Open Access Article
Open Access ArticleEnhanced antitumor activity of carbendazim on HeLa cervical cancer cells by aptamer mediated controlled release†
Bilge G.
Tuna
*a,
Pinar B.
Atalay
b,
Gamze
Kuku
 c,
E. Esma
Acar
d,
H. Kubra
Kara
d,
M. Deniz
Yilmaz
c,
E. Esma
Acar
d,
H. Kubra
Kara
d,
M. Deniz
Yilmaz
 d and
V. Cengiz
Ozalp
d and
V. Cengiz
Ozalp
 *d
*d
aDepartment of Biophysics, Yeditepe University School of Medicine, Yeditepe University, Istanbul, Turkey. E-mail: bilgeguv@gmail.com
bDepartment of Medical Biology and Genetics, Maltepe University Faculty of Medicine, Maltepe University, Istanbul, Turkey
cDepartment of Genetics and Engineering, Faculty of Engineering and Architecture, Yeditepe University, Istanbul, Turkey
dResearch and Development Center for Diagnostic Kits (KIT-ARGEM), Department of Bioengineering, Konya Food and Agriculture University, 42080, Konya, Turkey. E-mail: cengizozalp@gmail.com
First published on 5th November 2019
Abstract
Carbendazim, is a broad-spectrum fungicide and also a promising experimental antitumor drug as reproduction and developmental toxicant, which is currently under phase II preclinical trials. In this study, an approach based on controlled and targeted release with aptamers and mesoporous silica nanoparticles was investigated to improve the antitumor activity of carbendazim. To this end, we synthesized aptamer conjugated silica nanoparticles for testing cytotoxicity properties in vitro with human cervical adenocarcinoma (HeLa) cultured cells. Nucleolin (AS1411) binding aptamers were used to entrap carbendazim molecules inside nanopores of MCM-41 type silica nanoparticles to obtain a stimuli-dependent release system. The effect of carbendazim loaded aptamer silica complex was tested and compared to free carbendazim treatment on HeLa cells, demonstrating 3.3 fold increase of toxicity on targeted cells with our delivery system. In addition, cytotoxicity of the complex was determined to be mostly due to increased apoptosis and to a less extend necrosis related pathways.
Introduction
Cervical cancer is the fourth cancer type in frequency among women worldwide, corresponding to about 6.6% of all female cancers.1 Radical hysterectomy and chemotherapy which are associated with several complications and serious side effects are the most common clinical approaches.2 Despite the significant research and drug development studies and improvement in mortality rate due to early diagnosis for cervical cancer treatment, the prevalence and recurrence rates are still very high. As an alternative therapy, transport of the drugs with targeting ability is one of the major promising technologies for cancer therapy by increasing drug efficacy and limiting side effects.3 For example bevacizumab (Avastin®), which relies on an approved targeted drug delivery approach via monoclonal antibodies, targets vascular endothelial growth factor for cervical cancer treatment with less side effects.4 Although monoclonal antibodies are successful tools for targeted cancer therapy, it has still many complications such as auto-inflammatory toxicity and high cost. As an efficient alternative to all these limitations, aptamer targeting has been suggested for drug delivery systems due to its high affinity, specificity, safety, small size, lack of auto immune reactions and low cost to improve the therapeutic effects for cervical cancer.5 In addition, using nanoparticles as vehicles for safe drug transport might enhance drug efficacy at lower doses as desired.Aptamers, synthetic single-stranded nucleic acid oligonucleotides can specifically bind on biological targets both intercellular or cell surface. AS1411 aptamer (another name for AGRO100) is a guanine-rich 26-mer unmodified oligonucleotide currently under phase I and phase II clinical trials for acute myeloid leukemia therapy and metastatic renal cells.6,7 AS1411 was reported to initially bind to cell surface nucleolin protein and internalized for subsequent inhibition of DNA replication.8 Nucleolin is normally a nucleolus and cytoplasm located multifunctional protein and also expressed on plasma membrane.9 However, many types of cancer cells including cervical cancer, breast cancer and leukemia cells were reported to have higher expression level of nucleolin protein on cell membrane surface compared to normal cells.9,10
Carbendazim is a benzimidazole derivative drug in phase I clinical trials due to its antitumor activity on advanced solid tumors or lymphoma.11 Cytotoxic effects of carbendazim were characterized in 12 human cell lines including human breast cancer cells (MCF-7) and in primary cultures of patient tumor cells. In addition, carbendazim was shown to be more active in hematological malignancies compared to solid tumors and inhibit proliferation of drug and multidrug resistant tumor cells.12 However, human cervical adenocarcinoma (HeLa) cells were reported to be relatively resistant to the carbendazim treatment.12 Therefore, using aptamer targeting in combination with nanoparticles might exhibit a potential for cervical cancer therapy by reducing effective dose required for the antitumor activity of carbendazim. In agriculture, carbendazim has been used as pesticide to treat fungal diseases such as Seproria, Fusarium and Sclerotina. It is moderately persistent in soil and highly persistent in water. Carbendazim is known for its environmental toxicity to mammalian cells, honey bees and many aquatic animals.
In our previous reports, we have successfully synthesized and characterized aptamer-based specific bacterial cell or small molecule dependent release system conjugated to nanocapsules, which was capable of controlling the release of cargo molecule by changing conformation upon target binding on breast cancer cells.13–17 If the aptamers on the complex binds to the target, nanopores on silica nanoparticles holding cargo molecule opens and releases the cargo and closes according to the sufficient concentration. The advantage of this switchable aptamer nanomachine was real-time controlling the release of cargo addition to specific targeting.18 The aptamer gate as used in this study depends on designing a hairpin structure of aptamer sequence, which results in two alternating molecular conformation according to presence of target. Hairpin conformation was achieved by adding nucleotides at one end of the aptamer sequence. Since the added nucleotides were chosen as complementary to the other end of the molecule, the aptamer gate will assume a duplex structure in the hairpin form and a loop structure in the middle of the sequence (aptamer sequence). However, the hairpin structure switch to single stranded form when target molecules interacts with aptamer sequence in the loop region. Thus, covalently bound aptamer gate at one end of the sequence near nanopores will have duplex or single stranded form triggered by target molecule affinity binding, resulting in blocking (duplex form) or opening (single-strand form) of the pores on the nanoparticles. Mesoporous silica nanoparticles have been popular drug delivery systems with current attention.19,20
In this study, we adapted our research further on switchable AS1411 aptamer-based nano-carrier system to cervical cancer therapy with a promising new drug carbendazim. We aimed to increase antiproliferative efficiency of carbendazim on cervical cancer cells and optimize carbendazim therapy against cancer treatment. For this purpose, AS1411 aptamer based specific target release system integrated to nanocapsules were loaded with carbendazim, characterized, and the antitumor effects were investigated for cervical cancer cell culture (Scheme 1).
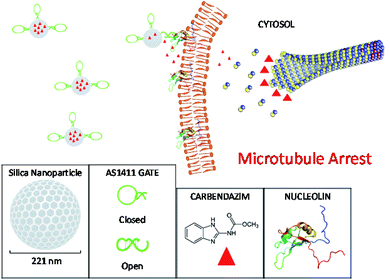 | ||
| Scheme 1 The principle of carbendazim delivery system. Mesoporous silica nanoparticles were loaded with carbendazim molecules and capped with AS1411 aptamer gates. The nucleolin specific aptamers targeted the particles to cell surface attached nucleolin proteins. The affinity binding of aptamers with nucleolin resulted in molecular conformation changes in aptamer gates, leading to open form structure, and hence release of carbendazim cargo onto cell membrane. This creates a microenvironment around cell surface with carbendazim at high concentrations.21 Carbendazim molecules diffuses to cytosol and interacts with microtubule integration/disintegration ends, interfere with microtubule dynamics and causes microtubule arrest related cell death. | ||
Experimental
Aptamer gate
AS1411 aptamer gate (CCA CCA CGG TGG TGG TGG TTG TGG TGC GTG GTG G)18 was synthesized by Sentromer Ltd (Istanbul, Turkey) with 5′ amino group functionalization by adding 7 nucleotide at 5′ end of nucleolin aptamer.9 Converting of an aptamer sequence to gate was described for Staphylococcus aureus aptamers as well as small molecules like ATP.13,14Carbendazim loaded aptamer silica nanoparticles
The particles were synthesized according to a sol–gel method. Briefly, N-cetyltrimethylammonium bromide (CTAB) solution of 2.74 mmol was mixed with 2 M sodium hydroxide. This mixture was heated and the temperature was kept 80 °C. Under constant stirring, 5 mL of 22.4 mmol tetraethoxysilane (TEOS) was added into the mixture dropwise within 5 min. After 2 h reaction, white precipitate was collected by filter, washed with water and methanol several times followed by drying under air. The surfactant template from 1.5 g synthesized product was removed by refluxing in HCl (1.5 mL, 37%)/methanol (150 mL) solution. After 6 h, the product was filtered, washed with methanol and water, finally surfactant free mesoporous silica nanoparticles were obtained after drying.The synthesized particles were incubated in carbendazim in PBS (100 µM) overnight as described for other small molecules with similar molecular weight to carbendazim. The particles were grafted with epoxy groups via silanization procedure with (3-glycidyloxypropyl)triethoxysilane. Finally, amine labelled nucleolin aptamer gate sequences (10 µM) were immobilized in a carbonate–bicarbonate buffer at pH = 9.4 and washed with PBS three times.22 Carbendazim at 100 µM was always present during all the preparation procedure except washing. Carbendazim loaded aptamer silica nanocapsules were characterized using UV-Vis spectroscopy (Biotek), dynamic light spectroscopy (DLS) (ZetaSizer Nano ZS, Malvern Instruments, Malvern, UK) and Transmission Electron Microscopy (TEM).
Cell culture
HeLa cervical cancer cell line (ATCC-CCL2) was grown and maintained in Dulbecco's modified Eagle's medium (DMEM, Sigma Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich, Germany) and 0.1% penicillin/streptomycin (Sigma-Aldrich, Germany) by incubating in a humidified 5% CO2 atmosphere at 37 °C and sub-cultured every 72 h when the cells reached about 80% confluent.AS1411 binding to HeLa cells
To confirm the AS1411 (nucleolin aptamer) uptake, 4 × 104 HeLa cells were added 24-well plates, and the next day incubated with 1 µM FAM-labelled AS1411 (FL-AS1411) for 4 h in the conditions as described above. After 4 h, cells were detached by trypsin treatment and washed with PBS to be instantly analysed on flow cytometer in triplicates (Cyte™ 5, Merck Millipore).WST-1 cell proliferation assay
To evaluate the extent of cytotoxicity a cell proliferation assay was used (Cell Counting Kit-8, Sigma) for counting live cells. 104 HeLa cells were added to 96-well plate. Upon 24 h exposure to carbendazim (Carb), silica nanoparticles (SNPs) or carbendazim aptamer silica nanoparticles (Carb-Apt-SNPs), CCK-8 reagent was added to each well and the conversion of tetrazolium salts to formazan dye was read on an ELISA microplate reader from the absorbance values at 440 nm wavelength.Apoptosis/necrosis analysis
Annexin V-FITC (Calbiochem, Merck Millipore) was used according to analyse the apoptotic or necrotic cells after treatments. Briefly, 4 × 104 HeLa cells were grown on 24-well plates. After incubation with free carbendazim or Carb-Apt-SNPs, cells were trypsinized and the detached cells in supernatants were collected in a new tube. After centrifugation, cells were stained with Annexin V-FITC and PI (Invitrogen). The samples were maintained at +4 °C throughout this procedure. For each sample, triplicates of more than 2.0000 counts were obtained on guava easy-Cyte™ 5 (Merck Millipore) flow cytometer.Statistical analysis
The measurements were given as mean ± standard deviation and analysed by using the statistical analysis package SPSS 25. Student's t-test was used to test similarities between groups pairs and one-way analysis of variance (ANOVA) was used for multiple groups with post hoc either Dunnett-t3 or Dunnett-t. A level of p < 0.05 was used as cut-off for statistical significance. IC50 values were calculated as 50% of the signal from relationship equations obtained by fitting curves to exponential growth 2 parameters of Sigmaplot 9.Results and discussions
Functional characterization of carbendazim loaded silica-aptamer nanoparticles
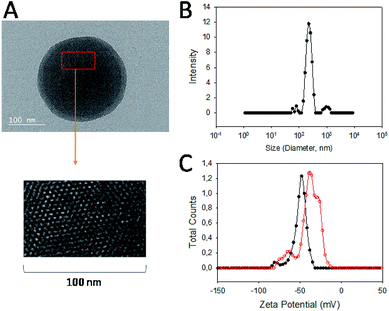
Carbendazim loaded and aptamer gated silica nanoparticles (Carb-Apt-SNPs) were prepared by our previous procedures. 13First, silica particles were synthesized by a sol–gel method to obtain MCM-41 type mesoporous nanoparticles. Fig. 1A shows a typical TEM image of the particles at around 200 nm with ordered hexagonal shaped nanopores (Fig. 1A, inset). BET analysis was in agreement with TEM images with the diameter size of nanopores at 3.4 ± 1.2 nm (data not shown). The Carb-Apt-SNPs were characterized further in terms of particle size as 221 nm ± 8.4 of hydrodynamic diameter by using dynamic light scattering (DLS) in phosphate buffer (pH = 7.5). The unmodified silica nanoparticles formed aggregates of 680 ± 24.6 nm diameter with −15 mV zeta potential value (Data not shown). The epoxy group functionalization reduced the zeta potentials to −38.3 mV with diameter of 238 nm ± 9.1 and the grafting with aptamer gates decreased the zeta potential value to −48.4 mV, increasing the stability of our particles (Fig. 1B). Zeta potential value of colloidal particles like silica is an indication for their stability as distinct particles, which is desired for drug delivery applications. Thus, we obtained carbendazim loaded and aptamer functionalized stable nanoparticles ready for functional characterizations.The particles were loaded with 53 ± 8.2 nmol carbendazim per mg of particles and tested for aptamer gate capping (Fig. 2, red line). There was minimum amount of carbendazim leakage up to 17.4 ± 3.1% in 24 hours of experiment. The release amount reached to 86.5 ± 5.8% in 24 hours upon treatment with aptamer gate target protein (1 µM nucleolin), demonstrating that delivery particles function as expected (Fig. 2, black line).
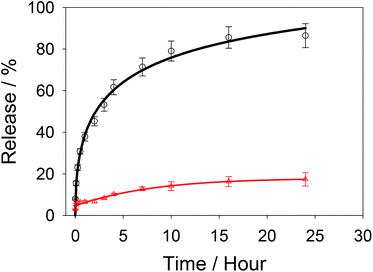 | ||
| Fig. 2 Release kinetics of Carb-Apt-SNPs in phosphate buffered saline (PBS) (red line) or with nucleolin addition at zero time (black line). | ||
The cytotoxicity of carbendazim loaded in silica particles were maintained for various cell types, including HeLa cells.23 The absorption and release kinetics under different physical conditions were reported for pH, ionic strength and temperature. Absorption efficiency was maximum around neutral pH, low ionic strength and low temperatures as expected.24
Targeting of nucleolin aptamer gate and its silica conjugates
Anti-nucleolin aptamer has been reported in numerous studies and well-proved for its specific ability to target tumor cells.9,25 Aptamer gate principle was also demonstrated for successful targeting bacterial cells as well as breast cancer cells.13,18 In this study, we checked first molecular switching ability by Förster Resonance Energy Transfer (FRET) analysis as applied previously for small molecules and bacterial cells.13,14 Fig. S2† shows that nucleolin protein can bind to aptamer gate and separates the two ends of the sequence away from each other. This decrease quenching as observed by increasing fluorescence signal. Then, targeting ability of aptamer AS1114 hairpin structure (aptamer gate) was investigated for HeLa cells by flow cytometry. Fluorophore-labelled aptamers (FL-Apt) were incubated with HeLa cells in a range of concentrations (0.01 to 1 µM) for a duration of 4 hours. 1 µM FL-Apt treated cells exhibited more fluorescence signal compared to lower doses (Fig. S1†). Thus, aptamer gate and nucleolin interactions resulted in attachment on the cell surface or intake of aptamers into HeLa cancer cells. In fact, studies on nucleolin aptamer cell intake mechanisms demonstrated that AS1411 sequences were taken by cancer cells within first two hours.26Effects of carbendazim and Carb-Apt-SNPs on viability of HeLa cells
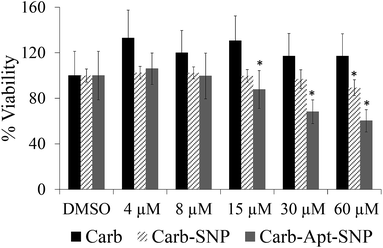
In order to investigate the effect of Carb-Apt-SNPs on the proliferation of HeLa cells, we treated the cells with 4, 8, 15, 30 and 60 µM carbendazim, aptamer functionalized silica nanoparticles without any cargo (Apt-SNP) or carbendazim loaded-aptamer functionalized silica nanoparticles for 24 hours and analyzed cell viability using CCK-8 based colorimetric cell proliferation assay. The nanoparticles treatments were adjusted according to release experiments (Fig. 3). Total released amount of carbendazim was calculated and corresponding amount of nanoparticles were used in the experiments. DMSO only treatments were included in the study as control experiments since nanoparticles were suspended DMSO solution. Equal amounts of silica particles (1.3 g mL−1 for 60 µM treatments) were used in the treatments with Apt-SNP and Carb-Apt-SNPs. CCK-8 assay revealed that administrations up to 15 µM concentrations of carbendazim or equal amounts of Apt-SNP and Carb-Apt-SNPs did not cause statiscally significant differences in cell viabilities (n = 3, p < 0.05). Carb-Apt-SNPs inhibited cell proliferation significantly to 68.17% viability at 30 µM and 60.26% at 60 µM treatments compared to the DMSO control. Apt-SNPs did not have a significant effect on proliferation of HeLa cells with respect to DMSO control except a slight decrease in 60 µM dose treatment group. However, as a single agent carbendazim treatment did not inhibited proliferation for any treatment concentration (Fig. 3). This is consistent with the previous reports of carbendazim on HeLa cells, indicating to resistance to carbendazim. A time-course analysis for 60 µM dose treatment demonstrated that Carb-Apt-SNPs decreased viability gradually (Fig. S3†). A comparison to carbendazim release amounts in Fig. 3 shows that viability decrease corresponds to the released amount of carbendazim from the particles.Apoptosis/necrosis
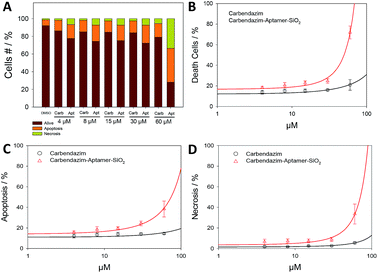
Next, Annexin/PI assay was performed for further investigate cell death mechanism (apoptotic, necrotic or live cells), resulting from targeted carbendazim treatments (Fig. 4). Flow cytometry analysis showed that 24 hours Carb-Apt-SNPs treatments significantly increased cell death compared to free carbendazim or DMSO control due to apoptosis (Fig. 4C). A time-course analysis for 60 µM dose treatment demonstrated that Carb-Apt-SNPs increased cell death gradually (Fig. S4†). Although carbendazim only treatments increased cell death by apoptosis and necrosis compared to DMSO control, aptamer targeted treatments showed significantly higher cell death (apoptosis and necrosis together). Fig. 4B represents that carbendazim-aptamer-nanoparticles lead to cell death in concentration dependent way with an IC50 value of 46.0 µM compared to predicted IC50 value of free carbendazim of 150.2 µM.In healthy organisms, an inherent mechanism, programmed cell death, removes unwanted cells like tumor cells by apoptosis and/or autophagy-related pathways. We have evaluated the anti-cancer toxicity mechanism of our delivery system for carbendazim by testing carbendazim-aptamer-nanoparticles for apoptosis and necrosis.
Some nanoparticles are known to stimulate necrosis, leading to unregulated cell death.27 Non-specific toxicity of nanomaterials can be evaluated via necrosis determination and the adverse effects can be overcome by specific targeting. Silica nanoparticles are among the most popular drug delivery research materials. Krętowski et al. reported about 10% necrosis and about 30% apoptosis in glioblastoma cells upon 24 hours of treatments with 100 µg silica nanoparticles of 5–15 nm size.28 We did not observe any necrosis or apoptosis under the experimental conditions as used in this manuscript, 4 hours of treatment with 213 nm silica nanoparticles-aptamer conjugates or silica nanoparticles, both not containing any cargo molecules (Fig. 3, DMSO). Schütz et. al. confirmed similar results with large silica nanoparticles (up to 245 nm size) with no necrotic or apoptotic effects on HeLa cells.29 Sgc8 aptamer functionalized silica nanoparticles loaded with fluorescein did not cause any necrosis or apoptosis in leukemia cells.30
Methyl benzimidazole carbamate fungicides (including carbendazim) binds to β-tubulin subunit in tubulin dimers, which prevents the integration of GTP-tubulin dimer to the growing end of microtubule. This results in microtubule instability and depolymerization, cell cycle arrest and eventually cell death. Although the exact mechanism of carbendazim action is not exactly known, it is suggested to induce apoptosis by arresting the cells at G2/M phase of cell cycle.31 In this study, apoptosis analysis showed that IC50 values improved from 266.8 µM to 73.9 µM in aptamer targeting silica nanoparticles treatments, resulting in 3.6 fold increase in anti-tumour activity. The same treatments resulted in lowering of IC50 values from 164.1 µM to 71.0 µM in aptamer targeting silica nanoparticles. Our results show that anti-tumor activity of carbendazim can be obtained at lower carbendazim treatments when targeted with aptamers, resulting from apoptosis and necrosis. In fact, encapsulation of carbendazim in solid lipid nanoparticles were shown to decrease human toxicity in agricultural applications.23 Deleterious effects of carbendazim on placental cells included decreasing cell viability and percentages of G0/G1 phase cells.32 HeLa cells are relatively resistant to carbendazim. Although the mechanism of action in resistance in HeLa cells are well characterized, the resistance mechanism for fungus cells has been investigated. Some point mutations in β-tubulin 2 gene (Tub2) were correlated with resistance to methyl benzimidazole carbamate type fungicides (including carbendazim). The mechanism behind the resistance were determined as weakening of binding between carbendazim and β-tubulin 2 binding region due to changes in the amino acids resulting from mutations.33 This suggests that high carbendazim concentration around tumour microenvironment achieved by targeted delivery could promote higher amount of binding in microtubules and result in arrest in resistant cells.
Conclusions
Carbendazim was implicated as promising potential cancer chemotherapeutic agent due to its anti-proliferative properties. Aptamer targeting of carbendazim inside silica mesopores can overcome the natural resistance of cervical tumor cells to carbendazim. Deleterious effects of carbendazim on human cells can be avoided by targeted encapsulated delivery. Our results could lead to further investigation of targeted delivery of carbendazim in nanocapsules for effective therapy development.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge Konya Food and Agriculture University, Research and Development Center for Diagnostic Kits (KITARGEM) project for use of the facilities.Notes and references
- W. H. O. (WHO), Cervical Cancer, 2018, accessed 31.12.2018 Search PubMed.
- P. T. Ramirez, M. Frumovitz, R. Pareja, A. Lopez, M. Vieira, R. Ribeiro, A. Buda, X. Yan, Y. Shuzhong, N. Chetty, D. Isla, M. Tamura, T. Zhu, K. P. Robledo, V. Gebski, R. Asher, V. Behan, J. L. Nicklin, R. L. Coleman and A. Obermair, N. Engl. J. Med., 2018, 379, 1895–1904 CrossRef PubMed.
- Y. Xin, M. Yin, L. Zhao, F. Meng and L. Luo, Cancer Biol. Med., 2017, 14, 228–241 CrossRef CAS.
- R. L. Pereira, I. C. Nascimento, A. P. Santos, I. E. Y. Ogusuku, C. Lameu, G. Mayer and H. Ulrich, Oncotarget, 2018, 9, 26934–26953 Search PubMed.
- J. P. Dassie, L. I. Hernandez, G. S. Thomas, M. E. Long, W. M. Rockey, C. A. Howell, Y. Chen, F. J. Hernandez, X. Y. Liu, M. E. Wilson, L.-A. Allen, D. A. Vaena, D. K. Meyerholz and P. H. Giangrande, Mol. Ther., 2014, 22, 1910–1922 CrossRef CAS.
- R. K. Stuart, K. Stockerl-Goldstein, M. Cooper, M. Devetten, R. Herzig, B. Medeiros, G. Schiller, A. Wei, G. Acton and D. Rizzieri, J. Clin. Oncol., 2009, 27, 7019 Search PubMed.
- J. E. Rosenberg, R. M. Bambury, E. M. Van Allen, H. A. Drabkin, P. N. Lara Jr, A. L. Harzstark, N. Wagle, R. A. Figlin, G. W. Smith, L. A. Garraway, T. Choueiri, F. Erlandsson and D. A. Laber, Invest. New Drugs, 2014, 32, 178–187 CrossRef CAS.
- C. R. Ireson and L. R. Kelland, Mol. Cancer Ther., 2006, 5, 2957 CrossRef CAS.
- P. J. Bates, D. A. Laber, D. M. Miller, S. D. Thomas and J. O. Trent, Exp. Mol. Pathol., 2009, 86, 151–164 CrossRef CAS.
- S. Soundararajan, L. Wang, V. Sridharan, W. Chen, N. Courtenay-Luck, D. Jones, E. K. Spicer and D. J. Fernandes, Mol. Pharmacol., 2009, 76, 984–991 CrossRef CAS.
- NIH, Carbendazim, in Treating Patients With Advanced Solid Tumors or Lymphoma, 2013, https://clinicaltrials.gov/ct2/show/NCT00023816 Search PubMed.
- D. Laryea, J. Gullboa, A. Isaksson, R. Larsson and P. Nygren, Anti-Cancer Drugs, 2010, 21, 33–42 CrossRef CAS PubMed.
- M. Kavruk, O. Celikbicak, V. C. Ozalp, B. A. Borsa, F. J. Hernandez, G. Bayramoglu, B. Salih and M. Y. Arica, Chem. Commun., 2015, 51, 8492–8495 RSC.
- V. C. Ozalp, F. Eyidogan and H. A. Oktem, Pharmaceuticals, 2011, 4, 1137–1157 CrossRef CAS.
- B. A. Borsa, B. G. Tuna, F. J. Hernandez, L. I. Hernandez, G. Bayramoglu, M. Y. Arica and V. C. Ozalp, Biosens. Bioelectron., 2016, 86, 27–32 CrossRef CAS.
- T. T. H. Thi, V. D. Cao, T. N. Q. Nguyen, D. T. Hoang, V. C. Ngo and D. H. Nguyen, Mater. Sci. Eng., C, 2019, 99, 631–656 CrossRef.
- E. Aznar, M. Oroval, L. Pascual, J. R. Murguia, R. Martinez-Manez and F. Sancenon, Chem. Rev., 2016, 116, 561–718 CrossRef CAS.
- F. J. Hernandez, L. I. Hernandez, A. Pinto, T. Schafer and V. C. Ozalp, Chem. Commun., 2013, 49, 1285–1287 RSC.
- F. Hu, B. Liu, H. Chu, C. Liu, Z. Li, D. Chen and L. Li, Nanoscale, 2019, 11, 9201–9206 RSC.
- T. Zhao, L. Chen, Q. Li and X. Li, J. Mater. Chem. B, 2018, 6, 7112–7121 RSC.
- B. Zhang, Y. Hu and Z. Pang, Front. Pharmacol., 2017, 8, 952 CrossRef.
- G. T. Hermanson, Bioconjugate Techniques, Elsevier, San Diego, USA, 2008 Search PubMed.
- E. V. R. Campos, J. L. de Oliveira, C. M. G. da Silva, M. Pascoli, T. Pasquoto, R. Lima, P. C. Abhilash and L. F. Fraceto, Sci. Rep., 2015, 5, 13809 CrossRef.
- S. laurella, C. M. P. Diez, I. D. Lick, P. E. Allegretti and M. F. Erben, International Journal of Engineering and Technical Research, 2015, 3, 96–101 Search PubMed.
- P. J. Bates, E. M. Reyes-Reyes, M. T. Malik, E. M. Murphy, M. G. O'Toole and J. O. Trent, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 1414–1428 CrossRef CAS.
- E. M. Reyes-Reyes, Y. Teng and P. J. Bates, Cancer Res., 2010, 70, 8617–8629 CrossRef CAS.
- M. Sharifi, S. H. Hosseinali, A. A. Saboury, E. Szegezdi and M. Falahati, J. Controlled Release, 2019, 299, 121–137 CrossRef CAS.
- R. Krętowski, M. Kusaczuk, M. Naumowicz, J. Kotyńska, B. Szynaka and M. Cechowska-Pasko, Nanomaterials, 2017, 7, 230 CrossRef.
- I. Schütz, T. Lopez-Hernandez, Q. Gao, D. Puchkov, S. Jabs, D. Nordmeyer, M. Schmudde, E. Rühl, C. M. Graf and V. Haucke, J. Biol. Chem., 2016, 291, 14170–14184 CrossRef PubMed.
- J. Tan, N. Yang, Z. Hu, J. Su, J. Zhong, Y. Yang, Y. Yu, J. Zhu, D. Xue, Y. Huang, Z. Lai, Y. Huang, X. Lu and Y. Zhao, Nanoscale Res. Lett., 2016, 11, 298 CrossRef.
- M. Yenjerla, C. Cox, L. Wilson and M. A. Jordan, J. Pharmacol. Exp. Ther., 2009, 328, 390 CrossRef CAS.
- J. H. Zhou, K. Xiong, Y. Yang, X. Q. Ye, J. Liu and F. X. Li, Reprod. Toxicol., 2015, 51, 64–71 CrossRef CAS.
- S. Xu, J. Wang, H. Wang, Y. Bao, Y. Li, M. Govindaraju, W. Yao, B. Chen and M. Zhang, BMC Genomics, 2019, 20, 115 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07974b |
| This journal is © The Royal Society of Chemistry 2019 |