface proteins on EVs in a single sample. As a proof-of-concept demonstration, their method was further tested in plasma-derived EVs in patients who have experienced PDAC.178 The magnetic separation method is suitable not only for EV detection, but can be implemented to concentrate and separate the EVs’ and exosomes’ protein biomarkers of interest, as has been done by Muhammad et al.213 The researchers developed an aptamer SERS-sandwich assay for highly sensitive detection of elevated exosomal miR-122. The biosensor was constructed with a SERS tag (silica-coated AuNP conjugated with aptamer and Raman reporter molecule) and magnetic capture substrate (Au-coated Fe3O4 NP functionalised with the target-recognition aptamer) (Fig. 10d). The researchers demonstrated that separated exosomal miR-122 can be recognized with the coexistence of other sequences of similar length with a slightly varying arrangement of DNA bases in a concentration as low as 8 fM.213
Label-aided SERS sensors can be integrated for multiplex and robust quantitative analysis with miniaturized devices for “at the point-of-care” testing. The conceptual design of such devices avoids the use of the complex protocols that hinder point-of-use of the developed techniques. As such, Reza et al.214 developed a SERS-based simple microfluidic device for screening of single CTC that enables the identification of the heterogeneous expression of CTC surface proteins (Fig. 11a). They have employed three types of Raman reporter molecules for the SERS-tags (DTNB, MBA, and TFMBA) for each type of biomarker analysed through the channel. CTC passing through the microfluidic channel was then analysed for the heterogeneous expression of multiple cell receptor proteins. The second part of the device consists of an electrochemical zone with nanoscopic fluid mixing for the detection of intracellular protein. By enhancing the antigen–antibody collision via nanoscopic mixing, the sensitivity of detection is significantly increased.214 Wang et al.215 adopted a similar nanoscopic fluid-mixing chip with SERS-based nanobarcode sorting for the detection of different EVs subpopulations (Fig. 11b). They have seen the elevated sensitivity of the platform due to the created nanoscopic flow. The reported detection limit was 103 particles per mL.215
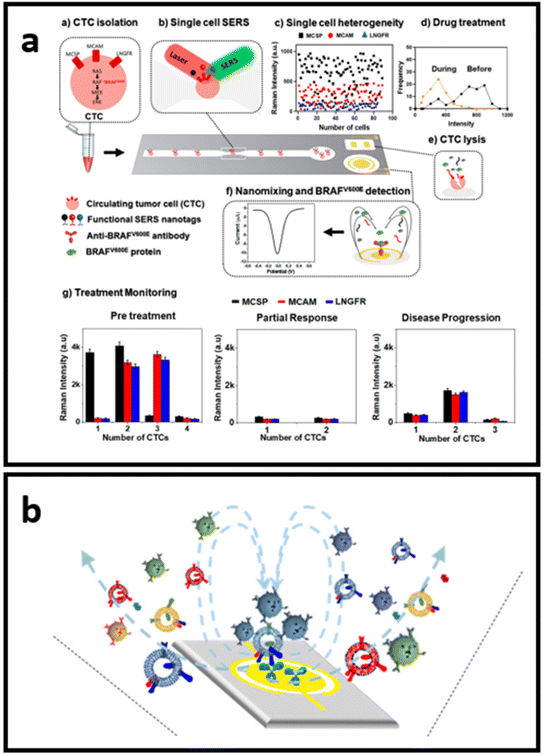 | ||
| Fig. 11 Schematics of the small devices with SERS-based sensors: (a) SERS-based microfluidic immunoassay for in situ characterization of single CTCs, adapted from ref. 214 with permission from the American Chemical Society, copyright 2021; (b) nano-mixing-enhanced EVs subpopulation characterization platform adapted from ref. 215 with permission from the American Chemical Society, copyright 2021. | ||
Overall, the described strategies have been developed in order to provide the most accurate analysis of CTCs or EVs in complex biological media, with a low limit of detection. A summary of the methods described above is presented in Table 2.
| SERS platform | Biomarkers | Media | Described detection ability | Ref. |
|---|---|---|---|---|
| Au–Ag nanohollows | T24 and RT4 bladder cancer cells | Urine | Detection limit up to 1250 cells | 209 |
| Porous CuFeSe2/Au heterostructured nanospheres | Gaseous aldehydes and lung cancer cells (A549) | Exhaled breath and cultured cells | Detection limit of gaseous aldehydes – 1.0 ppb; detection of folate receptor-positive cancer cells | 210 |
| AuAg NPs SERS tags with Fe3O4@TiO2 capturing substrate | EVs derived from A549 and normal human bronchial epithelial (BEAS-2B) cells | Human serum | Detection limit 1 particle per mL | 216 |
| AuNPs SERS tags with agarose beads capturing substrate | EVs derived from human acute lymphoblastic leukemia cells (CCRF-CEM) | Human serum | Detection limit 2.44 pg μL−1 | 212 |
| AuNPs SERS tags with magnetic beads capturing substrate | EVs derived from colorectal cancer (SW480), bladder cancer (C3) and pancreatic cancer (Panc-1) | Human serum | Phenotype profiling with low sample volume – 10 μL | 184 |
| AuNPs SERS tags with Fe3O4@TiO2 capturing substrate | miRNA inside EVs derived from Panc-1 cells | Human serum | In situ analysis with detection limit of 0.21 fM | 186 |
| SERS EpCAM-aptasensors | EVs derived from A549 | Isolated exosomes (cell medium of A549 cells) | Detection limit 2.4 particles per μL | 211 |
| SERS-based microfluidic immunoassay for in situ characterization of single CTCs | CTCs from melanoma cells (SK-MEL-28) | Isolated cells (patient's sample) | Single cell-level protein expression profiling | 214 |
| Aptamer functionalized SERS-sandwich assay | miR-122 from mice serum and human hepatoma (HepG2) cells’ EVs | Isolated EVs (mice serum and cell medium of HepG2 cells) | Detection limit 8 fM | 213 |
| Nano-mixing-enhanced subpopulation characterization platform | EVs derived from SK-MEL-28, SK-MES-1, NCI-H1703, NCI-H1650, and MCF7 cells | Isolated EVs (SK-MEL-28, SK-MES-1, NCI-H1703, NCI-H1650, and MCF7 cells culture media or plasma) | Detection limit 103 particles per mL | 215 |
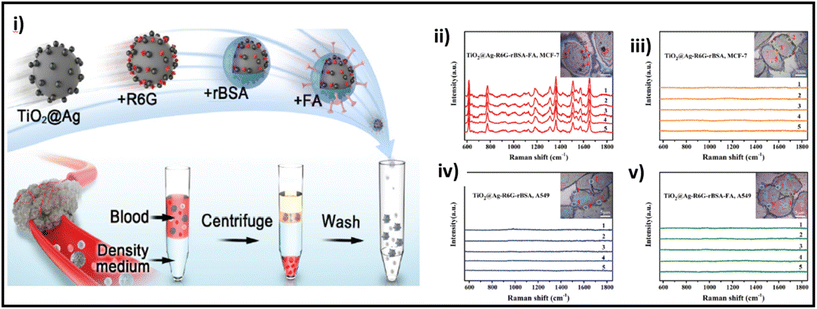 | ||
| Fig. 12 (i) Schematic of the preparation of TiO2@Ag-based SERS bioprobe; SERS detection of MCF-7 and A549 cells incubated with TiO2@Ag–R6G–rBSA–FA (ii and v, respectively) and TiO2@Ag–R6G–rBSA (iii and iv, respectively) SERS bioprobes. Adapted from ref. 217 with permission from the Royal Society of Chemistry, copyright 2022. | ||
Semiconductor materials provide another beneficial feature: photothermal (PT) conversion. The combination of SERS properties and local PT effects gives SERS platforms additional functionality as theranostic agents – targeting, detection/sensing and photothermal destruction of tumour cells. Hybrid SERS platforms with PT conversion provide sufficient EM signal enhancement for imaging and probe sensing.218–222 Semiconductor materials are also able to provide a strong SERS signal and PT effect without incorporation of Au.223,224 Lin et al. reported ultrahigh SERS activity (EF up to 105 for the nitro blue tetrazolium (NBT) probe molecule) of crystal–amorphous core–shell-structured black-TiO2 NPs.225 The designed core–shell structure enables high-efficiency exciton transition resulting in charge-separation that occurs due to interfacial band bending. Those effects induce photo exciton transition from the crystal core to the amorphous shell, further promoting photoinduced charge transfer between the substrate and probe molecule. Modified with SERS-active molecule (tag) and antibody (targeting molecule), the core–shell-structured TiO2 NPs can be used to quickly and accurately identify cancer cells in order to efficiently administer treatment with photothermal therapy.225 Hybrid SERS platforms are often used to incorporate additional chemical enhancements that increase overall SERS sensitivity. However, a typical constraint of such an approach is that chemical or physical binding of the analyte is required to produce effective chemical enhancement through charge transfer, limiting the choice as it requires the analyte to have either a thiol or aromatic binding group. Further, there are limited reports on the usage of hybrid systems to improve SERS through EM, which is a long path for development. Unfortunately, employing a particular linker to detect and identify disease-related CTC and EVs is rather difficult. The complex nature of cells and EVs including many biomolecules makes it difficult to identify particular biomarkers for selective labelling.226,227 As a result, identifying possible disease-related cells or EVs requires a more thorough strategy that takes into account signals from both known and undiscovered biomarkers to distinguish individuals at risk from healthy donors.
4.2. Label-free SERS
It is very important to understand that successful data analysis with a label-free SERS platform depends on both a strong, reliable and accurate Raman signal, of which the SERS substrate is in charge, and analysis of the signal. The Raman data acquired by the label-free SERS method contain huge amounts of information, especially when the analyte consists of multiple biomolecules that provide a complex pattern of signature peaks. The most common approach to interpreting SERS signals is to examine particular Raman bands attributed to the known vibrational modes and analyse the discernible differences in intensity and Raman shift compared with the control samples. Bioanalytes, such as proteins, nucleic acids and lipids, are usually analysed based on the spectral fingerprint areas (between 800 and 1800 cm−1) (Fig. 13).228 However, it is difficult to clearly identify the difference in signals from cells and EVs due to the interference from various substances. Therefore, to use the SERS signal of more complex structures as a fingerprint for disease diagnosis, more precise signal analysis methods are used such as multivariate statistical methods (principal component analysis, “PCA”),229–232 and the more recently developed methods of machine learning231,233 and neural networks.234,235 We will not discuss much about analysis techniques, focusing on the SERS platforms themselves, but the reader should take into consideration that label-free SERS analysis outcome depends 50% on data analysis tools.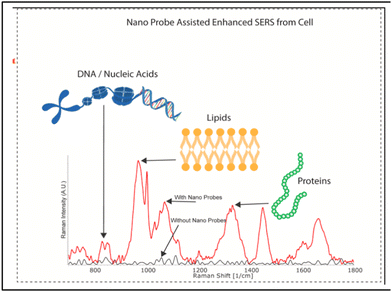 | ||
| Fig. 13 Characteristic Raman peaks of biomolecules, adapted from ref. 228 with permission from Elsevier, copyright 2021. | ||
Label-free SERS analysis is attractive to scientists due to the rich information outcome it provides. Apart from the importance of data analysis that we briefly mentioned, the direct SERS approach suffers multiple drawbacks that researchers are willing to tackle through the SERS platform designed to harvest the major benefit of analytes’ full structural information. In this section, we discuss the label-free SERS approach with the aim to deliver the advances in plasmon and chemical/hybrid enhancement approaches in the design of sensing platforms for the analysis of cells and EVs.
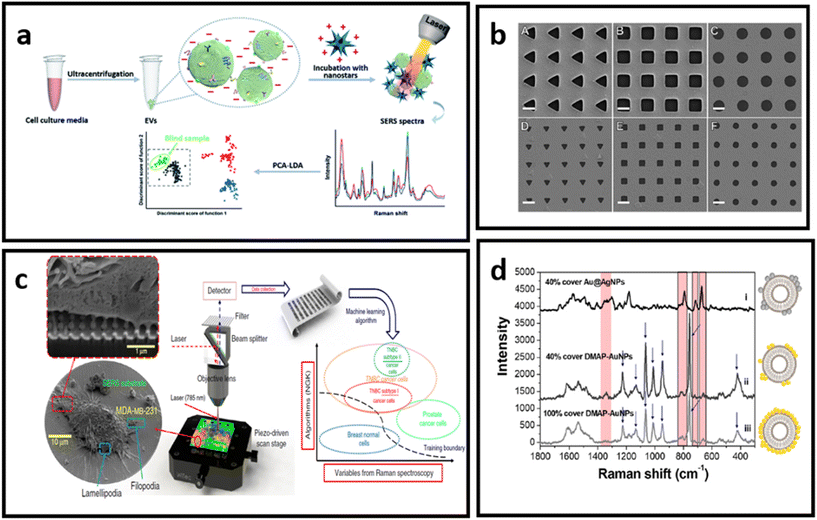 | ||
| Fig. 14 (a) Molecular characterisation of EVs with positively charged gold nanostars, adapted from ref. 232 with permission from the Royal Society of Chemistry, copyright 2020; (b) Au nanohole arrays of varying sizes and shapes for trapping single EVs and enhancing their vibrational signature, adapted from ref. 229 with permission from Springer-Nature Group, copyright 2021; (c) nanolaminated SERS multiwell cell culture assay, adapted from ref. 236 with permission from Springer-Nature Group, copyright 2020; (d) in situ EV-attached AuNPs with a grown over silver layer – to form a core–shell nanoparticle (Au@AgNPs) directly at the EV surface, adapted from ref. 231 with permission from the American Chemical Society, copyright 2019. | ||
Most reports demonstrate that label-free SERS necessitates the use of pre-treated samples to avoid signal detection from other molecules in the surrounding medium that are unrelated to the analyte. Indeed, the purity of the sample is critical, as it affects the reliability and repeatability of the SERS analysis. Using a label-free SERS platform, Koster et al.233 verified that common EV isolation methods (differential ultracentrifugation, density gradient ultracentrifugation, and size exclusion chromatography) yield variable lipoprotein content. For their tests, the research group developed a SERS platform based on a commercially available substrate composed of a quartz microfiber matrix embedded with AuNP clusters (solid Au spheres with an approximate diameter of 40–60 nm), grafted to a borosilicate glass microscope slide. After analysing different samples, the authors concluded that a dual-isolation method is necessary to isolate EVs from the major classes of lipoprotein for the better performance of the label-free SERS platform. The authors also reported that combined machine learning analysis and the developed SERS platform was sensitive enough to identify cancer-specific signatures that remained unaffected by the presence of additional co-isolated biomolecules, showing that the label-free diagnostic ability was effective across many different sample preparations.
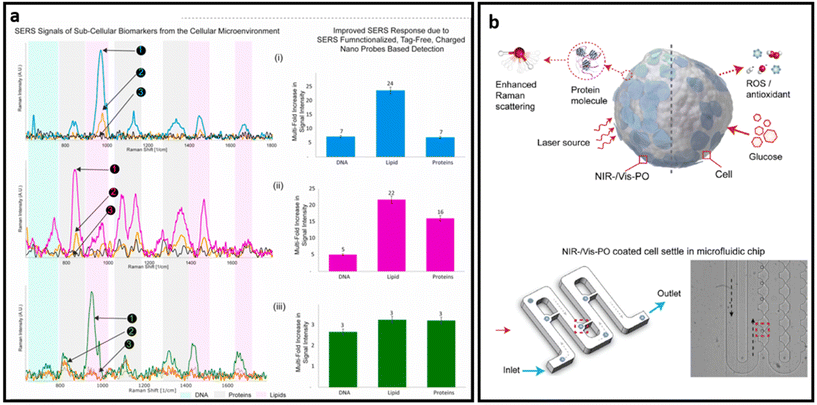 | ||
| Fig. 15 (a) SERS signals of sub-cellular biomarkers in cancer cell by self-functionalized tag-free nano sensor, adapted from ref. 228 with permission from Elsevier, copyright 2021; (b) schematic SERS detection via PO nanoprobe: PO-coated cell SERS sensing (top) and optical image of the PO-coated cells infused into a microfluidic chip and settled separately in the small grooves, highlighted in red (bottom), adapted from ref. 243 with permission from Elsevier, copyright 2022. | ||
Analyte-capturing techniques are also being extensively researched and developed, as an approach to achieving strong and accurate SERS signal rests on the analyte. Yin et al. demonstrated a facile polyethylene glycol (PEG)-based isolation method to classify and further analyse EVS with a label-free SERS platform.239 PEG is commonly used to bind and purify via centrifugation virus particles.240 The PEG-based method was adapted to isolate EVs,241 which were similar to viruses in physical size and membrane structure. EVs were wrapped in PEG films, and upon centrifugation formed PEG-based EV aggregates. The authors used a novel planar nanomaterial – amino molybdenum oxide (AMO) nanoflakes – as the SERS substrate.242 To prove the reliability of the platform, three types of common male cancer cell lines – leukemia, prostate cancer, and colorectal cancer – and one healthy male blood sample, were utilized to isolate and classify EVP. The obtained data were analysed by PCA and machine learning algorithm. As a result, analysis accuracies were higher than 85% and all the precisions and sensitivities were higher than 80% for all tested cell lines.239
Semiconductors exhibit plasmon-free SERS through the electron transferring between the semiconductor and adsorbed analytes.243 As such, Zhang et al. developed a dopant-driven oxide-based label-free SERS substrate with dual function. For the SERS measurement, cells were briefly (10 min) incubated with plasmonic oxide NPs. Then authors used a microfluidic chip system with grooves designed to hydrodynamically trap individual cells from the suspension and minimize possible interference on the target signals from the medium. The trapped single cells were then further analysed with a Raman spectrometer (Fig. 15b). The developed platform exhibits up to 90% accuracy in distinguishing single leukemia (THP1) cells from peripheral blood mononuclear cells (PBMC) and human fetal kidney (HEK 293) from a human macrophage cell line (U937). Based on the combined features of enhanced electromagnetic field-driven SERS and dopant-driven tunable plasmonics, this approach enables single-cell detection. In addition, the nanoprobe is triggered by the bio-redox response of individual cells to stimuli, enabling complementary colorimetric cell detection and achieving nearly uniform identification accuracy at the single cell level.243 A stronger Raman signal in label-free design is necessary to obtain distinct peaks from various biomarkers that exist on the surface of cells and EVs, leading to better analysis of samples through profiling the most possible biomarkers on their surfaces. Given that the combination of chemical enhancement and plasmon enhancement in the design of SERS substrates provides a higher SERS enhancement factor, many semiconductors and 2D nanomaterials have been combined with noble nanostructures to make ultra-sensitive SERS substrates.244,245
5. Conclusions, perspectives and future directions
SERS is one of the main approaches for ultrasensitive analysis of wide ranges of analytes for biomedical purposes. Various nanomaterials, ranging from plasmonic noble metal nanostructures to inorganic semiconductors and 2D semiconductor nanosheets, have been extensively used for the mechanistic study of SERS as well as in designing SERS systems in both colloidal and planar forms. One of the main challenges in comparing the SERS efficiencies of different nanomaterials is the inconsistencies in calculation of the SERS enhancement factor, where there is a variety of parameters (such as solid sample vs. colloidal solution). In mechanistic studies, researchers usually ignore one of the mechanisms and attribute the observed SERS solely to one of the main well-known mechanisms. Although one of the mechanisms is usually dominant – electromagnetic enhancement is dominant in metal nanoparticles and chemical enhancement is dominant in semiconductors – a comprehensive analysis should be performed to indicate the role of each mechanism in SERS of new nanomaterials.SERS biosensors have been designed for the analysis of various biomarkers in biological samples, and in particular, molecular signatures of cells and their extracellular vesicles. These SERS systems can be categorized into two main groups: label-aided and label-free SERS systems. Label-aided and label-free SERS present both great advantages and challenges. While the use of SERS tags allows ultra-sensitive detection of analytes and relatively simple manufacturing, it does not provide any structural information about the target molecules other than their presence. In addition, despite the development of SERS nanotags, there is still a need to develop SERS nanotags with high intensity and high colloidal stability in biological media, and reproducible SERS to make it more reliable in biomedical applications.
On the other hand, label-free SERS can provide structural information about the target sample. However, the analysis and interpretation of spectra from complex structures made up of many biomolecules are equally difficult, and necessitate the use of sophisticated data-processing techniques to enable reliable interpretation of complicated vibrational patterns. Although many machine learning-based techniques have been utilised to analyse the SERS spectra of complex samples, many methods still have not been investigated. Additionally, there is no comprehensive analysis comparing the accuracy of these techniques. The establishment of a standard method becomes very urgent and of significance for label-free SERS assays.
The further development of plasmonic and conductive materials would greatly benefit the SERS sensing technology, where there is a need to investigate the SERS performance of newly developed nanomaterials such as carbon dots and metal nanoclusters, and aggregation-induced emission luminogens which have shown great optical properties and, in particular, excellent charge/energy transfer ability. Additionally, the development of SERS biosensors should also move towards a more conceptual design of the platform. While most of the described techniques demonstrated reproducible and reliable results with low detection limits, the SERS platform compositions and protocols in the majority of the works are rather complicated for point-of-care use. The construction of small devices based on the powerful SERS platform, as well as reliable and powerful analysis tools, that would allow minimum effort from the user to perform analysis is one of the most important pathways for the development of this technology.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Australian Research Council (ARC) through Discovery Projects (DP200102004). A. T acknowledges funding support from the International Macquarie University Research Excellence scholarship (iMQRES).References
- A. Smekal, Naturwissenschaften, 1923, 11, 873–875 CrossRef CAS.
- R. Singh, Phys. Perspect., 2002, 4, 399–420 CrossRef.
- J. J. Laserna, Modern techniques in Raman spectroscopy, Wiley New York, 1996 Search PubMed.
- E. Smith and G. Dent, Modern Raman spectroscopy: a practical approach, John Wiley & Sons, 2019 Search PubMed.
- K. Kneipp, H. Kneipp, I. Itzkan, R. R. Dasari and M. S. Feld, Chem. Rev., 1999, 99, 2957–2976 CrossRef CAS PubMed.
- M. Fleischmann, P. J. Hendra and A. J. McQuillan, Chem. Phys. Lett., 1974, 26, 163–166 CrossRef CAS.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- C. Zong, M. Xu, L.-J. Xu, T. Wei, X. Ma, X.-S. Zheng, R. Hu and B. Ren, Chem. Rev., 2018, 118, 4946–4980 CrossRef CAS PubMed.
- X. Wang, E. Zhang, H. Shi, Y. Tao and X. Ren, Analyst, 2022, 147, 1257–1272 RSC.
- X. Liu, J. Guo, Y. Li, B. Wang, S. Yang, W. Chen, X. Wu, J. Guo and X. Ma, J. Mater. Chem. B, 2021, 9, 8378–8388 RSC.
- Y. Hang, J. Boryczka and N. Wu, Chem. Soc. Rev., 2022, 51, 329–375 RSC.
- S. Nie and S. R. Emory, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- M. L. Thomas and P. Marcato, Cancers, 2018, 10, 101 CrossRef PubMed.
- K. Ng, A. Stenzl, A. Sharma and N. Vasdev, Urol. Oncol., 2021, 39, 41–51 CrossRef CAS PubMed.
- J. Marrugo-Ramírez, M. Mir and J. Samitier, Int. J. Mol. Sci., 2018, 19, 2877 CrossRef PubMed.
- S. Chojnowska, I. Ptaszyńska-Sarosiek, A. Kępka, M. Knaś and N. Waszkiewicz, J. Clin. Med., 2021, 10, 517 CrossRef CAS PubMed.
- M. Russano, A. Napolitano, G. Ribelli, M. Iuliani, S. Simonetti, F. Citarella, F. Pantano, E. Dell'Aquila, C. Anesi, N. Silvestris, A. Argentiero, A. G. Solimando, B. Vincenzi, G. Tonini and D. Santini, J. Exp. Clin. Cancer Res., 2020, 39, 95 CrossRef CAS PubMed.
- G. Follain, D. Herrmann, S. Harlepp, V. Hyenne, N. Osmani, S. C. Warren, P. Timpson and J. G. Goetz, Nat. Rev. Cancer, 2020, 20, 107–124 CrossRef CAS PubMed.
- P. Lewczuk, P. Riederer, S. E. O'Bryant, M. M. Verbeek, B. Dubois, P. J. Visser, K. A. Jellinger, S. Engelborghs, A. Ramirez, L. Parnetti, C. R. Jack, Jr., C. E. Teunissen, H. Hampel, A. Lleó, F. Jessen, L. Glodzik, M. J. de Leon, A. M. Fagan, J. L. Molinuevo, W. J. Jansen, B. Winblad, L. M. Shaw, U. Andreasson, M. Otto, B. Mollenhauer, J. Wiltfang, M. R. Turner, I. Zerr, R. Handels, A. G. Thompson, G. Johansson, N. Ermann, J. Q. Trojanowski, I. Karaca, H. Wagner, P. Oeckl, L. van Waalwijk van Doorn, M. Bjerke, D. Kapogiannis, H. B. Kuiperij, L. Farotti, Y. Li, B. A. Gordon, S. Epelbaum, S. J. B. Vos, C. J. M. Klijn, W. E. Van Nostrand, C. Minguillon, M. Schmitz, C. Gallo, A. Lopez Mato, F. Thibaut, S. Lista, D. Alcolea, H. Zetterberg, K. Blennow and J. Kornhuber, World J. Biol. Psychiatry, 2018, 19, 244–328 CrossRef PubMed.
- B. Pardini, A. A. Sabo, G. Birolo and G. A. Calin, Cancers, 2019, 11, 1170 CrossRef CAS PubMed.
- B. Lei, Z. Tian, W. Fan and B. Ni, Int. J. Med. Sci., 2019, 16, 292–301 CrossRef CAS PubMed.
- E. Kilgour, D. G. Rothwell, G. Brady and C. Dive, Cancer Cell, 2020, 37, 485–495 CrossRef CAS PubMed.
- J. Müller Bark, A. Kulasinghe, B. Chua, B. W. Day and C. Punyadeera, Br. J. Cancer, 2020, 122, 295–305 CrossRef PubMed.
- W. Yu, J. Hurley, D. Roberts, S. K. Chakrabortty, D. Enderle, M. Noerholm, X. O. Breakefield and J. K. Skog, Ann. Oncol., 2021, 32, 466–477 CrossRef CAS PubMed.
- H. Shao, H. Im, C. M. Castro, X. Breakefield, R. Weissleder and H. Lee, Chem. Rev., 2018, 118, 1917–1950 CrossRef CAS PubMed.
- A. Tukova and A. Rodger, Emerging Top. Life Sci., 2021, 5, 61–75 CrossRef CAS PubMed.
- T. Huang and C.-X. Deng, Int. J. Biol. Sci., 2019, 15, 1–11 CrossRef PubMed.
- S. Wang, K. Zhang, S. Tan, J. Xin, Q. Yuan, H. Xu, X. Xu, Q. Liang, D. C. Christiani, M. Wang, L. Liu and M. Du, Mol. Cancer, 2021, 20, 13 CrossRef CAS PubMed.
- C. Alix-Panabières and K. Pantel, Cancer Discovery, 2021, 11, 858–873 CrossRef PubMed.
- J. Lin, J. Zheng and A. Wu, J. Mater. Chem. B, 2020, 8, 3316–3326 RSC.
- J. Perumal, Y. Wang, A. B. E. Attia, U. S. Dinish and M. Olivo, Nanoscale, 2021, 13, 553–580 CAS.
- D. Cialla-May, X. S. Zheng, K. Weber and J. Popp, Chem. Soc. Rev., 2017, 46, 3945–3961 RSC.
- L. K. Chin, T. Son, J.-S. Hong, A.-Q. Liu, J. Skog, C. M. Castro, R. Weissleder, H. Lee and H. Im, ACS Nano, 2020, 14, 14528–14548 CrossRef PubMed.
- Y. Zhang, X. Mi, X. Tan and R. Xiang, Theranostics, 2019, 9, 491–525 CrossRef CAS PubMed.
- J. Wang, K. M. Koo, Y. Wang and M. Trau, Adv. Sci., 2019, 6, 1900730 CrossRef CAS PubMed.
- S. D. Christesen, Appl. Spectrosc., 1988, 42, 318–321 CrossRef CAS.
- M. Käll, H. Xu and P. Johansson, J. Raman Spectrosc., 2005, 36, 510–514 CrossRef.
- S.-Y. Ding, J. Yi, J.-F. Li, B. Ren, D.-Y. Wu, R. Panneerselvam and Z.-Q. Tian, Nat. Rev. Mater., 2016, 1, 16021 CrossRef CAS.
- X. Wang and L. Guo, Angew. Chem., Int. Ed., 2020, 59, 4231–4239 CrossRef CAS PubMed.
- G. Mie, Ann. Phys., 1908, 330, 377–445 CrossRef.
- X. Fan, W. Zheng and D. J. Singh, Light: Sci. Appl., 2014, 3, e179–e179 CrossRef CAS.
- K. M. Mayer and J. H. Hafner, Chem. Rev., 2011, 111, 3828–3857 CrossRef CAS PubMed.
- M. Tavakkoli Yaraki, Y. Pan, F. Hu, Y. Yu, B. Liu and Y. N. Tan, Mater. Chem. Front., 2020, 4, 3074–3085 RSC.
- M. Tavakkoli Yaraki, F. Hu, S. Daqiqeh Rezaei, B. Liu and Y. N. Tan, Nanoscale Adv., 2020, 2, 2859–2869 RSC.
- M. Tavakkoli Yaraki, S. Daqiqeh Rezaei and Y. N. Tan, Phys. Chem. Chem. Phys., 2020, 22, 5673–5687 RSC.
- X. Zhang, H. Zhang, S. Yan, Z. Zeng, A. Huang, A. Liu, Y. Yuan and Y. Huang, Sci. Rep., 2019, 9, 17634 CrossRef PubMed.
- P.-P. Fang, J.-F. Li, Z.-L. Yang, L.-M. Li, B. Ren and Z.-Q. Tian, J. Raman Spectrosc., 2008, 39, 1679–1687 CrossRef CAS.
- R. Kumar, L. Binetti, T. H. Nguyen, L. S. M. Alwis, A. Agrawal, T. Sun and K. T. V. Grattan, Sci. Rep., 2019, 9, 17469 CrossRef PubMed.
- S. Hong and X. Li, J. Nanomater., 2013, 2013, 790323 Search PubMed.
- M. Tavakkoli Yaraki, S. Daqiqeh Rezaei, E. Middha and Y. N. Tan, Part. Part. Syst. Charact., 2020, 37, 2000027 CrossRef CAS.
- X. Liang, N. Li, R. Zhang, P. Yin, C. Zhang, N. Yang, K. Liang and B. Kong, NPG Asia Mater., 2021, 13, 8 CrossRef CAS.
- J. R. Lombardi and R. L. Birke, J. Chem. Phys., 2007, 126, 244709 CrossRef PubMed.
- J. R. Lombardi and R. L. Birke, J. Chem. Phys., 2012, 136, 144704 CrossRef PubMed.
- J. R. Lombardi and R. L. Birke, J. Phys. Chem. C, 2014, 118, 11120–11130 CrossRef CAS.
- L. E. Brus, J. Chem. Phys., 1984, 80, 4403–4409 CrossRef CAS.
- T. Takagahara and K. Takeda, Phys. Rev. B: Condens. Matter Mater. Phys., 1992, 46, 15578–15581 CrossRef CAS PubMed.
- G. Song, W. Gong, S. Cong and Z. Zhao, Angew. Chem., Int. Ed., 2021, 60, 5505–5511 CrossRef CAS PubMed.
- G. Song, S. Cong and Z. Zhao, Chem. Sci., 2022, 13, 1210–1224 RSC.
- L. Yang, Y. Peng, Y. Yang, J. Liu, H. Huang, B. Yu, J. Zhao, Y. Lu, Z. Huang, Z. Li and J. R. Lombardi, Adv. Sci., 2019, 6, 1900310 CrossRef PubMed.
- Y. Liu, H. Ma, X. X. Han and B. Zhao, Mater. Horiz., 2021, 8, 370–382 RSC.
- B. Yang, S. Jin, S. Guo, Y. Park, L. Chen, B. Zhao and Y. M. Jung, ACS Omega, 2019, 4, 20101–20108 CrossRef CAS PubMed.
- Z. Zheng, S. Cong, W. Gong, J. Xuan, G. Li, W. Lu, F. Geng and Z. Zhao, Nat. Commun., 2017, 8, 1993 CrossRef PubMed.
- S. Cong, X. Liu, Y. Jiang, W. Zhang and Z. Zhao, Innovation, 2020, 1, 100051 Search PubMed.
- K. H. Lee, H. Jang, Y. S. Kim, C.-H. Lee, S. H. Cho, M. Kim, H. Son, K. B. Bae, D. V. Dao, Y. S. Jung and I.-H. Lee, Adv. Sci., 2021, 8, 2100640 CrossRef CAS PubMed.
- X. Jiang, X. Sun, D. Yin, X. Li, M. Yang, X. Han, L. Yang and B. Zhao, Phys. Chem. Chem. Phys., 2017, 19, 11212–11219 RSC.
- Y. Peng, C. Lin, L. Long, T. Masaki, M. Tang, L. Yang, J. Liu, Z. Huang, Z. Li, X. Luo, J. R. Lombardi and Y. Yang, Nano-Micro Lett., 2021, 13, 52 CrossRef PubMed.
- Y. Peng, C. Lin, Y. Li, Y. Gao, J. Wang, J. He, Z. Huang, J. Liu, X. Luo and Y. Yang, Matter, 2022, 5, 694–709 CrossRef CAS PubMed.
- F. Avila, D. J. Fernandez, J. F. Arenas, J. C. Otero and J. Soto, Chem. Commun., 2011, 47, 4210–4212 RSC.
- M. T. Yaraki and Y. N. Tan, Chem. – Asian J., 2020, 15, 3180–3208 CrossRef CAS PubMed.
- H. Dong, Y. C. Chen and C. Feldmann, Green Chem., 2015, 17, 4107–4132 RSC.
- R. Purbia and S. Paria, Nanoscale, 2015, 7, 19789–19873 RSC.
- F. Fiévet, S. Ammar-Merah, R. Brayner, F. Chau, M. Giraud, F. Mammeri, J. Peron, J. Y. Piquemal, L. Sicard and G. Viau, Chem. Soc. Rev., 2018, 47, 5187–5233 RSC.
- H.-l. Liu, F. Nosheen and X. Wang, Chem. Soc. Rev., 2015, 44, 3056–3078 RSC.
- M. Tavakkoli Yaraki and Y. N. Tan, Sens. Int., 2020, 1, 100049 CrossRef.
- T. Zhang, X. Li, Y. Sun, D. Liu, C. Li, W. Cai and Y. Li, Chem. Sci., 2021, 12, 12631–12639 RSC.
- M. A. Tahir, N. E. Dina, H. Cheng, V. K. Valev and L. Zhang, Nanoscale, 2021, 13, 11593–11634 CAS.
- L. W. Yap, H. Chen, Y. Gao, K. Petkovic, Y. Liang, K. J. Si, H. Wang, Z. Tang, Y. Zhu and W. Cheng, Nanoscale, 2017, 9, 7822–7829 RSC.
- Y. N. Tan, J. Y. Lee and D. I. C. Wang, J. Phys. Chem. C, 2008, 112, 5463–5470 CrossRef CAS.
- Y.-N. Tan, J. Yang, J. Y. Lee and D. I. C. Wang, J. Phys. Chem. C, 2007, 111, 14084–14090 CrossRef CAS.
- X. T. Zheng, H. V. Xu and Y. N. Tan, in Advances in Bioinspired and Biomedical Materials Volume 2, American Chemical Society, 2017, vol. 1253, ch. 7, pp. 123–152 Search PubMed.
- Y. N. Tan, J. Y. Lee and D. I. C. Wang, J. Am. Chem. Soc., 2010, 132, 5677–5686 CrossRef CAS PubMed.
- Y. N. Tan, J. Y. Lee and D. I. C. Wang, J. Phys. Chem. C, 2009, 113, 10887–10895 CrossRef CAS.
- M. Mosaviniya, T. Kikhavani, M. Tanzifi, M. Tavakkoli Yaraki, P. Tajbakhsh and A. Lajevardi, Colloid Interface Sci. Commun., 2019, 33, 100211 CrossRef CAS.
- A. Nouri, M. Tavakkoli Yaraki, A. Lajevardi, Z. Rezaei, M. Ghorbanpour and M. Tanzifi, Colloid Interface Sci. Commun., 2020, 35, 100252 CrossRef CAS.
- K. Ijiro and H. Mitomo, Polym. J., 2017, 49, 815–824 CAS.
- A. Tukova, I. C. Kuschnerus, A. Garcia-Bennett, Y. Wang and A. Rodger, Nanomaterials, 2021, 11, 2565 CrossRef CAS PubMed.
- J. Li, G. Zhang, J. Wang, I. S. Maksymov, A. D. Greentree, J. Hu, A. Shen, Y. Wang and M. Trau, ACS Appl. Mater. Interfaces, 2018, 10, 32526–32535 CrossRef CAS PubMed.
- Y. Sun and Y. Xia, Science, 2002, 298, 2176–2179 CrossRef CAS PubMed.
- D. K. Smith and B. A. Korgel, Langmuir, 2008, 24, 644–649 CrossRef CAS PubMed.
- S. Zhou, J. Li, K. D. Gilroy, J. Tao, C. Zhu, X. Yang, X. Sun and Y. Xia, ACS Nano, 2016, 10, 9861–9870 CrossRef CAS PubMed.
- H. J. Kim, M. H. Lee, L. Mutihac, J. Vicens and J. S. Kim, Chem. Soc. Rev., 2012, 41, 1173–1190 RSC.
- Y. Liu, Z. Lu, X. Lin, H. Zhu, W. Hasi, M. Zhang, X. Zhao and X. Lou, RSC Adv., 2016, 6, 58387–58393 RSC.
- J. J. Mock, S. M. Norton, S. Y. Chen, A. A. Lazarides and D. R. Smith, Plasmonics, 2011, 6, 113–124 CrossRef CAS.
- J. Liu, L. Chen, B. Duan, Z. Gu, Q. Luo and C. Duan, RSC Adv., 2016, 6, 38690–38696 RSC.
- Q. He, A. Zhao, L. Li, H. Sun, D. Wang, H. Guo, M. Sun and P. Chen, New J. Chem., 2017, 41, 1582–1590 RSC.
- D. M. Solís, J. M. Taboada, F. Obelleiro, L. M. Liz-Marzán and F. J. García de Abajo, ACS Photonics, 2017, 4, 329–337 CrossRef PubMed.
- M. Hu, F. S. Ou, W. Wu, I. Naumov, X. Li, A. M. Bratkovsky, R. S. Williams and Z. Li, J. Am. Chem. Soc., 2010, 132, 12820–12822 CrossRef CAS PubMed.
- X. Zhang, Y. Zheng, X. Liu, W. Lu, J. Dai, D. Y. Lei and D. R. MacFarlane, Adv. Mater., 2015, 27, 1090–1096 CrossRef CAS PubMed.
- Y. Chen, K. J. Si, D. Sikdar, Y. Tang, M. Premaratne and W. Cheng, Adv. Opt. Mater., 2015, 3, 919–924 CrossRef CAS.
- C. Hanske, E. H. Hill, D. Vila-Liarte, G. González-Rubio, C. Matricardi, A. Mihi and L. M. Liz-Marzán, ACS Appl. Mater. Interfaces, 2019, 11, 11763–11771 CrossRef CAS PubMed.
- L. Song, J. Chen, B. B. Xu and Y. Huang, ACS Nano, 2021, 15, 18822–18847 CrossRef CAS PubMed.
- H. Liu, J. Zeng, L. Song, L. Zhang, Z. Chen, J. Li, Z. Xiao, F. Su and Y. Huang, Nanoscale Horiz., 2022, 7, 554–561 RSC.
- L. Song, B. B. Xu, Q. Cheng, X. Wang, X. Luo, X. Chen, T. Chen and Y. Huang, Sci. Adv., 2021, 7, eabk2852 CrossRef CAS PubMed.
- X. Lu, Y. Huang, B. Liu, L. Zhang, L. Song, J. Zhang, A. Zhang and T. Chen, Chem. Mater., 2018, 30, 1989–1997 CrossRef CAS.
- H. Yamada, Y. Yamamoto and N. Tani, Chem. Phys. Lett., 1982, 86, 397–400 CrossRef CAS.
- H. Yamada and Y. Yamamoto, Surf. Sci., 1983, 134, 71–90 CrossRef CAS.
- B. P. Majee, Bhawna and A. K. Mishra, Mater. Res. Express, 2020, 6, 1250j1251 Search PubMed.
- J. Lin, J. Yu, O. U. Akakuru, X. Wang, B. Yuan, T. Chen, L. Guo and A. Wu, Chem. Sci., 2020, 11, 9414–9420 RSC.
- E. Proniewicz, A. Tąta, A. Wójcik, M. Starowicz, J. Pacek and M. Molenda, Phys. Chem. Chem. Phys., 2020, 22, 28100–28114 RSC.
- C. Liu, Q. Song, J. Chen, X. Li, J. Cai, Z. Lu, W. Li, N. X. Fang and S.-P. Feng, Adv. Mater. Interfaces, 2019, 6, 1900534 CrossRef.
- L. Yang, M. Gong, X. Jiang, D. Yin, X. Qin, B. Zhao and W. Ruan, J. Raman Spectrosc., 2015, 46, 287–292 CrossRef CAS.
- L. Yang, Y. Peng, Y. Yang, J. Liu, Z. Li, Y. Ma, Z. Zhang, Y. Wei, S. Li, Z. Huang and N. V. Long, ACS Appl. Nano Mater., 2018, 1, 4516–4527 CrossRef CAS.
- X. Xue, W. Ji, Z. Mao, H. Mao, Y. Wang, X. Wang, W. Ruan, B. Zhao and J. R. Lombardi, J. Phys. Chem. C, 2012, 116, 8792–8797 CrossRef CAS.
- H. A. O. Wen, Mol. Phys., 1996, 88, 281–290 CrossRef CAS.
- H. Zhang, S. Huang, X. Yang, R. Yuan and Y. Chai, Sens. Actuators, B, 2021, 343, 130142 CrossRef CAS.
- L. Yang, Y. Wei, Y. Song, Y. Peng, Y. Yang and Z. Huang, Mater. Des., 2020, 193, 108808 CrossRef CAS.
- X. Yan, Y. Ai-Hua, J. Wen-Qi and W. De-Ping, Chin. J. Inorg. Chem., 2018, 34, 1392–1398 Search PubMed.
- Z. Tian, H. Bai, C. Chen, Y. Ye, Q. Kong, Y. Li, W. Fan, W. Yi and G. Xi, iScience, 2019, 19, 836–849 CrossRef CAS PubMed.
- Y. Li, R. Du, W. Li, J. Li, H. Yang, H. Bai, M. Zou and G. Xi, Anal. Chem., 2021, 93, 12360–12366 CrossRef CAS PubMed.
- Q. Zhang, X. Li, Q. Ma, Q. Zhang, H. Bai, W. Yi, J. Liu, J. Han and G. Xi, Nat. Commun., 2017, 8, 14903 CrossRef PubMed.
- M. K. Patil, S. H. Gaikwad and S. P. Mukherjee, J. Phys. Chem. C, 2020, 124, 21082–21093 CrossRef CAS.
- Y. Wang, W. Ruan, J. Zhang, B. Yang, W. Xu, B. Zhao and J. R. Lombardi, J. Raman Spectrosc., 2009, 40, 1072–1077 CrossRef CAS.
- Y. Wang, H. Hu, S. Jing, Y. Wang, Z. Sun, B. Zhao, C. Zhao and J. R. Lombardi, Anal. Sci., 2007, 23, 787–791 CrossRef CAS PubMed.
- A. Li, J. Lin, Z. Huang, X. Wang and L. Guo, iScience, 2018, 10, 1–10 CrossRef CAS PubMed.
- X. Wang, W. Shi, Z. Jin, W. Huang, J. Lin, G. Ma, S. Li and L. Guo, Angew. Chem., Int. Ed., 2017, 56, 9851–9855 CrossRef CAS PubMed.
- X. Wang, W. Shi, S. Wang, H. Zhao, J. Lin, Z. Yang, M. Chen and L. Guo, J. Am. Chem. Soc., 2019, 141, 5856–5862 CrossRef CAS PubMed.
- Y. Wang, Z. Sun, Y. Wang, H. Hu, B. Zhao, W. Xu and J. R. Lombardi, Spectrochim. Acta, Part A, 2007, 66, 1199–1203 CrossRef PubMed.
- M. Keshavarz, A. K. M. R. H. Chowdhury, P. Kassanos, B. Tan and K. Venkatakrishnan, Sens. Actuators, B, 2020, 323, 128703 CrossRef CAS.
- S. Huh, J. Park, Y. S. Kim, K. S. Kim, B. H. Hong and J.-M. Nam, ACS Nano, 2011, 5, 9799–9806 CrossRef CAS PubMed.
- D. Glass, E. Cortés, S. Ben-Jaber, T. Brick, W. J. Peveler, C. S. Blackman, C. R. Howle, R. Quesada-Cabrera, I. P. Parkin and S. A. Maier, Adv. Sci., 2019, 6, 1901841 CrossRef CAS PubMed.
- X. Zheng, F. Ren, S. Zhang, X. Zhang, H. Wu, X. Zhang, Z. Xing, W. Qin, Y. Liu and C. Jiang, ACS Appl. Mater. Interfaces, 2017, 9, 14534–14544 CrossRef CAS PubMed.
- Y. Wang, M. Zhang, H. Ma, H. Su, A. Li, W. Ruan and B. Zhao, ACS Appl. Mater. Interfaces, 2021, 13, 35038–35045 CrossRef CAS PubMed.
- J. Li, H. Bai, J. Zhai, W. Li, W. Fan and G. Xi, Chem. Commun., 2019, 55, 4679–4682 RSC.
- Samriti, V. Rajput, R. K. Gupta and J. Prakash, J. Mater. Chem. C, 2022, 10, 73–95 RSC.
- H. Guan, Z. Tian, Q. Kong and G. Xi, Chem. Commun., 2021, 57, 4815–4818 RSC.
- C. Gu, D. Li, S. Zeng, T. Jiang, X. Shen and H. Zhang, Nanoscale, 2021, 13, 5620–5651 RSC.
- Y. Peng, C. Lin, M. Tang, L. Yang, Y. Yang, J. Liu, Z. Huang and Z. Li, Appl. Surf. Sci., 2020, 509, 145376 CrossRef CAS.
- X. Pan, M.-Q. Yang, X. Fu, N. Zhang and Y.-J. Xu, Nanoscale, 2013, 5, 3601–3614 RSC.
- D. Luo, Z. Zhang, G. Li, S. Cheng, S. Li, J. Li, R. Gao, M. Li, S. Sy, Y.-P. Deng, Y. Jiang, Y. Zhu, H. Dou, Y. Hu, A. Yu and Z. Chen, ACS Nano, 2020, 14, 4849–4860 CrossRef CAS PubMed.
- Y. Cao, P. Liang, Q. Dong, D. Wang, D. Zhang, L. Tang, L. Wang, S. Jin, D. Ni and Z. Yu, Anal. Chem., 2019, 91, 8683–8690 CrossRef CAS PubMed.
- X.-D. Zheng, F. Ren, H.-Y. Wu, W.-J. Qin and C.-Z. Jiang, Nanotechnology, 2018, 29, 155301 CrossRef PubMed.
- L. Kang, J. Chu, H. Zhao, P. Xu and M. Sun, J. Mater. Chem. C, 2015, 3, 9024–9037 RSC.
- H. Lai, F. Xu, Y. Zhang and L. Wang, J. Mater. Chem. B, 2018, 6, 4008–4028 RSC.
- M. D. Stoller, S. Park, Y. Zhu, J. An and R. S. Ruoff, Nano Lett., 2008, 8, 3498–3502 CrossRef CAS PubMed.
- M. C. Dalfovo, G. I. Lacconi, M. Moreno, M. C. Yappert, G. U. Sumanasekera, R. C. Salvarezza and F. J. Ibañez, ACS Appl. Mater. Interfaces, 2014, 6, 6384–6391 CrossRef CAS PubMed.
- X. Ling, L. Xie, Y. Fang, H. Xu, H. Zhang, J. Kong, M. S. Dresselhaus, J. Zhang and Z. Liu, Nano Lett., 2010, 10, 553–561 CrossRef CAS PubMed.
- H. Xu, L. Xie, H. Zhang and J. Zhang, ACS Nano, 2011, 5, 5338–5344 CrossRef CAS PubMed.
- H. Xu, Y. Chen, W. Xu, H. Zhang, J. Kong, M. S. Dresselhaus and J. Zhang, Small, 2011, 7, 2945–2952 CrossRef CAS PubMed.
- X. Ling, J. Wu, W. Xu and J. Zhang, Small, 2012, 8, 1365–1372 CrossRef CAS PubMed.
- F. Yin, S. Wu, Y. Wang, L. Wu, P. Yuan and X. Wang, J. Solid State Chem., 2016, 237, 57–63 CrossRef CAS.
- E. Er, H.-L. Hou, A. Criado, J. Langer, M. Möller, N. Erk, L. M. Liz-Marzán and M. Prato, Chem. Mater., 2019, 31, 5725–5734 CrossRef CAS.
- D. Yan, W. Qiu, X. Chen, L. Liu, Y. Lai, Z. Meng, J. Song, Y. Liu, X.-Y. Liu and D. Zhan, J. Phys. Chem. C, 2018, 122, 14467–14473 CrossRef CAS.
- M. Li, Y. Gao, X. Fan, Y. Wei, Q. Hao and T. Qiu, Nanoscale Horiz., 2021, 6, 186–191 RSC.
- C. Weng, Y. Luo, B. Wang, J. Shi, L. Gao, Z. Cao and G. Duan, J. Mater. Chem. C, 2020, 8, 14138–14145 RSC.
- L. Tao, K. Chen, Z. Chen, C. Cong, C. Qiu, J. Chen, X. Wang, H. Chen, T. Yu, W. Xie, S. Deng and J.-B. Xu, J. Am. Chem. Soc., 2018, 140, 8696–8704 CrossRef CAS PubMed.
- P. Miao, J.-K. Qin, Y. Shen, H. Su, J. Dai, B. Song, Y. Du, M. Sun, W. Zhang, H.-L. Wang, C.-Y. Xu and P. Xu, Small, 2018, 14, 1704079 CrossRef PubMed.
- P. Karthick Kannan, P. Shankar, C. Blackman and C.-H. Chung, Adv. Mater., 2019, 31, 1803432 CrossRef PubMed.
- X. Ling, W. Fang, Y.-H. Lee, P. T. Araujo, X. Zhang, J. F. Rodriguez-Nieva, Y. Lin, J. Zhang, J. Kong and M. S. Dresselhaus, Nano Lett., 2014, 14, 3033–3040 CrossRef CAS PubMed.
- Y. Lee, H. Kim, J. Lee, S. H. Yu, E. Hwang, C. Lee, J.-H. Ahn and J. H. Cho, Chem. Mater., 2016, 28, 180–187 CrossRef CAS.
- L. Sun, H. Hu, D. Zhan, J. Yan, L. Liu, J. S. Teguh, E. K. L. Yeow, P. S. Lee and Z. Shen, Small, 2014, 10, 1090–1095 CrossRef CAS PubMed.
- Z. Liu, S. Li, C. Hu, W. Zhang, H. Zhong and Z. Guo, J. Raman Spectrosc., 2013, 44, 75–80 CrossRef CAS.
- J. C. Reed, H. Zhu, A. Y. Zhu, C. Li and E. Cubukcu, Nano Lett., 2012, 12, 4090–4094 CrossRef CAS PubMed.
- V. Kaushik, H. L. Kagdada, D. K. Singh and S. Pathak, Appl. Surf. Sci., 2022, 574, 151724 CrossRef CAS.
- E. Er, A. Sánchez-Iglesias, A. Silvestri, B. Arnaiz, L. M. Liz-Marzán, M. Prato and A. Criado, ACS Appl. Mater. Interfaces, 2021, 13, 8823–8831 CrossRef CAS PubMed.
- C. Lin, S. Liang, Y. Peng, L. Long, Y. Li, Z. Huang, N. V. Long, X. Luo, J. Liu, Z. Li and Y. Yang, Nano-Micro Lett., 2022, 14, 75 CrossRef CAS PubMed.
- W. Xu, X. Ling, J. Xiao, M. S. Dresselhaus, J. Kong, H. Xu, Z. Liu and J. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9281–9286 CrossRef CAS PubMed.
- P. Hoskin, T. Ajithkumar and V. Goh, Imaging for Clinical Oncology, Oxford University Press, 2021 Search PubMed.
- H. Brody, Nature, 2020, 579, S1 CrossRef CAS PubMed.
- M. Heinrich, L. Hofmann, H. Baurecht, P. M. Kreuzer, H. Knüttel, M. F. Leitzmann and C. Seliger, Nat. Med., 2022, 28, 852–859 CrossRef CAS PubMed.
- S. Haider, T. Azam, G. Leandro, H. Heinrich and H. Rumana, Eye, 2022 DOI:10.1038/s41433-022-02185-1.
- L. M. Almehmadi, S. M. Curley, N. A. Tokranova, S. A. Tenenbaum and I. K. Lednev, Sci. Rep., 2019, 9, 12356 CrossRef PubMed.
- V. M. Szlag, R. S. Rodriguez, J. He, N. Hudson-Smith, H. Kang, N. Le, T. M. Reineke and C. L. Haynes, ACS Appl. Mater. Interfaces, 2018, 10, 31825–31844 CrossRef CAS PubMed.
- Z. Wang, S. Zong, L. Wu, D. Zhu and Y. Cui, Chem. Rev., 2017, 117, 7910–7963 CAS.
- M. Bhamidipati, H.-Y. Cho, K.-B. Lee and L. Fabris, Bioconjugate Chem., 2018, 29, 2970–2981 CrossRef CAS PubMed.
- K. M. Koo, J. Wang, R. S. Richards, A. Farrell, J. W. Yaxley, H. Samaratunga, P. E. Teloken, M. J. Roberts, G. D. Coughlin, M. F. Lavin, P. N. Mainwaring, Y. Wang, R. A. Gardiner and M. Trau, ACS Nano, 2018, 12, 8362–8371 CrossRef CAS PubMed.
- Y. Yang, C. Song, J. Zhang, J. Chao, H. M. Luong, Y. Zhao and L. Wang, Nanoscale, 2022, 14, 4538–4547 RSC.
- Y. Liu, N. Lyu, A. Rodger and Y. Wang, in Principles and Clinical Diagnostic Applications of Surface-Enhanced Raman Spectroscopy, ed. Y. Wang, Elsevier, 2022, pp. 225–280, DOI:10.1016/B978-0-12-821121-2.00008-1.
- W. Zhang, L. Wang, D. Li, D. H. Campbell, B. J. Walsh, N. H. Packer, Q. Dong, E. Wang and Y. Wang, Anal. Methods, 2022, 14, 2255–2265 RSC.
- K. B. Shanmugasundaram, J. Li, A. I. Sina, A. Wuethrich and M. Trau, Mater. Adv., 2022, 3, 1459–1471 RSC.
- A. Khalifa and D. C. Rockey, Curr. Opin. Gastroenterol., 2020, 36, 184–191 CrossRef PubMed.
- B. Salzman, E. Collins and L. Hersh, Am. Fam. Physician, 2019, 99, 505–514 Search PubMed.
- J. Streicher, B. L. Meyerson, V. Karivedu and A. Sidana, Ther. Adv. Urol., 2019, 11, 1–8 Search PubMed.
- H. Liu, X. Gao, C. Xu and D. Liu, Theranostics, 2022, 12, 1870–1903 CrossRef CAS PubMed.
- W. Zhang, L. Jiang, R. J. Diefenbach, D. H. Campbell, B. J. Walsh, N. H. Packer and Y. Wang, ACS Sens., 2020, 5, 764–771 CrossRef CAS PubMed.
- J. Wang, Y.-C. Kao, Q. Zhou, A. Wuethrich, M. S. Stark, H. Schaider, H. P. Soyer, L. L. Lin and M. Trau, Adv. Funct. Mater., 2022, 32, 2010296 CrossRef CAS.
- S. Jiang, Q. Li, C. Wang, Y. Pang, Z. Sun and R. Xiao, ACS Sens., 2021, 6, 852–862 CrossRef CAS PubMed.
- F. Beffara, J. Perumal, A. Puteri Mahyuddin, M. Choolani, S. A. Khan, J. L. Auguste, S. Vedraine, G. Humbert, U. S. Dinish and M. Olivo, J. Biophotonics, 2020, 13, e201960120 CrossRef PubMed.
- H. Shin, D. Seo and Y. Choi, Molecules, 2020, 25, 5209 CrossRef CAS PubMed.
- L. Guerrini, E. Garcia-Rico, A. O'Loghlen, V. Giannini and R. A. Alvarez-Puebla, Cancers, 2021, 13, 2179 CrossRef CAS PubMed.
- S. C.-H. Tsao, J. Wang, Y. Wang, A. Behren, J. Cebon and M. Trau, Nat. Commun., 2018, 9, 1482 CrossRef PubMed.
- S. Schlücker, World Scientific Reference on Plasmonic Nanomaterials 2022, https://www.worldscientific.com/doi/10.1142/12236-vol5#t=aboutBook Search PubMed.
- G. Sanità, B. Carrese and A. Lamberti, Front. Mol. Biosci., 2020, 7, 587012 CrossRef PubMed.
- N. M. Cordina, W. Zhang, N. H. Packer and Y. Wang, Mol. Omics, 2020, 16, 339–344 RSC.
- N. Lyu, B. Pedersen, E. Shklovskaya, H. Rizos, M. P. Molloy and Y. Wang, Exploration, 2022, 2, 20210176 CrossRef.
- J. Li, A. Wuethrich, A. A. I. Sina, H.-H. Cheng, Y. Wang, A. Behren, P. N. Mainwaring and M. Trau, Nat. Commun., 2021, 12, 1087 CrossRef CAS PubMed.
- J. Wang, A. Wuethrich, A. A. I. Sina, R. E. Lane, L. L. Lin, Y. Wang, J. Cebon, A. Behren and M. Trau, Sci. Adv., 2020, 6, eaax3223 CrossRef CAS PubMed.
- J. Wang, W. Anderson, J. Li, L. L. Lin, Y. Wang and M. Trau, J. Colloid Interface Sci., 2019, 537, 536–546 CrossRef CAS PubMed.
- R. Mei, Y. Wang, X. Zhao, Q. Kang, D. Shen and L. Chen, Anal. Chem., 2021, 93, 16590–16597 CrossRef CAS PubMed.
- D. Song, R. Yang, S. Fang, Y. Liu, F. Long and A. Zhu, Microchim. Acta, 2018, 185, 491 CrossRef PubMed.
- S. Juergensen, P. Kusch and S. Reich, Phys. Status Solidi B, 2020, 257, 2000295 CrossRef CAS.
- H. Y. Lau, Y. Wang, E. J. H. Wee, J. R. Botella and M. Trau, Anal. Chem., 2016, 88, 8074–8081 CrossRef CAS PubMed.
- J. Ye, J. A. Hutchison, H. Uji-i, J. Hofkens, L. Lagae, G. Maes, G. Borghs and P. Van Dorpe, Nanoscale, 2012, 4, 1606–1611 RSC.
- A. Mao, X. Jin, X. Gu, X. Wei and G. Yang, J. Mol. Struct., 2012, 1021, 158–161 CrossRef CAS.
- C. H. Sun, M. L. Wang, Q. Feng, W. Liu and C. X. Xu, Russ. J. Phys. Chem. A, 2015, 89, 291–296 CrossRef CAS.
- C. Li, Y. Huang, K. Lai, B. A. Rasco and Y. Fan, Food Control, 2016, 65, 99–105 CrossRef CAS.
- Y. Zeng, J.-Q. Ren, A.-G. Shen and J.-M. Hu, J. Am. Chem. Soc., 2018, 140, 10649–10652 CrossRef CAS PubMed.
- G. Fan, X. Gao, S. Xu, X. Li, Q. Zhang, C. Dai, Q. Xue and H. Wang, Chem. Commun., 2022, 58, 407–410 RSC.
- D. Lu, M. Ran, Y. Liu, J. Xia, L. Bi and X. Cao, Anal. Bioanal. Chem., 2020, 412, 7099–7112 CrossRef CAS PubMed.
- Y. T. Yang, I. L. Hsu, T. Y. Cheng, W. J. Wu, C. W. Lee, T. J. Li, C. I. Cheung, Y. C. Chin, H. C. Chen, Y. C. Chiu, C. C. Huang and M. Y. Liao, Anal. Chem., 2019, 91, 8213–8220 CrossRef CAS PubMed.
- H. Wen, H. Wang, J. Hai, S. He, F. Chen and B. Wang, ACS Sustainable Chem. Eng., 2019, 7, 5200–5208 CrossRef CAS.
- C. Fan, N. Zhao, K. Cui, G. Chen, Y. Chen, W. Wu, Q. Li, Y. Cui, R. Li and Z. Xiao, ACS Sens., 2021, 6, 3234–3241 CrossRef CAS PubMed.
- M. Hou, D. He, H. Bu, H. Wang, J. Huang, J. Gu, R. Wu, H.-W. Li, X. He and K. Wang, Analyst, 2020, 145, 6232–6236 RSC.
- M. Muhammad, C.-S. Shao, C. Liu and Q. Huang, ACS Appl. Bio Mater., 2021, 4, 8386–8395 CrossRef CAS PubMed.
- K. K. Reza, S. Dey, A. Wuethrich, W. Jing, A. Behren, F. Antaw, Y. Wang, A. A. I. Sina and M. Trau, ACS Nano, 2021, 15, 11231–11243 CrossRef CAS PubMed.
- J. Wang, A. Wuethrich, R. J. Lobb, F. Antaw, A. A. Sina, R. E. Lane, Q. Zhou, C. Zieschank, C. Bell, V. F. Bonazzi, L. G. Aoude, S. Everitt, B. Yeo, A. P. Barbour, A. Möller and M. Trau, ACS Sens., 2021, 6, 3182–3194 CrossRef CAS PubMed.
- Y. Pang, J. Shi, X. Yang, C. Wang, Z. Sun and R. Xiao, Biosens. Bioelectron., 2020, 148, 111800 CrossRef CAS PubMed.
- Y. Xu, D. Zhang, J. Lin, X. Wu, X. Xu, O. U. Akakuru, H. Zhang, Z. Zhang, Y. Xie, A. Wu and G. Shao, Biomater. Sci., 2022, 10, 1812–1820 RSC.
- J. He, J. Dong, Y. Hu, G. Li and Y. Hu, Nanoscale, 2019, 11, 6089–6100 RSC.
- Q. Lv, H. Min, D.-B. Duan, W. Fang, G.-M. Pan, A.-G. Shen, Q.-Q. Wang, G. Nie and J.-M. Hu, Adv. Healthcare Mater., 2019, 8, 1801257 Search PubMed.
- J. Lu, W. Zhang, H. Wang, Y. Mao, D. Di, Q. Zhao and S. Wang, Colloids Surf., A, 2020, 603, 125212 CrossRef CAS.
- Y. Zhu, Q. Sun, Y. Liu, T. Ma, L. Su, S. Liu, X. Shi, D. Han and F. Liang, R. Soc. Open Sci., 2018, 5, 180159 CrossRef PubMed.
- M. Z. Quazi, U. Lee, S. Park, S. Shin, E. Sim, H. Son and N. Park, ACS Appl. Bio Mater., 2021, 4, 8377–8385 CrossRef CAS PubMed.
- A. Das, V. Arunagiri, H.-C. Tsai, A. Prasannan and R. S. Moirangthem, ACS Appl. Nano Mater., 2021, 4, 12278–12288 CrossRef CAS.
- P. T. Sujai, S. Shamjith, M. M. Joseph and K. K. Maiti, ACS Appl. Bio Mater., 2021, 4, 4962–4972 CrossRef CAS PubMed.
- J. Lin, W. Ren, A. Li, C. Yao, T. Chen, X. Ma, X. Wang and A. Wu, ACS Appl. Mater. Interfaces, 2020, 12, 4204–4211 CrossRef CAS PubMed.
- O. M. Elsharkasy, J. Z. Nordin, D. W. Hagey, O. G. de Jong, R. M. Schiffelers, S. E. Andaloussi and P. Vader, Adv. Drug Delivery Rev., 2020, 159, 332–343 CrossRef CAS PubMed.
- X. Zhang, H. Zhang, J. Gu, J. Zhang, H. Shi, H. Qian, D. Wang, W. Xu, J. Pan and H. A. Santos, Adv. Mater., 2021, 33, 2005709 CrossRef CAS PubMed.
- R. Haldavnekar, K. Venkatakrishnan and D. B. Tan, Biosens. Bioelectron., 2021, 190, 113407 CrossRef CAS PubMed.
- N. M. Ćulum, T. T. Cooper, G. I. Bell, D. A. Hess and F. Lagugné-Labarthet, Anal. Bioanal. Chem., 2021, 413, 5013–5024 CrossRef PubMed.
- S. K. Das, K. Pal, T. S. Bhattacharya, P. Karmakar and J. Chowdhury, Sens. Actuators, B, 2019, 299, 126962 CrossRef CAS.
- J. C. Fraire, S. Stremersch, D. Bouckaert, T. Monteyne, T. De Beer, P. Wuytens, R. De Rycke, A. G. Skirtach, K. Raemdonck, S. De Smedt and K. Braeckmans, ACS Appl. Mater. Interfaces, 2019, 11, 39424–39435 CrossRef CAS PubMed.
- Y. Liu, W. Zhang, T. H. Phan, W. Chrzanowski, A. Rodger and Y. Wang, Anal. Methods, 2020, 12, 5908–5915 RSC.
- H. J. Koster, T. Rojalin, A. Powell, D. Pham, R. R. Mizenko, A. C. Birkeland and R. P. Carney, Nanoscale, 2021, 13, 14760–14776 RSC.
- N. Cheng, J. Fu, D. Chen, S. Chen and H. Wang, NanoImpact, 2021, 21, 100296 CrossRef CAS PubMed.
- M. Erzina, A. Trelin, O. Guselnikova, B. Dvorankova, K. Strnadova, A. Perminova, P. Ulbrich, D. Mares, V. Jerabek, R. Elashnikov, V. Svorcik and O. Lyutakov, Sens. Actuators, B, 2020, 308, 127660 CrossRef CAS.
- X. Ren, W. Nam, P. Ghassemi, J. S. Strobl, I. Kim, W. Zhou and M. Agah, Microsyst. Nanoeng., 2020, 6, 47 CrossRef CAS PubMed.
- M. Jue, C.-G. Pack, S. Oh, B. Paulson, K. Lee and J. K. Kim, Materials, 2020, 13, 5321 CrossRef CAS PubMed.
- T.-H. Chang, K.-W. Chuang and Y.-C. Chang, J. Alloys Compd., 2022, 893, 162288 CrossRef CAS.
- P. Yin, G. Li, B. Zhang, H. Farjana, L. Zhao, H. Qin, B. Hu, J. Ou and J. Tian, Analyst, 2021, 146, 1949–1955 RSC.
- A. K. Ludwig, K. De Miroschedji, T. R. Doeppner, V. Börger, J. Ruesing, V. Rebmann, S. Durst, S. Jansen, M. Bremer, E. Behrmann, B. B. Singer, H. Jastrow, J. D. Kuhlmann, F. El Magraoui, H. E. Meyer, D. M. Hermann, B. Opalka, S. Raunser, M. Epple, P. A. Horn and B. Giebel, J. Extracell. Vesicles, 2018, 7, 1528109 CrossRef CAS PubMed.
- M. A. Rider, S. N. Hurwitz and D. G. Meckes, Sci. Rep., 2016, 6, 23978 CrossRef CAS PubMed.
- F. Haque, A. Zavabeti, B. Y. Zhang, R. S. Datta, Y. Yin, Z. Yi, Y. Wang, N. Mahmood, N. Pillai, N. Syed, H. Khan, A. Jannat, N. Wang, N. Medhekar, K. Kalantar-zadeh and J. Z. Ou, J. Mater. Chem. A, 2019, 7, 257–268 RSC.
- B. Y. Zhang, P. Yin, Y. Hu, C. Szydzik, M. W. Khan, K. Xu, P. Thurgood, N. Mahmood, C. Dekiwadia, S. Afrin, Y. Yang, Q. Ma, C. F. McConville, K. Khoshmanesh, A. Mitchell, B. Hu, S. Baratchi and J. Z. Ou, Biosens. Bioelectron., 2022, 198, 113814 CrossRef CAS PubMed.
- Z. Yan, S. Dutta, Z. Liu, X. Yu, N. Mesgarzadeh, F. Ji, G. Bitan and Y.-H. Xie, ACS Sens., 2019, 4, 488–497 CrossRef CAS PubMed.
- S. Adesoye and K. Dellinger, Sens. Biosensing Res., 2022, 37, 100499 CrossRef.
| This journal is © The Royal Society of Chemistry 2022 |



