Open Access Article
Open Access ArticleNanovector-mediated exogenous delivery of dsRNA induces silencing of target genes in very young tomato flower buds†
B.
Molesini
 *,
F.
Pennisi‡
,
C.
Cressoni‡
*,
F.
Pennisi‡
,
C.
Cressoni‡
 ,
N.
Vitulo
,
V.
Dusi
,
A.
Speghini
,
N.
Vitulo
,
V.
Dusi
,
A.
Speghini
 and
T.
Pandolfini
and
T.
Pandolfini

Department of Biotechnology, University of Verona, Strada Le Grazie, 15, 37134, Verona, Italy. E-mail: barbara.molesini@univr.it
First published on 14th September 2022
Abstract
RNA interference (RNAi) is a post-translational regulatory mechanism that controls gene expression in plants. This process can be artificially induced by double-stranded RNA (dsRNA) molecules with sequence homology to target mRNAs. Exogenously applied dsRNA on leaves has been shown to silence virulence genes of fungi and viruses, conferring protection to plants. Coupling dsRNA to nanoparticles has been demonstrated to prolong the silencing effect. The ability of exogenous dsRNA to silence endogenous genes in plants is currently under debate, mainly due to the difficulty in delivering dsRNA into plant tissues and organs. Our study aims to develop a method based on the exogenous application of dsRNA on tomato flowers for silencing endogenous genes controlling ovary growth. Two methods of dsRNA delivery into tomato flower buds (i.e., pedicel soaking and injection) were compared to test their efficacy in silencing the tomato Aux/IAA9 (SlIAA9) gene, which encodes for a known repressor of ovary growth. We examined the silencing effect of dsRNA alone and coupled to layered double hydroxide (LDHs) nanoparticles. We found that injection into the pedicel led to the silencing of SlIAA9 and the efficacy of the method was confirmed by choosing a different ovary growth repressor gene (SlAGAMOUS-like 6; SlAGL6). The coupling of dsRNA to LDHs increased the silencing effect in the case of SlIAA9. Silencing of the two repressors caused an increase in ovary size only when flower buds were treated with dsRNA coupled to LDHs. RNA-Seq of small RNAs showed that induction of RNAi was caused by the processing of injected dsRNA. In this work, we demonstrate for the first time that exogenous dsRNA coupled to LDHs can induce post-transcriptional gene silencing in the young tomato ovary by injection into the flower pedicel. This method represents a silencing tool for the study of the molecular changes occurring during the early stages of ovary/fruit growth as a consequence of downregulation of target genes, without the need to produce transgenic plants stably expressing RNAi constructs.
Introduction
RNA interference (RNAi) is an evolutionarily conserved sequence-specific gene inactivation process used by eukaryotes to regulate the expression of endogenous genes. This process also functions in plants as a defence mechanism against pests and pathogens.1–3 RNAi is triggered by small non-coding RNAs (sRNAs), which include micro RNAs (miRNAs) generated by Dicer-like endoribonuclease (DCL) activity on RNA precursors with stem-loop structures, and small interfering RNAs (siRNAs) derived from DCL activity on double-stranded RNA (dsRNA) molecules.2,4 Mature sRNAs are loaded into Argonaute-containing RNA-induced silencing complexes leading to the degradation or translational repression of homologous transcripts.2 RNAi-based technology is a tool employed for the discovery of gene function, and also applied in crops to confer disease protection and improve other traits of agronomic interest, including the nutritional value of fruits,5,6 and fruit production in the absence of fertilization.7–14In functional studies, the RNAi pathway can be activated by stably expressing dsRNA through hairpin or artificial miRNA-based constructs.15 However, the production of transgenic silenced plants, besides being a time-consuming process, is not feasible for those species and/or genotypes that are recalcitrant to transformation and/or regeneration. Alternatively, to obtain the first insights on the role of a gene, transient transformation can be used by infiltrating plant organs with Agrobacterium tumefaciens carrying viral or binary vectors containing the silencing construct.16 Recently, the existence of a communication mechanism based on sRNA trafficking between some fungal pathogens and their respective host plants, called bidirectional cross-kingdom RNAi, has been demonstrated.17 During infection, the fungal pathogen delivers sRNAs into the host plant and uses the host RNAi machinery to suppress defence genes.18 The host plant, in turn, produces sRNAs that, when internalized by the fungus, silence virulence genes.17–21 This discovery led to the development of a strategy for crop disease management based on the exogenous application of sRNAs or dsRNAs on plant surfaces.18,22 This new generation of RNAi-based biopesticides represents an environmentally sustainable system as an alternative to chemical pesticides.3,23–27
For practical application in crop protection, it is crucial to improve the stability of dsRNA and thus prolong the silencing effect. The use of layered double hydroxide (LDH) clay nanosheets as RNA nanovectors, represents an effective solution to promote long-term silencing effects.25 Indeed, the application of naked dsRNAs targeted to viral genes, protected plants for 5 days, whereas treatment with dsRNAs coupled to LDHs extended protection for up to 20 days.25,28 Furthermore, coupling RNA to nanoparticles would facilitate their penetration into plant cells.25,29 There is evidence that nanoparticles can passively enter natural plant openings (e.g., stomata) and those of reduced size can also pass through the cell wall.29 Exogenous RNA treatment may be used to silence endogenous genes, but, so far, a limited number of reports demonstrate the effectiveness of this approach for downregulating plant genes30–34 or transgenes.35 Delivery of RNA molecules within plant tissues can be achieved using different methods such as high-pressure spraying on leaves, injection into the trunk, foliar abrasion, root and leaf pedicel soaking.26,31,36–38 Once inside the cell, sRNAs can move over short distances most likely through plasmodesmata, and over long distances through the vasculature.39–41 Setting the optimal mode of application for each plant organ is a critical parameter for promoting dsRNA entry. To the best of our knowledge, there is only one study in the literature concerning the delivery of exogenous dsRNA into reproductive organs, describing the silencing of the MYB1 gene in flower buds of Dendrobium hybrida.33 Except for this example, the potential use of exogenous dsRNAs for targeting genes implicated in the ovary to fruit transition has never been explored. In this work, we applied exogenous dsRNA to very young tomato flower buds to silence endogenous genes expressed in the ovary. To test the silencing efficacy, we selected two target transcripts encoding for known ovary growth repressors (i.e., SlIAA9 and SlAGL6), whose silencing by stable transformation resulted in the initiation of fruit growth in the absence of fertilization.7,42 We compared two forms of dsRNA administration (i.e., flower pedicel soaking and injection) and investigated the effects produced by naked and nanovector-coupled dsRNAs. Reduced expression of SlIAA9 and SlAGL6 and the presence of target-specific small RNAs demonstrated that dsRNAs delivered by flower pedicel injection were able to trigger the RNAi mechanism in flower buds. Notably, the coupling of dsRNAs to LDHs stabilized the effects of the RNA molecules resulting in the relief of ovary growth repression in unfertilized flower buds. Optimization of this strategy could potentially open new opportunities for the future use of dsRNA treatment on flower buds for qualitative and quantitative crop-yield improvement.
Experimental
Plant material
Tomato (Solanum lycopersicum) cv UC82 plants were used in this study. After germination in soil, seedlings were transferred in pots (25 cm diameter) and grown in the glasshouse with a 12 h light/dark cycle (22–28 °C day, 17–18 °C night). For treatments, flower buds were obtained from a population of 8–10 plants.In vitro synthesis of dsRNA
Target sequences for dsRNA production were amplified from a cDNA template using primers containing the T7 RNA polymerase promoter sequence at both ends. After purification with the “Nucleospin Gel and PCR purification kit” (Macherey-Nagel), 0.5 μg of the DNA fragments were used as templates for in vitro transcription using “HiScribe™ T7 Quick High Yield RNA Synthesis Kit” (BioLabs). The following forward (F) and reverse (R) primers were used: for dsIAA9 (F, 5′-TAATACGACTCACTATAGGGAGATTGGCCACCCATTCGATCTTTTAG-3′ and R, 5′-TAATACGACTCACTATAGGGAGAAGACAAACTCCAATATCAAACGG-3′), for dsAGL6 (F, 5′-TAATACGACTCACTATAGGGAGAGAACAAATCAACCGTCAAGT-3′ and R, 5′-TAATACGACTCACTATAGGGAGAGTTACTCTCAACAGCCATGTTCC-3′). The synthesized RNA was purified using the ‘‘Monarch RNA Clean-up Kit’’ (BioLabs) and subsequently quantified by NanoDrop. The solution containing purified RNAs was denatured at 70 °C for 10 min, and then allowed to cool slowly to room temperature to obtain dsRNA.Synthesis details and physico-chemical characterization of the nanoparticles
The layered double hydroxide (LDHs) nanoparticles were prepared using a coprecipitation method followed by a microwave-assisted hydrothermal treatment, by adapting a method by Dong and collaborators.43 In brief, 3 mmol of Mg(NO3)2·6H2O (Sigma-Aldrich, 99.9%) and 1 mmol of Al(NO3)3·9H2O (Sigma-Aldrich, 99.9%) were dissolved in 10 ml of absolute ethanol (Sigma-Aldrich, 99%). A separate solution was prepared dissolving 8 mmol of NaOH (Sigma-Aldrich, 99.9%) in 10 ml of absolute ethanol. The metal-ion precursor solution was then added to the NaOH solution (1 ml min−1) under vigorous magnetic stirring. The resulting white dispersion was stirred for 40 min and then the solid product was separated by centrifugation (10 min, 6000×g). The obtained precipitate was dispersed in 15 ml of absolute ethanol and heat-treated for 15 min at 100 °C in a microwave reactor (Monowave400, Anton Paar). After the hydrothermal procedure, the resulting white product was centrifuged and washed twice with deionized water. Finally, the LDHs were dispersed in 40 ml of deionized water and left at room temperature for 6 days. After this aging, the LDHs were collected by centrifugation and the pellet was stable over several months. This procedure has been repeated several times and the reproducibility was demonstrated to be very high. To analyze the crystal structure of the LDHs, XRPD patterns were measured, on powdered samples. The LDHs were dried in an oven at 80 °C for a few hours and the obtained powders were finely homogenized in a mortar and deposited on a low background Si sample stage. The X-ray power diffraction patterns were measured with a Thermo ARL X’TRA powder diffractometer equipped with a Cu-anode X-ray source (λ = 1.5418 Å) and a Peltier Si(Li) cooled solid state detector. The XRPD patterns were collected with a scan rate of 0.04° s−1, with a measurement time of 1.0 s per step. The analysis of the patterns was carried out using the Inorganic Crystal Structure Database (ICSD). Hydrodynamic size and ζ-potential of the LDHs in water colloidal dispersion (0.5 mg ml−1) were measured using the Dynamic Light Scattering (DLS) technique with a Malvern Zetasizer Nano ZS instrument, operating with a HeNe laser at 633 nm. The particle size distribution and the morphology of the LDHs were investigated using a Transmission Electron Microscope (FEI TECNAI G2, operating at 80 kV). The samples were prepared by depositing the nanoparticles on a Formvar support film on copper grids (300 square mesh).Raman spectra of the LDHs in powder form were obtained using a Raman spectrometer (DXR2, Thermo Fisher Scientific) operating in microprobe mode with a 50× objective. The spectrometer was equipped with a holographic notch filter, which concurrently allowed high-efficiency and high-resolution spectral acquisitions. The sample was excited with a 632.8 nm He–Ne laser source at a power of 3 W. The Raman scattered radiation was collected using a single monochromator connected with an air-cooled charge-coupled device (CCD) detector.
Coupling of dsRNAs to LDHs and analysis of nuclease resistance
To define the optimal loading of dsRNA into LDHs, the procedure described by Mitter and collaborators (2017) was followed introducing slight modifications.25 Different mass ratios of dsRNA targeting the SlIAA9 transcript (dsIAA9) and LDHs were assayed. Briefly, 500 ng of dsIAA9 was coupled to LDHs at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 1
50, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 1
100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 (w/w). The coupling was performed in a volume of 10 μl at room temperature for 1 h using an orbital shaker. To confirm coupling, the mixtures were loaded on a 2% agarose gel. To test the stability of dsRNAs coupled to LDHs against cellular degrading enzymes, dsIAA9 alone and dsIAA9-LDHs were incubated in the presence of flower bud cell lysate. Fifty milligrams of flower buds were ground using 300 μl of CelLytic™ P Cell Lysis Reagent (Sigma-Aldrich). Ten microliters of the lysate were added to 10 μl of the dsIAA9-LDHs and the mixture was incubated at room temperature for 4, 6, and 24 h. As controls, dsIAA9 alone and dsIAA9-LDHs were incubated in CelLytic™ P Cell Lysis Reagent. As a positive control for ribonuclease activity, RNAse A was added to dsIAA9-LDHs samples at a final concentration of 20 μg ml−1. At each timepoint, an aliquot of the mixture was sampled and stored at −70 °C before loading onto a 2% agarose gel.
200 (w/w). The coupling was performed in a volume of 10 μl at room temperature for 1 h using an orbital shaker. To confirm coupling, the mixtures were loaded on a 2% agarose gel. To test the stability of dsRNAs coupled to LDHs against cellular degrading enzymes, dsIAA9 alone and dsIAA9-LDHs were incubated in the presence of flower bud cell lysate. Fifty milligrams of flower buds were ground using 300 μl of CelLytic™ P Cell Lysis Reagent (Sigma-Aldrich). Ten microliters of the lysate were added to 10 μl of the dsIAA9-LDHs and the mixture was incubated at room temperature for 4, 6, and 24 h. As controls, dsIAA9 alone and dsIAA9-LDHs were incubated in CelLytic™ P Cell Lysis Reagent. As a positive control for ribonuclease activity, RNAse A was added to dsIAA9-LDHs samples at a final concentration of 20 μg ml−1. At each timepoint, an aliquot of the mixture was sampled and stored at −70 °C before loading onto a 2% agarose gel.
Exogenous application of dsRNAs to the flower buds by pedicel soaking and syringe injection
For pedicel soaking, flower buds (1.0–1.1 cm long) were collected at pre-anthesis, leaving a pedicel segment of approximately 0.8–1.0 cm, and immediately emasculated, also removing the petals. After sterilization in 5% diluted commercial bleach for 10 min, flower buds were rinsed 3 times for 10 min in sterile water, blotted on sterile paper, and the pedicel was further trimmed at a final length of 0.5 cm. Drops (5 μl) of dsRNA-LDHs solution (500 ng/100 μg; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 w/w) were deposited under sterile conditions in the wells of a 96-well plate and then a single sterile flower bud was placed in each well. Uncoupled dsRNA and LDHs alone were also used to treat the flower buds. The plate was incubated for 15 min under a laminar flow hood since the uptake could be facilitated under air stress.38 After incubation, flower buds were transferred in crystal-clear polypropylene Eco2BOX vessels (Duchefa Biochemie) containing agar medium not supplemented with hormones (4.3 g l−1 Murashige and Skoog basal salts, 60 g l−1 sucrose, 100 mg l−1 myoinositol, 3 mg l−1 glycine, 0.1 mg l−1 thiamine, 0.1 mg l−1 pyridoxine, 0.5 mg l−1 nicotinic acid, 6 g l−1 agar, pH 5.7). Cefotaxime at a final concentration of 300 mg l−1 was added to the medium to hinder bacterial infection. Flower buds were cultivated in vitro for 14 days in a climatic chamber at a constant temperature of 25 °C during a 10/14 h light/dark cycle, with an average irradiance of 120 μmol m−2 s−1 of photosynthetically active radiation. After two weeks of in vitro cultivation, flower buds were transferred for additional 14 days on a fresh medium and the pedicels were trimmed removing approximately 0.1 cm of tissue.
200 w/w) were deposited under sterile conditions in the wells of a 96-well plate and then a single sterile flower bud was placed in each well. Uncoupled dsRNA and LDHs alone were also used to treat the flower buds. The plate was incubated for 15 min under a laminar flow hood since the uptake could be facilitated under air stress.38 After incubation, flower buds were transferred in crystal-clear polypropylene Eco2BOX vessels (Duchefa Biochemie) containing agar medium not supplemented with hormones (4.3 g l−1 Murashige and Skoog basal salts, 60 g l−1 sucrose, 100 mg l−1 myoinositol, 3 mg l−1 glycine, 0.1 mg l−1 thiamine, 0.1 mg l−1 pyridoxine, 0.5 mg l−1 nicotinic acid, 6 g l−1 agar, pH 5.7). Cefotaxime at a final concentration of 300 mg l−1 was added to the medium to hinder bacterial infection. Flower buds were cultivated in vitro for 14 days in a climatic chamber at a constant temperature of 25 °C during a 10/14 h light/dark cycle, with an average irradiance of 120 μmol m−2 s−1 of photosynthetically active radiation. After two weeks of in vitro cultivation, flower buds were transferred for additional 14 days on a fresh medium and the pedicels were trimmed removing approximately 0.1 cm of tissue.
For injection into the distal part of the flower truss pedicel, a 0.5 ml insulin syringe with 30 G needle-0.3 × 8 mm was employed. For each injection, a volume of 50 μl of dsRNA-LDHs (5 μg:1 mg; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 w/w) or dsRNA alone was used. Prior to injection, flowers that were not at the correct developmental stage were removed from each truss, leaving only those at pre-anthesis (1–2 per truss, generally). After 6 h, flower buds were emasculated and either bagged to avoid pollination and left on plants for 5 days or detached, sterilized, and placed in vitro for 15 days following the cultivation conditions described above. As controls, injections using H2O and LDHs were performed. For phenotypical analysis, ovary weight was measured.
200 w/w) or dsRNA alone was used. Prior to injection, flowers that were not at the correct developmental stage were removed from each truss, leaving only those at pre-anthesis (1–2 per truss, generally). After 6 h, flower buds were emasculated and either bagged to avoid pollination and left on plants for 5 days or detached, sterilized, and placed in vitro for 15 days following the cultivation conditions described above. As controls, injections using H2O and LDHs were performed. For phenotypical analysis, ovary weight was measured.
RT-qPCR analysis
Total RNA extraction was performed using the “NucleoSpin RNA Plant” kit (Macherey-Nagel). Purified RNA was treated with DNase I, then first-strand cDNA was synthesized starting from 1 μg of RNA using the ‘‘ImProm-II Reverse Transcription System’’ (Promega) in a 20 μl reaction volume. Three/four cDNA samples derived from independent RNA extractions were synthesized. cDNA was amplified using the “Luna Universal qPCR Master mix” (Biolabs) on the QuantStudio3 Real-Time PCR system (Applied Biosystem). Melting curves from 60 °C to 98 °C were used to confirm the presence of a single amplification product. The 2−ΔΔCt method was used44 to calculate the difference between the expression level of the target gene and that of the housekeeping SAND gene (Solyc03g115810) as the internal control.10 Transcript levels of genes coding for SlIAA9, SlAGL6, and SAND were determined using the following primers: SlIAA9, F 5′-CCCTTGCACCCTTCCAAAGA-3′ and R 5′-GCACACGTGAGGTCTCCTTT-3′; SlAGL6 F 5′-TATTGCTAACCCCACCTCCC-3′ and R 5′-TTCCACTCTCCCTCTCCCCA-3′; and SAND F 5′- TTTGCTTGGAGGAACAGACG-3′ and R 5′-GCAAACAGAACCCCTGAATC-3′.Fluorescein labelling of LDHs
LDHs were coupled to fluorescein dye (100 μM); the fluorescein–LDHs were prepared by incubation of the negatively charged dye with positively charged LDHs in a weight ratio of fluorescein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LDH = 2
LDH = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The two mixtures of fluorescein and LDHs were mixed and kept under vigorous magnetic stirring for 2 h. At the end of the incubation, the resulting fluorescein–LDHs nanocomposite was precipitated by centrifugation (10 min, 6000×g) and washed three times with water to remove residual dye. The emission properties of fluorescein–LDHs in water dispersion were characterized through JASCO FP-8200 fluorimeter. The colloidal properties of fluorescein–LDHs nanocomposite were investigated by DLS technique, measuring the hydrodynamic diameter and ζ-potential. Optical images of dissected flower buds were acquired with a Leica MZ16F fluorescence stereomicroscope or a LEICA TCS-SP5 Upright Confocal-Multiphoton Microscope.
1. The two mixtures of fluorescein and LDHs were mixed and kept under vigorous magnetic stirring for 2 h. At the end of the incubation, the resulting fluorescein–LDHs nanocomposite was precipitated by centrifugation (10 min, 6000×g) and washed three times with water to remove residual dye. The emission properties of fluorescein–LDHs in water dispersion were characterized through JASCO FP-8200 fluorimeter. The colloidal properties of fluorescein–LDHs nanocomposite were investigated by DLS technique, measuring the hydrodynamic diameter and ζ-potential. Optical images of dissected flower buds were acquired with a Leica MZ16F fluorescence stereomicroscope or a LEICA TCS-SP5 Upright Confocal-Multiphoton Microscope.
sRNA sequencing
Preparation and sequencing of small RNA libraries were performed by Novogene (United Kingdom) starting from 3 μg of total RNA isolated from flower buds 5 days after injection with dsIAA9-LDHs, dsAGL6-LDHs, and LDHs. Libraries were constructed by ligating 3′ and 5′ adapters to the 3′ and 5′ ends of small RNAs, respectively; after cDNA synthesis, double-stranded cDNA libraries were generated by PCR enrichment. After purification and size selection, libraries with insertions between 18 and 40 bp were sequenced with the NovaSeq 6000 platform. The length distribution was analyzed to determine the composition of the three small RNA samples. Small RNA reads were mapped to the S. lycopersicum reference genome using Bowtie v1-3-0 (ref. 45) with 0 mismatches, to analyze their expression level and distribution. Small RNA reads were aligned to mRNA exons of SlIAA9 (GenBank accession NM_001278959) and SlAGL6 (GenBank accession NM_001361530) genes to find degraded mRNA fragments in the small RNA reads. The alignment was performed using blastn algorithm,46 setting the word–size parameter to 17. Only the reads mapping with an e-value equal or lower to 1 × 10−5 were considered for further analysis. The blastn output was parsed using a home-made Perl script to calculate the sequence coverage. Briefly, for each reference gene, the mapping coverage was calculated by counting the number of reads at each single position. The coverage plot was generated in R using the ggplot library.Results
Selection of target genes implicated in tomato ovary growth and double-stranded RNA synthesis
Fruit set is defined as the transition between the quiescent ovary before anthesis and the resumption of its growth after successful fertilization of the ovules. Before fertilization, the inhibition of ovary growth is imposed by the activity of repressor proteins; silencing of these repressors by stable RNAi allowed ovary growth before and/or independently of fertilization.47 To test the efficacy of exogenous dsRNAs to silence genes expressed in flower buds, we selected two repressors of ovary growth, Indole Acetic Acid 9 (SlIAA9) and SlAGAMOUS-LIKE 6 (SlAGL6). SlIAA9 is a negative regulator of the auxin response pathway belonging to the Aux/IAA family transcription factors,7 and SlAGL6, which is a MADS-box protein.42 Downregulation or mutation of SlIAA9 and SlAGL6 resulted in early ovary growth and parthenocarpic fruit development.7,42 If exogenously applied dsRNAs designed to target SlIAA9 (dsIAA9) and SlAGL6 (dsAGL6) reach the flower buds and generate small interference RNAs (siRNAs), we expect to observe downregulation of the respective transcripts and an increase in ovary size. The expression of SlIAA9 and SlAGL6 is quite constant in unfertilized flower buds, whereas it declines after fertilization.7,42 Therefore, we used emasculated flower buds collected before anthesis to be sure that the downregulation of target transcripts is solely due to exogenous dsRNAs. For the design of dsIAA9, we chose a DNA fragment corresponding to the portion of SlIAA9 (NM_001278959.3; 717 bp covering part of the 3′ coding sequence and part of the 3′ untranslated terminal region) used by Wang and co-workers (2005) to achieve SlIAA9 silencing (ESI file 1†). Concerning dsAGL6, we selected the portion of SlAGL6 (NM_001361530.1; 702 bp of the CDS) comprising the guide sequence for CRISPR/Cas9 editing used by Klap and collaborators (2017) (ESI file 1†). For in vitro transcription of dsRNAs, the DNA sequences were amplified using primers containing the T7 RNA polymerase promoter at both ends.Physico-chemical characterization of the LDH nanoparticles
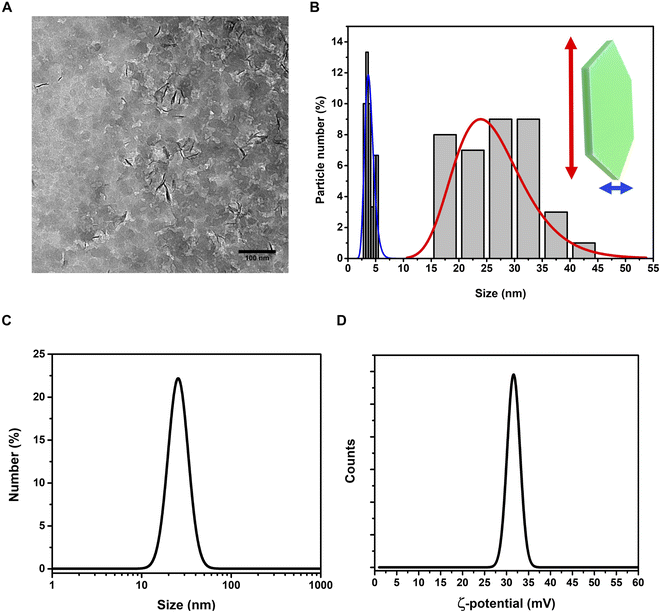
Layered Double Hydroxides (LDHs) or hydrotalcites are anionic clays that present a layered structure. This structure consists of positively-charged OH-layers, similar to those in brucite, Mg(OH)2, in which some of the Mg ions are substituted by a trivalent metal ion, such as Al3+, separated by hydrated anions in the interlayer domains.48 A broad range of compositions is possible, of the type [M1−x2+Mx′3+(OH)2][An−]x/n·yH2O, where M2+ and M′3+ are divalent and trivalent metal ions in octahedral sites within the OH-layers, with typical 0.17 < x < 0.33. The counterbalancing An− anions are virtually unlimited. Among the most common An−, are carbonates, nitrates, or sulfates. The X-ray Powder Diffraction (XRPD) pattern for the obtained LDHs is shown in Fig. 1A.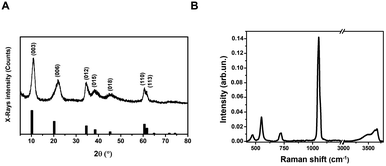 | ||
| Fig. 1 Structural and spectroscopic characterization of the LDH nanostructure. (A) XRPD pattern (vertical bars: from card n. 00-062-0583, PDF-4+ database); (B) Raman spectrum (λexc = 633 nm). | ||
Very broad features are observed, typical of LDH clay systems.49–52 In particular, the (003) reflection is a typical feature of hydrotalcite-like materials and its intensity is related to the degree of crystallinity of the material.53 The XRD pattern for LDHs was carefully analyzed using the PDF-4+ database54 and it was found that the pattern is typical of Mg/Al-hydrotalcites, compatible with a Mg6Al2(OH)16(NO3)2 composition.
The (003), (006), (110) and (113) reflections appear at 2θ values of 11.4°, 22.8°, 60° and 62°, respectively. These four basal reflections can also be ascribed to the lamellar structure. The reflection at (003) is particularly strong, indicating that the LDHs have a very good crystal structure.55
On the other hand, the (012), (015), and (018) relatively weak reflections are characteristic of LDH rhombohedral structures, containing NO3− as the interlayer anion.56 From the diffraction pattern, the d-spacing corresponding to the (003) reflection calculated from Bragg's law results of 8.1 Å. This value is in perfect agreement with that reported by Kloprogge and collaborators,57 for an Mg/Al-hydrotalcite containing nitrate ions as interlayer anionic species. To shed light on the structural properties of the obtained LDHs, we measured the Raman spectrum using excitation radiation of 633 nm (Fig. 1B). The observed Raman bands peak at 473, 553, 718, and 1050 cm−1; in addition, a very broad band is present at Raman shifts extending from 3200 to 3700 cm−1. These features are in very good agreement with those reported by Kloprogge et al. (2002) for a similar Mg/Al-hydrotalcite containing NO3− anions.57 As reported by Kloprogge and co-workers,57 the corresponding Raman bands are located at 470, 551, 716, and 1049 cm−1. This experimental evidence confirms the presence of nitrate ions in the LDH structure. TEM images of LDHs are shown in Fig. 2A and B; from the figures, it can be noted that the nanoparticles have a round platelet shape, confirmed by the evident presence of some nanoparticles accommodated in a vertical position with respect to the TEM grid.
The average size of the platelet flat face is approximately 26.2 ± 6.3 nm and the average platelet thickness is estimated to be around 3.2 ± 2.5 nm. The hydrodynamic size of LDHs in a colloidal dispersion, using water as a solvent, was 28.1 ± 9.2 nm (PDI = 0.249) (Fig. 2C), highlighting a good monodispersion of the nanoparticles. The ζ-potential of the LDHs in water dispersion was found to be positive, 31.4 ± 6.1 mV (Fig. 2D), confirming the excellent colloidal stability of the LDHs.
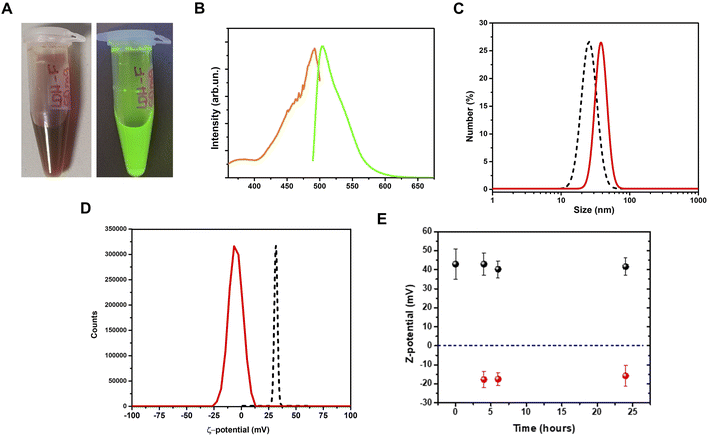
Adsorption of the fluorescein dye on LDHs
After the adsorption procedure, the fluorescein–LDHs were separated from the solvent by centrifugation. After proper washing with water, the fluorescein–LDHs were redispersed in water and the resulting colloidal dispersions showed very strong fluorescence under UV excitation with a Wood's lamp (Fig. 3A and B).By a DLS analysis on the colloidal fluorescein–LDHs dispersion we observed an increase of the hydrodynamic size of about 15 nm while the ζ-potential switched from positive to negative, reaching the average value of 13.1 ± 6.5 mV (Fig. 3C and D). These changes in the surface potential demonstrate that the negatively charged dye is strongly coupled to the LDHs. The colloidal stability of the nanoparticles was also tested by measuring the ζ-potential of both LDHs and fluorescein–LDHs in aqueous dispersions at successive times (Fig. 3E). Both LDH and fluorescein–LDH samples showed excellent colloidal stability within 24 h of dispersion.
Formation and stability of dsRNA–LDH complexes
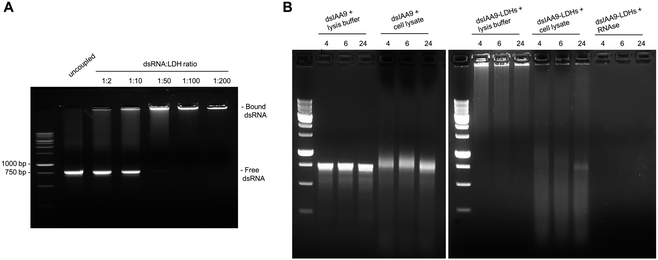
Conjugation of dsRNA to LDHs was monitored by agarose gel electrophoresis analysis (Fig. 4A). In vitro synthesized dsRNA designed to target SlIAA9 was coupled to increasing concentrations of LDHs. We observed a progressive inhibition of RNA migration with increasing dsRNA-LDH mass ratio ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 to 1
2 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. A dsRNA-LDH mass ratio of 1
200. A dsRNA-LDH mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 (Fig. 4A) resulted in total retention of dsRNA in the well with no visible smeared product in the gel; thus, this mass ratio was used for subsequent experiments. To test whether the coupling of dsRNA to LDHs could protect RNA against endogenous plant degrading enzymes, dsRNA alone or coupled to LDHs was incubated at room temperature for 4, 6, and 24 h in the presence of cell lysate obtained from flower buds (Fig. 4B). Spontaneous degradation of dsRNA was assessed by incubation with cell lysis buffer. As expected, dsRNA, either naked or coupled to LDHs, is less stable in the presence of cell lysate than the respective controls (i.e. samples incubated with lysis buffer) (Fig. 4B). However, even in the presence of cell lysate, part of dsRNA-LDHs remained coupled to LDHs and protected from degradation, as indicated by RNA staining visible in the wells of the gel at all incubation times (Fig. 4B).
200 (Fig. 4A) resulted in total retention of dsRNA in the well with no visible smeared product in the gel; thus, this mass ratio was used for subsequent experiments. To test whether the coupling of dsRNA to LDHs could protect RNA against endogenous plant degrading enzymes, dsRNA alone or coupled to LDHs was incubated at room temperature for 4, 6, and 24 h in the presence of cell lysate obtained from flower buds (Fig. 4B). Spontaneous degradation of dsRNA was assessed by incubation with cell lysis buffer. As expected, dsRNA, either naked or coupled to LDHs, is less stable in the presence of cell lysate than the respective controls (i.e. samples incubated with lysis buffer) (Fig. 4B). However, even in the presence of cell lysate, part of dsRNA-LDHs remained coupled to LDHs and protected from degradation, as indicated by RNA staining visible in the wells of the gel at all incubation times (Fig. 4B).
Delivery of exogenous dsRNA to flower buds by pedicel soaking
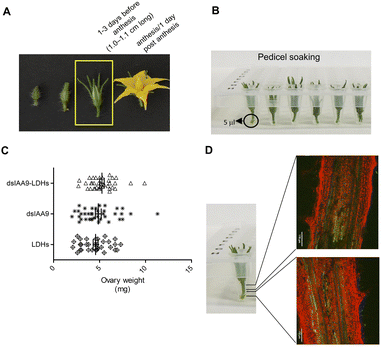
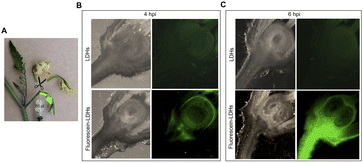
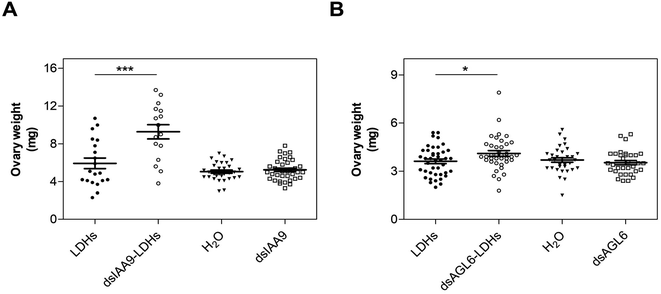
To ascertain whether dsRNA alone or coupled to LDHs was able to induce silencing of endogenous genes expressed in flower buds, pedicels of detached flower buds collected at the pre-anthesis stage (Fig. 5A) were soaked with dsIAA9 or dsIAA9-LDHs (Fig. 5B).The flower buds were placed in a 5 μl drop of dsRNA or dsRNA-LDHs solution for 15 min. They were then grown in vitro for 28 days on MS agar medium not supplemented with hormones. Under these in vitro conditions, the ovary is normally in a quiescent state and its growth is almost completely blocked. If repressors of the ovary growth (i.e. SlIAA9) are silenced, the ovary is expected to maintain its viability and increase its size at least partially. As shown in Fig. 5C, no significant differences in ovary weights were observed in flower buds treated with dsIAA9 or dsIAA9-LDHs, compared to those treated with LDHs alone. To investigate whether the lack of effect on ovary growth was due to an inefficient delivery system, we soaked the flower bud pedicels in fluorescein-labelled LDHs. After 24 h of application, the fluorescent signal remained confined to a few millimeters above the application point, without reaching the basal part of the flower buds (Fig. 5D).
Delivery of exogenous dsRNA by syringe injection into the pedicel of the flower truss
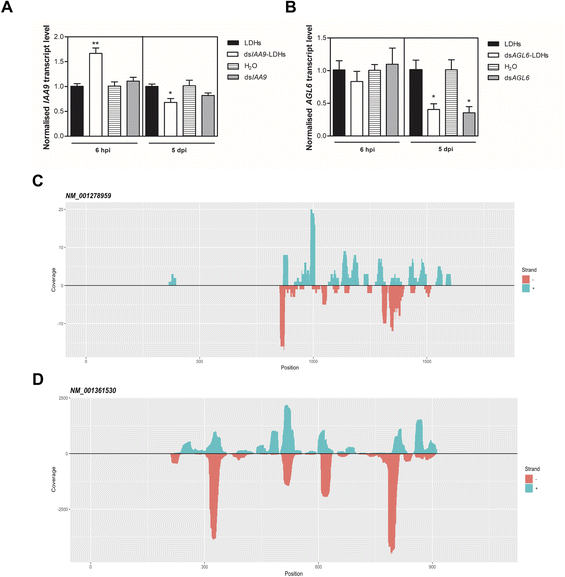
We applied exogenous dsIAA9 into the distal part of a flower truss pedicel by syringe injection (Fig. 6A). Prior to injection, we left in each truss only the flowers at 1–3 days before anthesis (Fig. 5A and 6A). Differently from the pedicel soaking, where exogenous molecules had to move approximately 0.5 cm to reach the ovary, RNAs injected in the pedicel needed to take a longer route, which was about 1.5 cm from the application point (Fig. 6A). We monitored the movement of fluorescein-labelled LDHs at 4 and 6 h post injection (hpi). At 4 hpi, the fluorescent signal was visible in the pedicel and the basal part of the flower buds (Fig. 6B); while at 6 hpi, a strong fluorescence signal was visible in the flower receptacle and inside the ovary (Fig. 6C). Therefore, after injecting dsIAA9 alone and coupled to LDHs, we monitored the downstream effect on SlIAA9 expression in flower buds at 6 hpi and 5 days post injection (dpi), a temporal window compatible with dsRNA processing into siRNAs.35,58 As negative controls, we injected LDHs and H2O. At 6 hpi, we did not observe a reduction in SlIAA9 expression in all treatments, whereas at 5 dpi, dsIAA9-LDHs only caused a statistically significant reduction (i.e. approximately 40%) in SlIAA9 mRNA level (Fig. 7A).The uncoupled dsIAA9 appeared less effective in causing SlIAA9 silencing as compared to dsIAA9 coupled to LDHs (Fig. 7A). Unexpectedly, dsIAA9-LDHs treatment at 6 hpi causes a transient increase in SlIAA9 expression. To confirm the efficacy of this delivery method, we repeated the experiment using a target gene coding for a different ovary growth repressor (SlAGL6). At 6 hpi, SlAGL6 expression remained unaltered, whereas after 5 dpi both dsAGL6 alone and dsAGL6-LDHs caused a statistically significant decrease in SlAGL6 transcript level of approximately 60% (Fig. 7B). Expression of both genes assayed after 15 dpi returned to baseline levels (i.e., expression in control samples) as expected for transient silencing (ESI file 2A†). To verify whether the dsRNA structure could interfere with the silencing of target transcripts, we analysed in dsRNA- and dsRNA-LDHs treated flower buds the transcript level of the non-intended target (i.e. SlAGL6 in dsIAA9-treated and SlIAA9 in dsAGL6-treated flower buds, respectively) (ESI file 2B†). These analyses revealed that the dsRNA specifically designed for a target mRNA was not able to silence unintended transcripts.
Analysis of sRNA populations generated by LDH-mediated delivery of exogenous dsRNAs
To evaluate if dsRNA was processed by Dicer, giving rise to silencing effector molecules (siRNAs), we performed the sequencing of sRNA populations in ovaries 5 days after dsIAA9 and dsAGL6 injection, using ovaries treated with LDHs alone as a comparative control. The length distribution of sRNAs revealed that 21–24 nt-long small RNAs are the most abundant categories in all 3 samples (ESI file 3 and 4†). dsIAA9-LDH- and LDH- treated ovaries displayed a similar sRNA length distribution; however, in dsIAA9-LDHs the abundance of 22 nt and 24 nt species was higher compared to that in LDH treated ones (ESI file 3A and 3B†). In the dsAGL6 treated sample, we observed the highest percentage (approximately 44%) of 24 nt-long sRNAs (ESI file 3C†). We mapped the sRNAs (21–24 nt range) obtained from dsIAA9-LDH and dsAGL6-LDH ovaries to their respective transcripts (Table 1).| Treatment | Target | #reads (+) | #reads (−) | Coverage (+) | Coverage (−) |
|---|---|---|---|---|---|
| a #reads: number of reads in the range 21-24 nt mapping on target transcript. Coverage: total abundance of the reads. +/−: reads mapping to sense or antisense strand of the target transcript. | |||||
| dsAGL6-LDHs | AGL6 | 555 | 502 | 5689 | 3979 |
| IAA9 | 1 | 9 | 2 | 12 | |
| dsIAA9-LDHs | AGL6 | 1 | 0 | 1 | 0 |
| IAA9 | 50 | 67 | 71 | 100 | |
| LDHs | AGL6 | 0 | 1 | 0 | 1 |
| IAA9 | 2 | 6 | 4 | 12 | |
We observed that when ovaries were treated with LDHs alone, the sRNAs mapping to SlIAA9 and SlAGL6 transcripts were almost absent. The same in the case of treatment with dsIAA9-LDHs and dsAGL6-LDHs, when sRNAs derived from non-cognate dsRNAs were considered (Table 1). In the dsAGL6-LDH treated sample, a total of 1057 unique reads with a coverage of 9668 were mapped to the SlAGL6 transcript (Table 1). A lower number (i.e. 117 reads with total coverage of 171) of cognate sRNAs was observed in the dsIAA9-LDH treated sample (Table 1). In both cases (Fig. 7C and D), the sRNAs were mapped to the portion of the sequence chosen for dsRNA production, with some areas generating more sRNAs than others. Following the identification of dsRNA-derived sRNAs, we tested whether the transient silencing led to increased ovary growth.
Hence, 6 hpi flower buds were excised from plants and grown in vitro for 15 days. The ovaries of flower buds treated with dsIAA9- and dsAGL6-coupled to LDHs displayed an increased weight compared to those treated with LDHs only (Fig. 8). On the other hand, the application of uncoupled dsRNA did not cause any effect on ovary growth (Fig. 8).
Discussion
The exogenous application of RNA molecules to plants has recently received increasing attention as an alternative strategy to transgenic plants expressing RNAi-based constructs to control traits of agronomic interest. In particular, numerous studies have shown that topical application of dsRNAs to leaves and fruits of various species can significantly reduce the symptoms caused by pests and pathogens.18,24,28,59 Once applied to leaves, dsRNAs artificially designed to target virulence genes are internalized by the pathogen where they trigger the RNAi of homologous transcripts compromising infection.18 In the near future, the development of RNAi-based biopesticides will represent an environmentally sustainable alternative to chemical pesticide use.27,60 In agriculture, the main problem with applying exogenous dsRNA is the low stability of the RNA molecule, which is highly sensitive to nuclease activity and/or environmental conditions such as excessive sunlight. To overcome these limitations, RNA molecules can be coupled to nanostructures of various shapes and sizes such as carbon nanotubes and clay nanosheets.25,61,62The use of nanovectors resulted in protection of RNA from degradation, promoting long-term silencing effects, and facilitating transport into plant cells.63,64 Furthermore, being as biodegradable as RNAs, these adjuvants are field compatible because they are environmentally friendly.25 Exogenous RNA can also be used to control the expression of endogenous genes and transgenes.
However, the examples documented in the literature are limited. What has emerged from these studies is that exogenous RNAs are more efficient at suppressing transgenes than endogenous ones.36,58,61,62,65 It has been suggested that the high level of transgene transcription, which is quite often regulated by strong promoters, can cause aberrant transcripts that produce secondary dsRNAs.58 Furthermore, transgenic constructs are generally produced using mature transcript sequences that lack introns and untranslated regions, sequences that may interfere with silencing efficiency.66–68 In studies describing the use of exogenous RNAs for silencing endogenes, varied RNA forms (i.e., siRNAs, miRNAs, and dsRNAs) and delivery modes have been exploited. Dubrovina et al. (2021) modulated anthocyanin biosynthesis in Arabidopsis by targeting a biosynthetic gene (i.e., chalcone synthase) and two transcription factors (i.e., MybL2 and ANAC032) that act as repressors of anthocyanin biosynthesis.35 Thirty-five micrograms of dsRNAs were applied to 4 week-old plant leaves by soft brushing, resulting in a marked reduction in target mRNA levels. Hendrix et al. (2021) demonstrated that foliar application of siRNAs caused silencing of three endogenes (magnesium cheletase subunit I, magnesium cheletase subunit H, and GENOMES UNCOUPLED4) in 14- to 21 day-old Nicotiana benthamiana plants.65 Another successful example of foliar application is that reported by Marcianò et al. (2021), where dsRNA targeting a downy mildew susceptibility gene (VviLBDIf7) was applied to fully developed leaves of Vitis vinifera.30 In addition, several studies have shown that roots and young seedlings can absorb both dsRNAs and miRNAs when supplied exogenously.31,32,69,70 A few reports describe the use of nanomaterials coupled to RNA cargos to silence endogenes. Jiang et al. (2014) demonstrated that applying dsRNAs mixed with cationic fluorescent nanoparticles to Arabidopsis root caused a significant reduction in expression of two developmental genes (SHOOT MERISTEMLESS and WHEREWOLF).70
Demirer and colleagues (2020) achieved silencing of ROQ1, implicated in disease resistance, after infiltrating N. benthamiana leaves with siRNAs loaded on single-walled carbon nanotubes.62 The use of exogenous RNA to silence genes expressed in reproductive organs has been poorly investigated, although it could provide a rapid way to study the function of genes involved in economically important processes such as fruit set and growth.
To our knowledge, there has been only one example of successful exogenous delivery of dsRNA to flowers.33 Repeated applications to flower buds of the orchid Dendrobium hybrida of a crude bacterial lysate containing dsRNA by gentle rubbing resulted in downregulation of DhMYB1 transcription factor and alterations in floral epidermal cell morphology.33
One of the most important challenges related to the topical application of dsRNA to reproductive organs, in particular to ovules/ovary, can be attributed to anatomical features. The pistil is the innermost whorl of the flower; therefore, it is not easily accessible to external treatments, and it has also a thick waxy coating that hinders the penetration of organic molecules. In addition, stomata, that are natural ways for RNA to penetrate plant tissues, are abundant in leaves but scarce in flowers. For instance, in tomato flowers, stomata have been reported to be scattered on the style but appeared to be absent on the ovary.71 Since flowers are sink organs, to which nutrients are actively transported, we decided to vehiculate dsRNA to flower buds via the vascular system instead of spraying dsRNA on the entire surface of the flowers.
We evaluated the efficiency of two methods for delivering dsRNA coupled to LDHs nanovectors to the flower pedicel, by soaking and direct injection. We found that soaking the pedicel of detached flower buds for 15 min with dsRNA-LDHs was ineffective in silencing the SlIAA9 mRNA within the ovary. Even though excised flower buds were maintained under air stress conditions to facilitate dsRNA-LDH movement,38 inefficient silencing seemed to be due to poor dsRNA-LDH penetration. Indeed, we observed that fluorescein-labelled LDHs did not reach the ovary even after 24 h of pedicel soaking. On the other hand, fluorescein-labelled LDHs directly injected in the pedicel of flower truss were transported to the ovary already after 6 h. The efficacy of RNA delivery by injection was confirmed by the downregulation of two target genes (i.e., SlIAA9 and SlAGL6) in the ovary. The effect of treatment was more pronounced for SlAGL6 than for SlIAA9; this could be due to several factors. For instance, it is known that the silencing efficacy may depend on the target gene, reflecting relative abundance and accessibility to the RNA silencing effector complex (i.e., RISC) of its transcript.72 The RNAi mechanism implies that dsRNA is processed by Dicer-like enzymes into 21–24 nt siRNAs, which guide the RISC to the target mRNA.73 Sequencing of small RNAs demonstrated that the injected dsRNA was processed in the ovaries, generating siRNAs of 21–24 nt. In agreement with the greater effect of injected dsRNA on SlAGL6 expression, the number of reads mapping to the SlAGL6 transcript was ten-fold higher than those specific for SlIAA9. This work shows the numerous advantages of using nanovectors to deliver RNA molecules in reproductive organs. We observed with in vitro experiments, conducted by incubating dsRNA-LDHs with cell lysate, that LDH nanovectors delayed RNA degradation. This could explain why in the case of SlIAA9, only RNA coupled to nanoparticles produced a statistically significant reduction in the target transcript level. Further evidence that nanovectors stabilize and thus potentiate the effect of dsRNA, came from the observation that ovary growth was stimulated, as a consequence of silencing the two repressors, only when dsRNA was coupled to LDHs.
Conclusions
Our work demonstrates that exogenously delivered dsRNA coupled to LDH-based nanovectors by floral pedicle injection, triggers RNA silencing of endogenes expressed in the tomato ovary. This method represents a rapid transient silencing tool for studying molecular events controlling fruit set. In addition, we show that the use of nanovectors as RNA cargos can be advantageous mainly to prolong and stabilize endogene silencing. The findings derived from this study may provide the basis for future applications to improve crop productivity and/or resilience.Author contributions
B. M., A. S, and T. P. conceived and designed this work; F. P. and V. D. carried out the molecular analyses; C. C., and A. S. synthesised LDHs and performed the physicochemical characterization, N.V. conducted the bioinformatic analysis; B. M., F. P., C. C., A. S, and T. P. wrote the manuscript. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
All the authors thank the Centre for Technological Platforms (CPT) of the University of Verona for instrumental setup and technical support. This research was supported by a MIUR grant (PRIN2017, number 20173LBZM2) given to T. P. University of Verona, Italy, is gratefully acknowledged for financial support in two “Joint Projects” grants provided to B. M. and A. S.References
- A. Eamens, M. B. Wang, N. A. Smith and P. M. Waterhouse, Plant Physiol., 2008, 147, 456–468 CrossRef CAS PubMed.
- A. E. Martínez de Alba, E. Elvira-Matelot and H. Vaucheret, Biochim. Biophys. Acta, 2013, 1829, 1300–1308 CrossRef.
- A. Dalakouras, M. Wassenegger, E. Dadami, I. Ganopoulos, M. L. Pappas and K. Papadopoulou, Plant Physiol., 2020, 182, 38–50 CrossRef CAS PubMed.
- M. Ghildiyal and P. D. Zamore, Nat. Rev. Genet., 2009, 10, 94–108 CrossRef CAS PubMed.
- G. R. Davuluri, A. Van Tuinen, P. D. Fraser, A. Manfredonia, R. Newman, D. Burgess, D. A. Brummell, S. R. King, J. Palys, J. Uhlig, P. M. Bramley, H. M. J. Pennings and C. Bowler, Nat. Biotechnol., 2005, 23, 890–895 CrossRef CAS.
- E. Butelli, L. Titta, M. Giorgio, H. P. Mock, A. Matros, S. Peterek, E. G. W. M. Schijlen, R. D. Hall, A. G. Bovy, J. Luo and C. Martin, Nat. Biotechnol., 2008, 26, 1301–1308 CrossRef CAS PubMed.
- H. Wang, B. Jones, Z. Li, P. Frasse, C. Delalande, F. Regad, S. Chaabouni, A. Latché, J.-C. Pech and M. Bouzayen, Plant Cell, 2005, 17, 2676–2692 CrossRef CAS.
- M. de Jong, M. Wolters-Arts, R. Feron, C. Mariani and W. H. Vriezen, Plant J., 2009, 57, 160–170 CrossRef CAS PubMed.
- L. Du, C. Bao, T. Hu, Q. Zhu, H. Hu, Q. He and W. Mao, Mol. Genet. Genomics, 2016, 291, 93–105 CrossRef CAS PubMed.
- S. Matsuo, K. Miyatake, M. Endo, S. Urashimo, T. Kawanishi, S. Negoro, S. Shimakoshi and H. Fukuoka, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 12784–12790 CrossRef CAS.
- F. Mounet, A. Moing, M. Kowalczyk, J. Rohrmann, J. Petit, V. Garcia, M. Maucourt, K. Yano, C. Deborde, K. Aoki, H. Bergès, A. Granell, A. R. Fernie, C. Bellini, C. Rothan and M. Lemaire-Chamley, J. Exp. Bot., 2012, 63, 4901–4917 CrossRef CAS.
- B. Molesini, T. Pandolfini, G. L. Rotino, V. Dani and A. Spena, Plant Physiol., 2009, 149, 534–548 CrossRef CAS PubMed.
- L. Martínez-Bello, T. Moritz and I. López-Díaz, J. Exp. Bot., 2015, 66, 5897–5910 CrossRef PubMed.
- C. Martí, D. Orzáez, P. Ellul, V. Moreno, J. Carbonell and A. Granell, Plant J., 2007, 52, 865–876 CrossRef PubMed.
- B. Molesini, Y. Pii and T. Pandolfini, Trends Biotechnol., 2012, 30, 80–88 CrossRef CAS PubMed.
- B. Molesini and T. Pandolfini, in RNAi for plant improvement and protection., ed. B. Mezzetti, J. Sweet and L. Burgos, CAB International, London, UK, 2021, pp. 14–24 Search PubMed.
- A. Weiberg, M. Bellinger and H. Jin, Curr. Opin. Biotechnol., 2015, 32, 207–215 CrossRef CAS PubMed.
- M. Wang, A. Weiberg, F. M. Lin, B. P. H. J. Thomma, H. Da Huang and H. Jin, Nat. Plants, 2016, 2, 1–10 Search PubMed.
- T. Higashiyama and W.-C. Yang, Plant Physiol., 2017, 173, 112–121 CrossRef CAS PubMed.
- J. Zeng, V. Gupta, Y. Jiang, B. Yang, L. Gong and H. Zhu, Cells, 2019, 8, 371 CrossRef CAS.
- C. Y. Huang, H. Wang, P. Hu, R. Hamby and H. Jin, Cell Host Microbe, 2019, 26, 173–182 CrossRef CAS.
- Q. Cai, B. He, A. Weiberg, A. H. Buck and H. Jin, PLoS Pathog., 2019, 15, 1–13 Search PubMed.
- D. Gan, J. Zhang, H. Jiang, T. Jiang, S. Zhu and B. Cheng, Plant Cell Rep., 2010, 29, 1261–1268 CrossRef CAS PubMed.
- F. Tenllado, B. Martínez-García, M. Vargas and J. R. Díaz-Ruíz, BMC Biotechnol., 2003, 3, 3 CrossRef PubMed.
- N. Mitter, E. A. Worrall, K. E. Robinson, P. Li, R. G. Jain, C. Taochy, S. J. Fletcher, B. J. Carroll, G. Q. Lu and Z. P. Xu, Nat. Plants, 2017, 3, 16207 CrossRef CAS PubMed.
- A. S. Dubrovina and K. V. Kiselev, Int. J. Mol. Sci., 2019, 20, 2282 CrossRef CAS PubMed.
- S. J. Fletcher, P. T. Reeves, B. T. Hoang and N. Mitter, Front. Plant Sci., 2020, 11, 1–10 CrossRef PubMed.
- N. Mitter, E. A. Worrall, K. E. Robinson, Z. P. Xu and B. J. Carroll, Curr. Opin. Virol., 2017, 26, 49–55 CrossRef CAS PubMed.
- I. Sanzari, A. Leone and A. Ambrosone, Front. Bioeng. Biotechnol., 2019, 7, 1–12 CrossRef PubMed.
- D. Marcianò, V. Ricciardi, E. Marone Fassolo, A. Passera, P. A. Bianco, O. Failla, P. Casati, G. Maddalena, G. De Lorenzis and S. L. Toffolatti, Front. Plant Sci., 2021, 12, 1–14 Search PubMed.
- F. Betti, M. J. Ladera-carmona, D. A. Weits, G. Ferri, S. Iacopino, G. Novi, B. Svezia, A. B. Kunkowska, A. Santaniello, A. Piaggesi, E. Loreti and P. Perata, Nat. Plants, 2021, 7, 1379–1388 CrossRef CAS PubMed.
- H. Li, R. Guan, H. Guo and X. Miao, Plant Cell Environ., 2015, 38, 2277–2285 CrossRef CAS.
- S. E. Lau, T. Schwarzacher, R. Y. Othman and J. A. Harikrishna, BMC Plant Biol., 2015, 15, 1–14 CrossRef CAS PubMed.
- K. Numata, M. Ohtani, T. Yoshizumi, T. Demura and Y. Kodama, Plant Biotechnol. J., 2014, 12, 1027–1034 CrossRef CAS PubMed.
- K. V. Kiselev, A. R. Suprun, O. A. Aleynova, Z. V. Ogneva, A. V. Kalachev and A. S. Dubrovina, Int. J. Mol. Sci., 2021, 22, 6749 CrossRef CAS.
- A. S. Dubrovina, O. A. Aleynova, A. R. Suprun, Z. V. Ogneva and K. V. Kiselev, Appl. Microbiol. Biotechnol., 2020, 104, 2125–2135 CrossRef CAS.
- A. Dalakouras, W. Jarausch, G. Buchholz, A. Bassler, M. Braun, T. Manthey, G. Krczal and M. Wassenegger, Front. Plant Sci., 2018, 9, 1–11 CrossRef PubMed.
- P. C. Faustinelli, I. L. Power and R. S. Arias, Plant Biol., 2018, 20, 444–449 CrossRef CAS PubMed.
- C. W. Melnyk, A. Molnar and D. C. Baulcombe, EMBO J., 2011, 30, 3553–3563 CrossRef CAS PubMed.
- C. A. Brosnan and O. Voinnet, Curr. Opin. Plant Biol., 2011, 14, 580–587 CrossRef CAS PubMed.
- J. O. Brunkard and P. C. Zambryski, Curr. Opin. Plant Biol., 2017, 35, 76–83 CrossRef PubMed.
- C. Klap, E. Yeshayahou, A. M. Bolger, T. Arazi, S. K. Gupta, S. Shabtai, B. Usadel, Y. Salts and R. Barg, Plant Biotechnol. J., 2017, 15, 634–647 CrossRef CAS.
- H. Dong, M. Chen, S. Rahman, H. S. Parekh, H. M. Cooper and Z. P. Xu, Appl. Clay Sci., 2014, 100, 66–75 CrossRef CAS.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- B. Langmead, C. Trapnell, M. Pop and S. L. Salzberg, Genome Biol., 2009, 10, R25 CrossRef PubMed.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, J. Mol. Biol., 1990, 215, 403–410 CrossRef CAS.
- T. Pandolfini, Nutrients, 2009, 1, 168–177 CrossRef CAS PubMed.
- L. X. Zhang, J. Hu, Y. B. Jia, R. T. Liu, T. Cai and Z. P. Xu, Nanoscale, 2021, 13, 7533–7549 RSC.
- V. R. L. Constantino and T. J. Pinnavaia, Inorg. Chem., 1995, 34, 883–892 CrossRef CAS.
- L. Ma, Q. Wang, S. M. Islam, Y. Liu, S. Ma and M. G. Kanatzidis, J. Am. Chem. Soc., 2016, 138, 2858–2866 CrossRef CAS PubMed.
- S. Ribet, D. Tichit, B. Coq, B. Ducourant and F. Morato, J. Solid State Chem., 1999, 142, 382–392 CrossRef CAS.
- R. E. Johnsen, Q. Wu, A. O. Sjåstad, Ø. B. Vistad, F. Krumeich and P. Norby, J. Phys. Chem. C, 2008, 112, 16733–16739 CrossRef CAS.
- K. Nejati, A. Mokhtari, F. Khodam and Z. Rezvani, Can. J. Chem., 2015, 94, 66–71 CrossRef.
- S. Gates-Rector and T. Blanton, Powder Diffr., 2019, 34, 352–360 CrossRef CAS.
- C. E. Ciocan, E. Dumitriu, T. Cacciaguerra, F. Fajula and V. Hulea, Catal. Today, 2012, 198, 239–245 CrossRef CAS.
- E. M. Seftel, P. Cool and D. Lutic, Mater. Sci. Eng. C, 2013, 33, 5071–5078 CrossRef CAS PubMed.
- J. T. Kloprogge, D. Wharton, L. Hickey and R. L. Frost, Am. Mineral., 2002, 87, 623–629 CrossRef CAS.
- K. V. Kiselev, A. R. Suprun, O. A. Aleynova, Z. V. Ogneva and A. S. Dubrovina, Plants, 2021, 10, 1–13 CrossRef PubMed.
- A. Koch, D. Biedenkopf, A. Furch, L. Weber, O. Rossbach, E. Abdellatef, L. Linicus, J. Johannsmeier, L. Jelonek, A. Goesmann, V. Cardoza, J. McMillan, T. Mentzel and K. H. Kogel, PLoS Pathog., 2016, 12, 1–22 Search PubMed.
- C. N. T. Taning, S. Arpaia, O. Christiaens, A. Dietz-Pfeilstetter, H. Jones, B. Mezzetti, S. Sabbadini, H. G. Sorteberg, J. Sweet, V. Ventura and G. Smagghe, Pest Manag. Sci., 2020, 76, 841–845 CrossRef CAS PubMed.
- H. Zhang, G. S. Demirer, H. Zhang, T. Ye, N. S. Goh, A. J. Aditham, F. J. Cunningham, C. Fan and M. P. Landry, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 7543–7548 CrossRef CAS PubMed.
- G. S. Demirer, H. Zhang, N. S. Goh, R. L. Pinals, R. Chang and M. P. Landry, Sci. Adv., 2020, 6, eaaz0495 CrossRef CAS PubMed.
- F. J. Cunningham, N. S. Goh, G. S. Demirer, J. L. Matos and M. P. Landry, Trends Biotechnol., 2018, 36, 882–897 CrossRef CAS PubMed.
- G. S. Demirer, H. Zhang, J. L. Matos, N. S. Goh, F. J. Cunningham, Y. Sung, R. Chang, A. J. Aditham, L. Chio, M. J. Cho, B. Staskawicz and M. P. Landry, Nat. Nanotechnol., 2019, 14, 456–464 CrossRef CAS PubMed.
- B. Hendrix, W. Zheng, M. J. Bauer, E. R. Havecker, J. T. Mai, P. H. Hoffer, R. A. Sanders, B. D. Eads, A. Caruano-Yzermans, D. N. Taylor, C. Hresko, J. Oakes, A. B. Iandolino, M. J. Bennett and J. Deikman, Planta, 2021, 254, 1–17 CrossRef PubMed.
- L. Vermeersch, N. De Winne and A. Depicker, Plant J., 2010, 64, 392–401 CrossRef CAS PubMed.
- Z. Luo and Z. Chen, Plant Cell, 2007, 19, 943–958 CrossRef CAS PubMed.
- E. Dadami, A. Dalakouras, M. Zwiebel, G. Krczal and M. Wassenegger, RNA Biol., 2014, 11, 934–941 CrossRef PubMed.
- N. D. Warnock, L. Wilson, J. V. Canet-Perez, T. Fleming, C. C. Fleming, A. G. Maule and J. J. Dalzell, Int. J. Parasitol., 2016, 46, 473–477 CrossRef CAS PubMed.
- L. Jiang, L. Ding, B. He, J. Shen, Z. Xu, M. Yin and X. Zhang, Nanoscale, 2014, 6, 9965–9969 RSC.
- D. Cooper, Bot. Gaz., 1927, 83, 399–411 CrossRef.
- C. Helliwell and P. Waterhouse, Methods Enzym., 2005, 392, 24–35 CrossRef CAS.
- P. M. Waterhouse, M. W. Graham and M. B. Wang, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 13959–13964 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2na00478j |
| ‡ These authors equally contributed. |
| This journal is © The Royal Society of Chemistry 2022 |