Open Access Article
Open Access ArticleMetal-detection based techniques and their applications in metallobiology†
Ying
Zhou
 *a,
Hongyan
Li
*a,
Hongyan
Li
 *a,
Eric
Tse
*a,
Eric
Tse
 *b and
Hongzhe
Sun
*b and
Hongzhe
Sun
 *a
*a
aDepartment of Chemistry, CAS-HKU Joint Laboratory of Metallomics for Health and Environment, The University of Hong Kong, Pokfulam Road, Hong Kong SAR, P. R. China. E-mail: zy2014@connect.hku.hk; hylichem@hku.hk; hsun@hku.hk
bDepartment of Medicine, LKS Faculty of Medicine, The University of Hong Kong, Pokfulam Road, Hong Kong SAR, P. R. China. E-mail: ewctse@hku.hk
First published on 6th June 2024
Abstract
Metals are essential for human health and play a crucial role in numerous biological processes and pathways. Gaining a deeper insight into these biological events will facilitate novel strategies for disease prevention, early detection, and personalized treatment. In recent years, there has been significant progress in the development of metal-detection based techniques from single cell metallome and proteome profiling to multiplex imaging, which greatly enhance our comprehension of the intricate roles played by metals in complex biological systems. This perspective summarizes the recent progress in advanced metal-detection based techniques and highlights successful applications in elucidating the roles of metals in biology and medicine. Technologies including machine learning that couple with single-cell analysis such as mass cytometry and their application in metallobiology, cancer biology and immunology are also emphasized. Finally, we provide insights into future prospects and challenges involved in metal-detection based techniques, with the aim of inspiring further methodological advancements and applications that are accessible to chemists, biologists, and clinicians.
1. Introduction
Metals play a crucial role in human health and are involved in a wide range of biological processes.1–3 Maintaining metal homeostasis is essential for human health and dysregulation of metal homeostasis has been linked to several diseases. The exploration of metals as both diagnostic and therapeutic agents has emerged as a prominent area of research in the field of bioinorganic chemistry and medicinal chemistry.4 Numerous comprehensive reviews are readily accessible, including the development of metallo-pharmacophores as anticancer, antimicrobial and antiviral agents,5–9 as well as the mechanisms underlying their therapeutic functions.4,7 Recently, there has been a growing recognition of the potential of metal-based pharmaceuticals in effectively combating viruses and addressing the challenge of antimicrobial resistance (AMR).9,10 Moreover, as the immune modulation role of metals is being increasingly acknowledged, metal-based immunotherapy has emerged as a promising field within pharmaceuticals.11–13Metals have also been extensively utilized as contrast agents in various diagnostic imaging techniques, including magnetic resonance imaging (MRI), X-ray imaging, and computed tomography (CT). These techniques have found broad applications in clinical settings, playing a critical role in the diagnosis, treatment, and management of various diseases. Moreover, there has been a rapid development of advanced techniques (e.g., mass cytometry, imaging mass cytometry (IMC), Nano-SIMS, and multiplexed ion beam imaging (MIBI)) based on metal detection in recent years, opening up new opportunities for metallobiology. Additionally, the rapid development of novel metallomics/metalloproteomics techniques,4 along with integrative multi-omics, has provided valuable tools for our understanding of the mechanisms of action of metal-based complexes and their therapeutic activities. In this perspective, we aim to summarize the latest progress in advanced metal-detection based techniques along with examples of successful applications of these methodologies as drivers for novel insights for elucidating the roles of metals in biology and medicine. The cutting-edge single-cell techniques that can aid in the development of metallo-pharmaceuticals are also highlighted.4,14 Finally, we provide our view on future prospects and challenges involved in metal-detection based techniques, aiming to inspire further advancements in the approach for cancer biology, immunology and potentially clinical diagnosis.
2. Recent advances in metal-detection based techniques
Given the increasingly recognized significance of metals in disease biology and therapeutics, numerous research groups globally have strived to comprehend their underlying mechanism of action at cellular and molecular levels.14–16 Consequently, metallomics/metalloproteomics has emerged as a research area, aiming to uncover metal–protein interactions/associations or metal variations on a metallomic and proteome-wide scale. Over the past two decades, various metallomics/metalloproteomics approaches have been developed for the application of metal relevant research,4 for instance, the use of different metal-detection based fluorescence probes to visualize17 and track metal-binding proteins with weak/transient binding,18 immobilized metal affinity chromatography to enrich and separate low abundance metalloproteins,19 and gel electrophoresis-inductively coupled plasma mass spectrometry (ICP-MS) to identify metal-binding proteins with strong binding affinities.20 The relevant technical development of metallomics/metalloproteomics approaches has been summarized in a recent review.4 The rapid advancement of metal-tagging strategies has sparked growing interest in metal-detection based techniques for both clinical and fundamental research. This is particularly evident with the emergence of mass cytometry. In this section, some of the emerging metal-detection based approaches, with a special focus on single cell techniques, will be discussed (Table 1).| Technologies | Applications | Advantages | Limitations |
|---|---|---|---|
| ICP-MS | Metallome and proteome profiling in bulk solution21,22,24 | Wide range of analytes | Matrix interference |
| Wide dynamic range | Relatively low sensitivity for light element quantification | ||
| High sensitivity | |||
| High multiplexity | |||
| GE-ICP-MS | Tracking metal-binding proteome35,36 | Capable of identifying target proteins of a wide range of metals | Only identifies metal-binding proteins with strong binding affinity |
| TR-ICP-MS | Single cell element and protein quantification28,29 | Wide range of analytes | Lack of multiplexity |
| Single cell resolution | |||
| CyTOF | Single cell proteome profiling38 | High multiplexity | Only detects element signals with atomic weights between 75 and 209 |
| High sensitivity | |||
| Low background | |||
| High throughput | |||
| LA-ICP-MS | Element and protein imaging32,33 | Wide range of analytes | Slow acquisition rate |
| Wide dynamic range | Relatively low resolution | ||
| IMC | Multiplexed imaging42 | High multiplexity | Slow acquisition rate |
| High resolution | Only maps elements with atomic weights between 75 and 209 | ||
| MIBI | Multiplexed imaging54 | High sensitivity | Low multiplexity |
| Super high resolution (10 nm) | Relatively slow acquisition rate | ||
| High cost | |||
| MIBI-TOF | Multiplexed imaging55 | High multiplexity | High cost |
| High sensitivity | |||
| Super high resolution (10 nm) | |||
| High throughput |
2.1 ICP-MS-based metallomic and metalloproteomic approaches
ICP-MS is an exceptionally robust analytical technique for precise quantification of metals and metalloids across a wide range of samples. Metallome profiling of serum21,22 and saliva23 with ICP-MS has emerged as a valuable tool for monitoring metallome response in various diseases, as well as identifying biomarkers and evaluating the role of different metals played in health and disease.21,22,24,25 The establishment of these profiling techniques has enabled the comprehensive analysis of metal ions in biological fluids, shedding light on their potential implications for disease pathology and progression.With the advancements in metal tagging strategies,26 the application of ICP-MS has expanded beyond direct metallome profiling to encompass the analysis of various biomolecules (Fig. 1A), e.g. proteins and nucleic acids, opening up a new opportunity for multiplexed integrative-omics profiling by ICP-MS.21,22 This integrative-omics profiling by ICP-MS offers a powerful tool for unraveling complex biological processes, identifying biomarkers and elucidating the intricate relationships between metal ions and different molecular components (Fig. 1B).24 The evolution of this technique platform is propelled by the development of diverse metal-tagging strategies, which have progressed from the use of single lanthanide isotope tags to the application of multiple lanthanide polymers, metal nanoparticles, and multifunctional metal tags with various detection modes.27 The advancements in metal tagging have significantly enhanced the sensitivity and multiplexity of ICP-MS-based integrative-omics profiling.
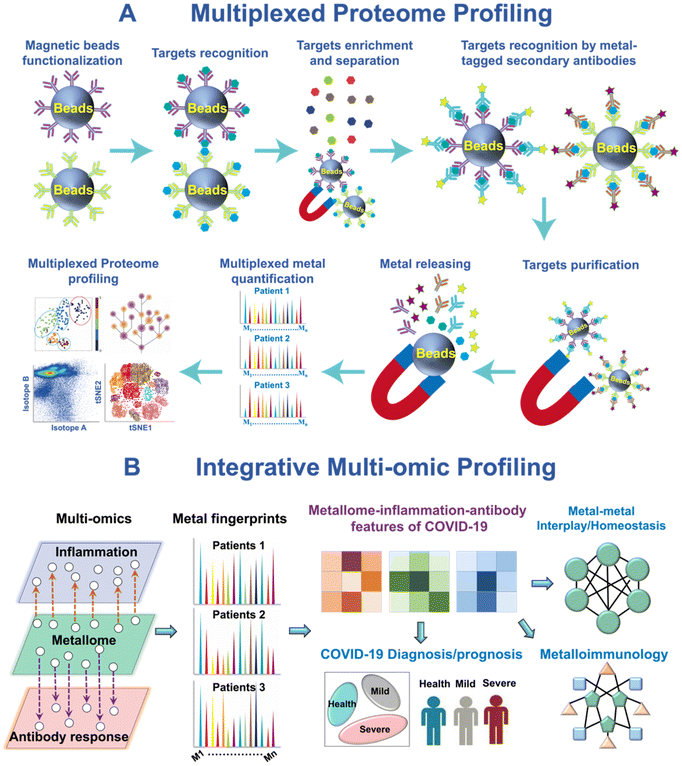 | ||
| Fig. 1 ICP-MS-based metallomic and multi-omic approaches. (A) Multiplexed serological proteomic profiling of COVID-19 patients. (B) Integrative metallomic and immunoproteomic profiling showing the role of metals in COVID-19 immunity.21,22 | ||
In spite of the extensive application of ICP-MS for elemental or biomolecule quantification in bulk solution, a single cell/particle analysis based on time-resolved ICP-MS (TR-ICP-MS) has also been established by modulating the sampling system and analyzing in time-resolved mode.28,29 This technique allows for the measurement of elements or proteins on a cell-by-cell basis, which has been successfully utilized to track the uptake of metallodrugs or endogenous metals at the single-cell level in different cell lines,28 in cells at different states29 or with varied drug sensitivity,30 and single bacteria.31
The combination of laser ablation (LA) and ICP-MS (LA-ICP-MS) represents a powerful technique for the in situ analysis and imaging of metals or metallodrugs in biological samples.32 It provides exceptional spatial resolution, high sensitivity, and the ability to perform quantitative imaging. In a recent study, Schaier and coworkers33 developed a single-cell metallomics imaging method through integration of low-dispersion laser ablation and ICP-TOF-MS, enabling the rapid mapping of endogenous elements at the single cell level. To further enhance the information obtained, the researchers introduced metal-labeled antibodies, enabling the visualization of specific proteins along with the endogenous cellular ionome (Fig. 2A). This integrated assay signifies a notable breakthrough in the field of single-cell metallomics, as it allows for the simultaneous correlation of metal accumulation with multi-dimensional characterization of cells and cell populations.
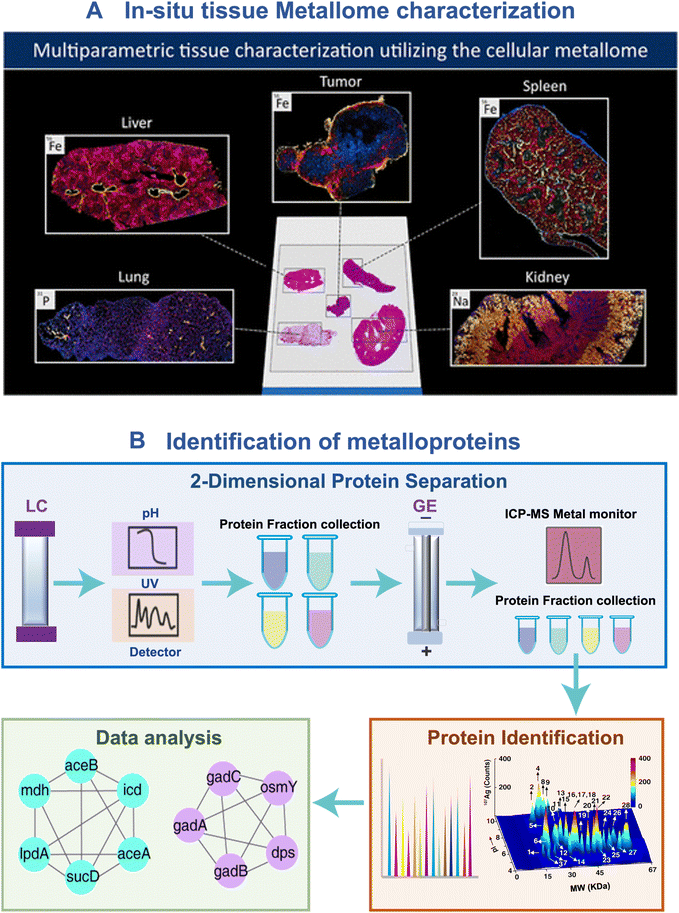 | ||
| Fig. 2 ICP-MS-based approaches for in situ metallome imaging and metalloproteome identification. (A) Characterization of the metallomic profile in tissue sites.33 (B) General scheme of GE-ICP-MS for tracking metalloproteins. | ||
The integration of ICP-MS with protein separation techniques such as liquid chromatography (LC), high-performance liquid chromatography (HPLC), capillary electrophoresis (CE), and gel electrophoresis (GE), along with identification methods like matrix-assisted laser desorption/ionization mass spectrometry (MALDI), has emerged as an invaluable method in metalloproteomics for the identification of metal-binding proteins. One of the most prevalent approaches is the combination of gel electrophoresis, ICP-MS, and MALDI-MS, known as GE-ICP-MS.20 The GE-ICP-MS approach employs gel electrophoresis to separate proteins based on their molecular weight and charge, while ICP-MS enables the monitoring of metal content in these separated proteins. Subsequently, the metal-binding proteins are identified by MALDI-MS. GE-ICP-MS has been revolutionized from one dimensional GE-ICP-MS to two dimensional GE-ICP-MS by introducing liquid chromatography (LC), i.e. LC-GE-ICP-MS, further improving protein separation efficiency (Fig. 2B). This method has been utilized to profile different metal-binding proteomes in cells34 and microbes.35,36
2.2 Mass cytometry
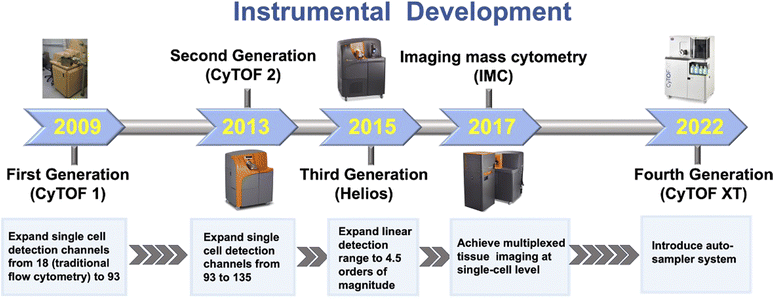
Mass cytometry, also known as CyTOF (Cytometry by Time of Flight), is a cutting-edge technology that combines flow cytometry with ICP-TOF-MS.37 For mass cytometry, multiple proteins of interest are coupled by their complementary antibodies or secondary antibodies, which are fused with lanthanide chelates.38 The use of lanthanide tags in mass cytometry has significantly expanded its detection dimensionality, allowing for the quantification of over 100 markers simultaneously. This makes mass cytometry the most highly multiplexed technique currently available for proteome profiling in single cells. Mass cytometry has undergone significant advancements over the years, characterized by successive evolution in detection channels, linear detection range, higher sensitivity, wider application, and automation, leading to improved analytical capabilities and more efficient sample handling and analysis.Advancements in instrument design have played a critical role in the development of mass cytometry technology (Fig. 3). In 2009, the advent of the first-generation mass cytometry instrument revolutionized single-cell analysis by expanding the number of detection channels from the limited 18 in traditional flow cytometry to 93.39 This breakthrough opened the “post-fluorescence” era, where scientists could monitor a much larger number of parameters in individual cells. The evolution of mass cytometers continued with the development of second- and third-generation (Helios) instruments. These advancements further expanded the number of detection channels to 135, allowing for even more comprehensive analysis of cellular markers and molecules. Additionally, the linear detection range was extended to 4.5 orders of magnitude, enhancing the dynamic range of detection and enabling more accurate quantification of high and low expression levels of cellular markers.40 The fourth-generation mass cytometry (CyTOF XT™) introduced an auto-sampler system, which has transformed the sample handling and analysis process by automating various tasks, further improving the analysis efficiency and consistency.41 A significant breakthrough in mass cytometry is the integration of a laser ablation system into mass cytometry, i.e. imaging mass cytometry (IMC) (Fig. 4),42 which allows for the highly multiplexed imaging of tissues at the single-cell level. By preserving the spatial context and cellular microenvironment, IMC allows for a more accurate representation of tissue architecture and function, enabling researchers to better understand the complex biomolecular networks and dynamics within the cellular microenvironment.42
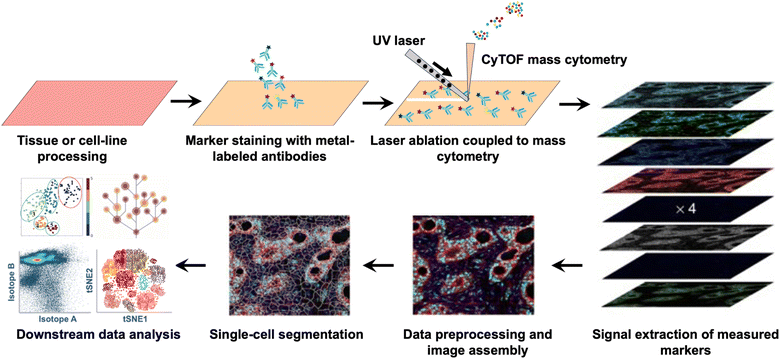 | ||
| Fig. 4 Workflow of imaging mass cytometry: highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry.42 | ||
The sample throughput of mass cytometry was significantly enhanced with the introduction of the mass-tag cellular barcoding (MCB) technique.43 The MCB technique allows for the unique coding of individual samples in a 96-well plate, which are later combined into a single tube for antibody staining, allowing the analysis of multiple samples simultaneously, while still maintaining the ability to differentiate between individual cells within each sample. The introduction of MCB significantly reduces data acquisition time by eliminating sample loading and between-sample wash steps. Additionally, it enhances staining consistency and reduces reagent consumption by concentrating samples prior to antibody staining. These advantages make mass cytometry highly suitable for high-content and high-throughput drug screening, providing researchers with a powerful tool to efficiently analyze large datasets while conserving valuable resources.38
In addition to proteome profiling of single cells using mass cytometry, Frei and colleagues44 expanded the capabilities of mass cytometry for multiplexed transcript quantification by introducing the concept of proximity ligation. They developed a method called proximity ligation assay for RNA (PLAYR), which involves the design of two DNA oligonucleotide probes. These probes have regions that hybridize with target transcripts and serve as templates for the binding and circularization of two additional oligonucleotides called insert and backbone oligonucleotides. The insert and backbone oligonucleotides are ligated and amplified upon hybridization to the adjacent DNA oligonucleotide probes. The amplified products can be detected by mass cytometry using an oligonucleotide labelled with metal tags. This approach has been successfully used for cytokine transcript profiling in lipopolysaccharide-stimulated peripheral blood mononuclear cells (PBMCs), enabling further functional study of different molecules within individual cells.44
The development of metal tags represents a crucial research aspect that drives the advancement of mass cytometry.27 The bifunctional metal-chelating polymers (MCPs) are commercially available tags that are currently used in mass cytometry (Fig. 5A).45 These MCPs contain multiple metal chelator groups, such as DTPA or DOTA, and functional groups that can interact with antibodies, such as N-hydroxysuccinimide (-NHS) or -maleimide (-MAL).27 The metal chelator groups enable MCP tags to bind to different metal isotopes, while the functional groups allow for the conjugation of metal tags onto antibodies. The metal content of metal tags plays a critical role in determining the detection limit of mass cytometry. In recent years, there has been a surge of interest worldwide to improve the sensitivity of mass cytometry by increasing the metal content of metal tags.27 One prominent strategy for improving the sensitivity of mass cytometry involves the utilization of metal nanoparticle tags.46 These tags offer a distinct advantage over traditional metal tags, primarily due to their significantly higher metal load. Lin et al.46 has reported that the incorporation of NaHoF4 nanoparticles can enhance the sensitivity of mass cytometry by two orders of magnitude compared to traditional metal-conjugated polymers (MCPs). Recently, Wu and his team47 have introduced a novel bifunctional lanthanide nano-polymer that possesses both fluorescent and metal properties, making it suitable for both flow and mass cytometry analyses. Furthermore, Dang and coworkers48 have synthesized a nanometal organic framework (NMOF) (Fig. 5B), specifically Zr-NMOF, which has been successfully utilized in mass cytometry. This innovative application allows for the utilization of four additional channels and provides a fivefold signal amplification. Additionally, they have developed a range of multicolor lanthanide-doped dual-functional tags that serve as both fluorescence and mass dual-imaging probes (Fig. 5C).49 This approach significantly reduces the scanning time of imaging mass cytometry (IMC) by approximately 90%, greatly improving the overall imaging efficiency.
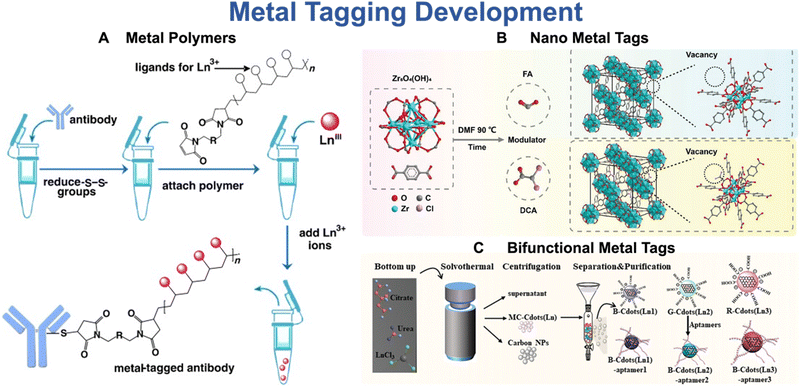 | ||
| Fig. 5 Representatives of metal polymers (A), nano-metal tags (B), and bifunctional metal tags (C) as reporters of mass cytometry. Reproduced with permission from ref. 45 and 48. | ||
2.3 Nano-SIMS
Nanoscale Secondary Ion Mass Spectrometry (Nano-SIMS) is a powerful analytical technique that enables the imaging and quantification of elements and isotopes with a resolution of up to 50 nm.50 It works by focusing a beam of primary ions onto the sample, causing the emission of secondary ions, which are then analyzed using a mass spectrometer. Nano-SIMS can be operated using two primary ion sources, i.e. Cs+ or O−. These primary ions are rastered over the sample surface with an impact energy of 16 keV and a probe current typically falls within the picoampere range. Over several years of development, Nano-SIMS has undergone remarkable improvement in terms of spatial resolution, sensitivity, multiplexity, and the breadth of analytes.50 With the advent of Nano-SIMS 50L, which incorporates a co-linear optics system consisting of an electrostatic sector and a magnetic sector in a co-axial configuration, Nano-SIMS is now capable of simultaneously detecting up to seven ionic species from the same sputtered volume.50 This significant advancement allows for enhanced analysis and precise measurement of isotope ratios in diverse samples.51 Nano-SIMS has a broad range of applications in tracking both endogenous elements, including Cu, Zn, N, S, and P, and metallodrugs such as platinum-52 and ruthenium-based53 anticancer drugs. The application of Nano-SIMS for a wide range of elemental analysis provides information on the distribution, localization, and interaction of metallodrugs with cellular components at the nanoscale,52,53 providing a deeper understanding of the behavior of these drugs within cells. Moreover, Nano-SIMS has the capability to map the distribution of specific elements that are naturally present in biological systems, such as 12C, 14N, 31P, and 32S. These elements can be effectively employed to trace various cell organelles, as well as visualize and characterize specific biomolecules within biological samples.2.4 Multiplexed ion beam imaging (MIBI) techniques
Given the highly heterogeneous components and complex microenvironment of biological samples, there is an increasing demand for analytical techniques that can simultaneously quantify and visualize multiple targets.40 Multiplexed ion beam imaging (MIBI) has emerged as a cutting-edge technique to address this challenge by integration of multiple ion beams, time-of-flight secondary ion mass spectrometry and high-throughput imaging, enabling highly multiplexed imaging of various metal isotopes in tissue samples (Fig. 6).54 By facilitating multiplexed quantification and visualization, MIBI holds great promise in revolutionizing the field of biological imaging and expediting discoveries across various critical areas, including diagnostics, therapeutics, and personalized medicine.54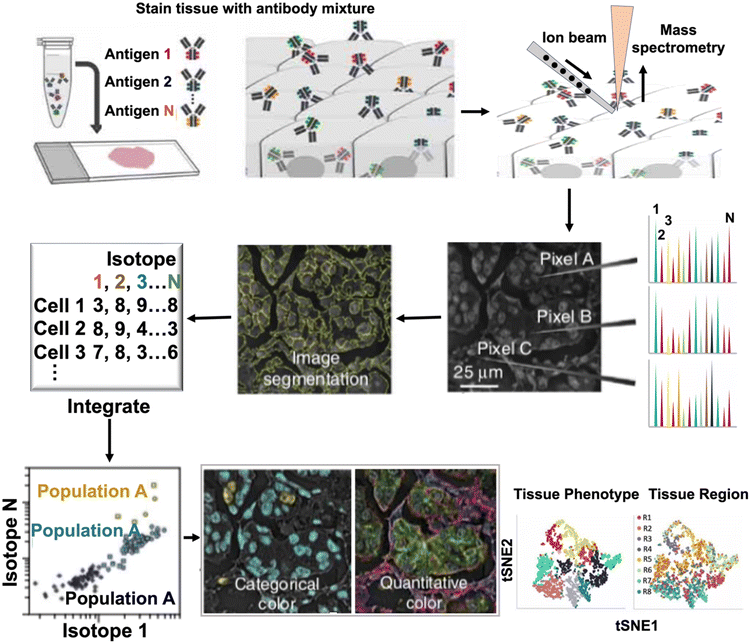 | ||
| Fig. 6 Experimental design of multiplexed ion beam imaging (MIBI) techniques: multiplexed imaging for cellular phenotypes and tissue structures by MIBI. | ||
The workflow of the first generation of MIBI is illustrated in Fig. 6.54 Typically, biological specimens are initially incubated with multiple primary antibodies, each labeled with a distinct stable lanthanide tag. These specimens are then mounted on a sample holder and subjected to a rasterized oxygen duoplasmatron primary ion beam. As a result, the lanthanide reporters and tissue-endogenous elements are sputtered and liberated as secondary ions. Subsequently, these secondary ions are analyzed using a magnetic sector mass spectrometer equipped with multiple detectors, enabling the simultaneous detection of various lanthanide isotopes (metal-based reporters). The intensities of these isotopes are directly proportional to the corresponding antibodies and their associated specific targets. By constructing a two-dimensional map of the elemental distribution of each lanthanide, researchers can gain valuable insights into the spatial distribution and abundance of the target molecules within the biological sample. MIBI signifies a groundbreaking advancement in the field of single-cell imaging, offering the remarkable capability to simultaneously analyze up to 100 targets with a wide dynamic range of five logs. Significantly, MIBI achieves imaging resolutions as low as 10 nm, exceeding the capabilities of IMC by over 100-fold. This remarkable improvement in imaging resolution enables researchers to capture intricate details and molecular interactions with unmatched precision and clarity.
For the first generation of MIBI, the utilization of a magnetic sector imposes constraints on the simultaneous detection of masses, which is limited to the number of ion detectors available.55 In commercial instruments, the number of ion detectors typically does not exceed seven due to cost and complexity constraints. Additionally, the mass calibration, primary and secondary ion optics alignment, and sample loading process in MIBI are intricate and time-consuming, further contributing to the overall complexity of operating the instrument. Unlike the first-generation MIBI, which uses a magnetic sector to separate ions by their mass-to-charge ratio, the second-generation MIBI, i.e. MIBI-TOF,55 employs a linear time-of-flight (TOF) mass analyzer, which remarkably increases the multiplexity and sensitivity of MIBI. Furthermore, there is no need for mass calibration or ion optics alignment in MIBI-TOF, making it more accessible and user-friendly. MIBI-TOF significantly enhances the throughput of MIBI by 36-fold, resulting in a significant reduction in acquisition time during experiments. The throughput could be further improved by introducing a nanobarcoding platform that is based on isotopically encoded silica nanotags.56 MIBI-TOF represents the most powerful elemental imaging technique currently available, as it enables the simultaneous quantification of all naturally occurring elements ranging from hydrogen to uranium. As a result, it provides subcellular quantification and visualization of both elementally labeled antibodies and native biological elements (e.g., P and Fe) with extremely high multiplexity. Furthermore, MIBI-TOF allows for repeat scanning of a single section, making it possible to conduct intelligible study designs at different scales of throughput and resolution and even acquire multiplexed three-dimensional images.55
Recently, Rovira-Clavé and coworkers made a significant breakthrough in MIBI technology with the development of high-definition multiplex ion beam imaging (HD-MIBI). This innovative technique utilizes a positively charged cesium primary ion beam with a small spot size, allowing for the pseudo-3D super-resolution visualization of subcellular features with exceptional lateral (XY) and axial (Z) resolutions of around 30 nm and 5 nm, respectively. Additionally, they have designed a powerful toolkit that utilizes multiple isotope-tagged antibodies for specific protein imaging. This enables the accurate determination of the precise subcellular locations of various small molecules, proteins, and nucleic acids, ultimately providing a comprehensive understanding of the intricate interactions among biomolecules within cells. HD-MIBI represents the most advanced and powerful multiplexed imaging technique, providing ultra-high resolution, exceptional sensitivity, and the capacity to analyze a diverse range of analytes.57
3. Metal-detection based techniques for the elucidation of the role of metals in biology and medicine
The rapid development of metal-detection based and metallomic/metalloproteomic approaches has greatly accelerated our understanding of the vital roles that metals play in biology and medicine.4 These advancements have provided valuable insights into the intricate mechanisms and functions of metals in biological systems, leading to significant progress in the field. This section highlights the latest findings derived from these advanced metal-detection based and metallomic/metalloproteomic techniques and provides insights into their future potential applications.3.1 The involvement of metals in disease development and progression
Metals play a crucial role in many important biological and physiological pathways.58,59 Increasing clinical evidence suggests that imbalances in the metallome are associated with different kinds of diseases.21,22,60 The identification of specific metals and their roles in disease pathogenesis is crucial for understanding the underlying mechanisms and developing targeted therapeutic interventions.ICP-MS is the most widely used method for the determination of metal contents in clinical samples, such as serum, urine, and saliva. Metallome profiling by ICP-MS has provided valuable insights into the associations between specific metals and diseases. It enables the identification of different metals associated with disease status and uncovers metal–metal interplays in disease development and progression. For instance, metallome profiling using ICP-MS uncovered a significant correlation between serum copper (Cu) levels and bladder cancer.62 Additionally, selenium (Se), zinc (Zn), and iron (Fe) have shown significant links to bipolar disorder and schizophrenia.61 These findings underscore the potential of metallome profiling in uncovering the role of metals in the development and progression of various diseases. Recently, we have comprehensively evaluated metallome modulation in individuals infected with SARS-CoV-2 and with disease progression.21,22 The study revealed distinct metallome features in COVID-19 patients compared to non-infected control subjects, serving as a potential biomarker for disease diagnosis. Additionally, the serum levels of Se and Zn were significantly reduced in COVID-19 patients, particularly in more severe cases, indicating their potential role in the progression of the disease. Furthermore, the study also revealed the modulation of metal–metal interplay with the disease progression. This comprehensive investigation provides a holistic view of the host response to SARS-CoV-2 infections through modulation of the metallome. Jiang and coworkers discovered that bladder cancer (BCa) can lead to a significant alteration in the ratio of natural copper isotopes (65Cu/63Cu) in the blood of patients compared to benign and healthy controls.62 The identification of inherent copper isotopic signatures in bladder cancer patients has yielded valuable insights into the molecular mechanisms responsible for copper imbalance during cancer development. Furthermore, these copper isotopic signatures have emerged as valuable biomarkers for the diagnosis of bladder cancer, offering significant potential for early detection and improved clinical management of the disease.
3.2 The mode of action behind the pharmaceutical activities of metal complexes
In recent years, significant progress has been made in understanding the mode of action of metal-based pharmaceuticals with the aid of innovative metal-detection based techniques. Systematic identification of metal-binding proteins is particularly important toward unveiling their roles in biology, given that metallodrugs often bind and functionally disrupt the biological functions of metalloproteins.36,63,64 To elucidate the mechanisms of action of metallo-pharmaceuticals as antibacterial and anticancer agents, GE-ICP-MS was utilized to identify different metal-binding proteins in bacterial and cancer cells, including bismuth (Bi) binding proteins in H. pylori65,66 and arsenic (As) binding proteins in leukemia cells.34 To further enhance the resolution of protein separation, two-dimensional GE-ICP-MS, i.e., LC-GE-ICP-MS, was developed by incorporating Liquid Chromatography (LC) with the GE-ICP-MS technique.35 This platform was utilized to successfully identify 34 Ag+-binding proteins in Escherichia coli (E. coli). By the integration of metalloproteomics, metabolomics, bioinformatics, and system biology, the antimicrobial action of Ag+ was successfully decoded at the system level.35 Furthermore, Ag+ was found to induce systematic damage and bacterial death primarily by disrupting glycolysis and the TCA cycle, followed by the destruction of the adaptive glyoxylate pathway and suppression of cellular oxidative stress responses. Key enzymes such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and malate dehydrogenase were validated as critical targets of Ag+.67,68 This approach has also been utilized to uncover the molecular mechanism underlying the antimicrobial activity of Ag+ against S. aureus (Fig. 7).36 Unlike its mode of action against E. coli, Ag eradicates S. aureus primarily by targeting glycolysis through the inhibition of multiple essential enzymes and inducing elevated levels of reactive oxygen species (ROS) by disrupting the redox homeostasis system at a late stage. This leads to the upregulation of oxidative pentose phosphate pathway (oxPPP) enzymes as a response to alleviate Ag+ stress. However, the activation of oxPPP is ultimately ineffective due to the inhibition of key oxPPP enzymes by Ag+. This mode of action enables Ag to enhance the efficacy of various conventional antibiotics and restore the susceptibility of methicillin-resistant S. aureus to antibiotics.36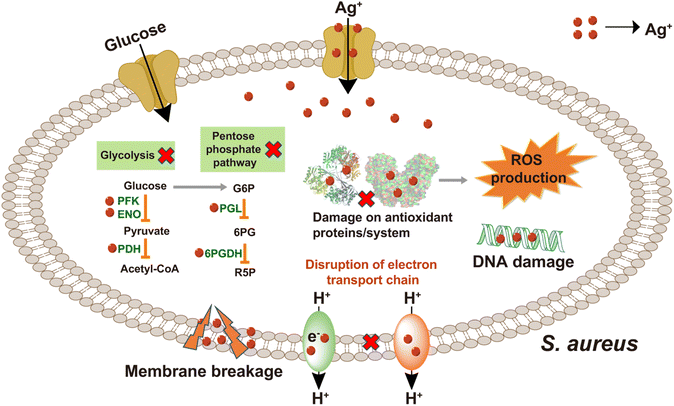 | ||
| Fig. 7 Mode of action of silver: LC-GE-ICP-MS reveals that the multi-target mode of action of silver suppresses its antibiotic selection effect on S. aureus. | ||
Given the inherent cellular heterogeneity and intricate diversified functions, the simultaneous investigation of multiple parameters at the single-cell level is crucial for comprehending the impact of metal-based pharmaceuticals on diverse cellular processes. As a powerful tool for studying complex biological systems, mass cytometry has found wide application in various fields, such as immunology, cancer diagnosis and prognosis, cellular signaling, and drug discovery.38 In recent years, there has been increasing recognition of the potential of mass cytometry in studying metal-based pharmaceuticals, particularly in the investigation of their uptake in tumor cells and corresponding cellular response.69,70 For instance, mass cytometry has been employed to quantify the intracellular accumulation of cisplatin in ovarian cancer to investigate the impact of ABCC2 overexpression on cisplatin resistance. It was found that certain ABCC2 inhibitors effectively counteracted cisplatin resistance by enhancing the accumulation of platinum within the cancer cells. Interestingly, these inhibitors maintained a significant portion of calcein efflux activity, indicating the existence of distinct efflux mechanisms mediated by ABCC2 that are independent of calcein.71 Furthermore, by utilizing mass cytometry, Chang et al.69 simultaneously quantified the levels of intracellular cisplatin and assessed cell cycle progression and cell proliferation biomarkers (IdU), unveiling a dose- and time-dependent cell cycle arrest triggered by cisplatin in BxPC-3 and ME-180 xenografts. Such insights into the dynamics of cell cycle arrest provide valuable information for understanding the mechanisms of action of cisplatin and optimizing its therapeutic efficacy in specific tumor models.69
In addition to intracellular quantification of metallodrugs, advanced multiplex imaging techniques such as IMC and Nano-SIMS have emerged as powerful tools for visualizing the biodistribution of metallodrugs and studying their interactions within tissues at the site of origin. These multiplexed imaging techniques offer valuable insights into the localization and impact of metallodrugs within biological systems. Through the utilization of IMC, Cao et al.72 have found the long-term retention of platinum in the skin following oxaliplatin administration, suggesting a potential association with oxaliplatin-induced peripheral neuropathy. Chang et al.73 utilized IMC to visualize the localization of platinum and found the extensive binding of platinum to collagen fibers in both tumor and normal mouse tissues (Fig. 8A, top row).
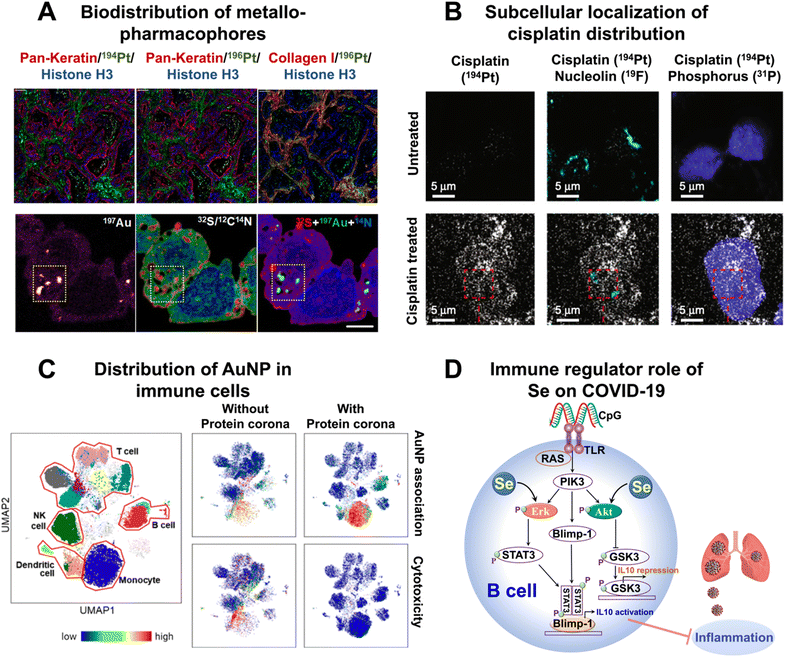 | ||
| Fig. 8 Applications of metal-detection based techniques in the elucidation of the role of metals in medicine. (A) Biodistribution of cisplatin (top row) and AuMesoIX (bottom rows) analyzed by IMC and NanoSIMS, respectively. (B) Representative HD-MIBI images of TYK-nu ovarian cancer cells treated with DMSO (top row) or cisplatin (bottom rows) and colocalized with nucleolin and phosphorus. (C) Heterogeneities of the cellular distribution of AuNPs revealed by mass cytometry. (D) Immune regulatory effect of Se on COVID-19 uncovered by the metal-coding assisted multiplexed multi-omics profiling platform.21 | ||
Apart from visualizing metallodrugs within tissues, NanoSIMS offers the unique capability of simultaneously visualizing ligands labeled with metals and isotopes. This enables the comprehensive study of the dynamic transformation of metallodrugs within cells, as well as the visualization of interactions between metallodrugs and various cellular components.50 NanoSIMS analysis has provided valuable insights into the accumulation and distribution of platinum (Pt) in various cellular structures and organelles following treatment with cisplatin.52 In human colon cancer cells, Pt was found to predominantly accumulate in sulfur (S)-rich structures within the nucleus and cytoplasm, indicating its affinity for DNA.52 Through the integration of confocal laser scanning microscopy and NanoSIMS, it was observed that Pt–N bonds partially dissociate during the accumulation process, which may contribute to the mechanism of action of cisplatin.52 The different processing of Pt drugs in acidic organelles appears to be linked to their detoxification and modes of action.52 The combination of LA-ICP-MS, TEM, and NanoSIMS imaging techniques demonstrated the accumulation of Pt in cytoplasmic S-rich organelles in both kidney and malignant cells of mice treated with investigational Pt(IV) anticancer compounds.74 NanoSIMS also revealed the preferential aggregation of oxaliplatin and other new Pt(II) anticancer compounds in cytoplasmic organelles.75 Recently, the integration of NanoSIMS and electron microscopy (EM) techniques has enabled the discovery that a gold-based compound (AuMesoIX) selectively targets specific cytosolic cancer cell proteins that are rich in thiols (Fig. 8A, bottom rows).76 Benjamin et al.77 employed a multi-omics approach complemented by NanoSIMS and TEM to investigate the molecular events associated with the ER stress response induced by KP1339, a ruthenium-based complex. Their research revealed that KP1339 could directly interact with ribosomal proteins, specifically RPL10 and/or RPL24, leading to the inhibition of proliferation and disruption of ER function. The use of NanoSIMS allowed the visualization of a predominantly homogeneous distribution of ruthenium (KP1339) within the cellular interior, providing further evidence to support the direct interaction between KP1339 and cytoplasmic ribosomes. Confocal laser scanning microscopy and NanoSIMS analysis showed that Pt–N bonds partially dissociate during the accumulation process, and this finding may shed light on the mechanism of action of cisplatin.
In a recent study, Wang et al.78 developed a dual-modal microscopy imaging strategy to investigate the formation of ternary binding complexes involving the transcription cofactor HMGB1, transcription factor Smad3, and cisplatin-crosslinked DNA in single cells. Confocal microscopy was used to map EYFP-fused HMGB1 and fluorescent dye-stained DNA, followed by ToF-SIMS imaging to visualize cisplatin. This integrated imaging approach revealed the formation of HMGB1–Pt–DNA ternary complexes in the cells and the disruption of the interactions between Smad3 and DNA by cisplatin lesions at Smad-binding elements.
The simultaneous visualization of multiple biomolecules associated with their ligands or small molecules within cells may enhance our understanding of biological processes. Recently, Rovira-Clavé and coworkers57 introduced HD-MIBI to visualize cisplatin and five subnuclear structures (Fig. 8B). They found that cisplatin exhibited a preferential enrichment in nuclear speckles while being excluded from closed-chromatin regions, suggesting that cisplatin may play a role in the active regions of chromatin. Furthermore, by evaluating cells that survived multi-drug treatment with cisplatin and the BET inhibitor JQ1, they showed a near-complete exclusion of cisplatin from the nucleus. This finding indicates that selective subcellular relocalization of cisplatin could potentially modulate resistance to cisplatin treatment.
3.3 Interactions between nanoparticles and cells
The utilization of nanoparticles in medicine has become increasingly popular due to their versatility in serving as drug delivery vehicles, imaging agents, radiosensitizers, and even therapeutic agents.79 However, a better understanding of their interactions with different types of cells is critical to advancing research in the field of nanomedicine and developing more effective and targeted nanoparticle-based treatments.Mass cytometry has emerged as a powerful tool for nano-related research in recent years, with rapid advancements in its application for nanoparticle biodistribution analysis, functional characterization, and toxicity assessment. For instance, mass cytometry has been employed to track the in vivo fate of AuNPs by combining gold quantification with comprehensive cellular phenotyping.80 This approach allows for a detailed assessment of how AuNPs interact with different cell types and provides valuable insights into their distribution and behavior within the body.80 In a recent study by Park et al.,81 it was discovered that the presence of a protein corona has a profound impact on the cellular associations and cytotoxicity of AuNPs in human immune cells (Fig. 8C). Interestingly, the effects of the protein corona varied significantly depending on the type of immune cell and their respective subsets. Especially, it was observed that naive T killer cells displayed greater heterogeneity with unique cellular association patterns compared to memory T killer cells. These findings open up possibilities for the application of precisely engineered nanoparticles (NPs) in nanomedicine, offering potential opportunities for targeted drug delivery and vaccine development in diverse clinical settings. Through the use of mass cytometry, it was revealed that AgNPs exhibit heterogeneous biodistribution among different types of immune cells, with a notable accumulation in monocytes and B cells.84 By incorporating scRNA-seq analysis, it was found that exposure to AgNPs triggers NRF2-mediated oxidative stress in B cells, as well as NFAT-regulated immune response and Fcγ-mediated phagocytosis in monocytes.82 These findings provide a deeper understanding of how AgNPs can perturb the immune system at a molecular level. Chen's group83 demonstrated that peripheral nerve fibers can act as direct conduits for the translocation of silver nanomaterials (Ag NMs) from the gut to the central nervous system in both mice and rhesus monkeys. Interestingly, after oral administration, Ag NMs were found to be significantly enriched in the brain and spinal cord of mice, despite not efficiently entering the bloodstream. This study also revealed that enterocytes and enteric nerve cells take up substantial amounts of Ag NMs, which are subsequently transferred to the connected peripheral nerves.83 These findings provide compelling evidence for the existence of a previously undocumented gut-CNS axis facilitated by peripheral nerves, highlighting the occurrence of nanoparticle transfer through this pathway. In a recent study, Malysheva et al.84 investigated the cellular binding, uptake, and intracellular transformation of AgNPs through the integration of mass cytometry, TR-ICP-MS, and synchrotron X-ray absorption spectrometry. It showed that the intracellular and extracellularly bound AgNPs undergo significant transformation, which is dependent on their primary size and surface coating. Additionally, sulfidation was found to dominate the biotransformation of AgNPs, which was involved in the cellular detoxification pathways for Ag. This integrative approach provides a general strategy to explore the cell–NP interactions and biotransformation in different systems.84
3.4 The mechanisms underlying metal-mediated immunity
The importance of metals in immunology has been increasingly recognized. Several metals, including Zn, Cu, Fe, Se, and Mn, are essential for various aspects of immune cell development, activation, and function.85 For example, zinc is involved in the maturation and activity of immune cells,86 while copper helps regulate immune cell proliferation and function.87 Iron is crucial for the growth and differentiation of immune cells,88 and selenium is important for the optimal function of immune cells, including T cells89 and natural killer (NK) cells.90 Understanding the role of metals in immunity is crucial for developing strategies to modulate immune responses and combat immune-related disorders.Very recently, to uncover the intrinsic mechanisms of action of metals in immunity and pathogenesis, a metal-coding assisted multiplexed multi-omics profiling platform, i.e., metallome–metalloproteome–immuneproteome, by using ICP-MS was developed.21,22,24 This approach provides a general platform to identify the correlation between different metals and the immune response of different kinds of disease (Fig. 8D). By taking COVID-19 patient serum samples as a showcase, a correlation network between the host metallome and immunoproteomes was constructed, revealing a significant association between selenium and interleukin-10 (IL-10). Moreover, by using mass cytometry to identify and characterize immune cell modulation in COVID-19 patients, the impact of selenium on the functions of various immune cell types was explored. The discovered association between selenium and IL-10 is partially attributable to the activation of ERK and Akt pathways in B cells by selenium.21 Furthermore, using similar approach, the same research group further uncovered the critical role of liver-mediated iron dysregulation in COVID-19 disease severity, providing solid evidence on the involvement of iron-related proteins in COVID-19 pathophysiology and immunity.22 The integration of a metal-coding assisted multiplexed multi-omics profiling platform with mass cytometry represents a powerful and versatile method for metalloimmunity study.
Alongside the immune modulation role of endogenous metals in immune cells, there has been growing interest in exploring metal complexes for their potential use in cancer immunotherapy. Researchers have been actively investigating these metal complexes with the goal of leveraging their immune-modulating capabilities to enhance the efficacy of immunotherapeutic strategies in cancer treatment. For instance, platinum complexes have been extensively studied as potential inhibitors of immune checkpoints, specifically targeting checkpoints like TDO and PD-L1, which have the ability to restore the immune surveillance of cancer cells.91 Additionally, cisplatin has shown the ability to enhance antigen presentation through MHC I, leading to increased levels of tumor antigen presentation and activation of specific T lymphocytes.92 Moreover, Pt and Ru-based complexes have been studied for their potential to interfere with the immunosuppressive tumor microenvironment (TME), thereby inhibiting cancer progression and immune evasion.93–95 Iridium-based complexes have demonstrated the ability to regulate immune functions in cancer cells by inhibiting the release of pro-inflammatory cytokines and cyclooxygenase (COX).96 Rhenium complexes have shown promise in stimulating the maturation and antigen presentation of dendritic cells (DCs), leading to the activation of adaptive immunity.97
The immense potential of metal-based complexes in immunotherapy necessitates a deeper understanding of the intrinsic mechanisms underlying their immune modulation roles. Mass cytometry and IMC hold great potential for elucidating the mechanisms of the immune modulation role of metal-based complexes by tracking the uptake and assessing the influence of different metal complexes across diverse cell subsets. However, there is currently a lack of studies utilizing mass cytometry and IMC for metalloimmunology research. This direction should be given more attention in future research to fully explore the potential of metal-based complexes in immunotherapy.
4. Machine learning-assisted data interpretation of metal-detection based techniques
In the past few decades, the rapid advancements in metal-detection based techniques have led to increasingly complex data derived from multiplexed metal-detection based assays and images. While effectively clarifying these data offers a unique opportunity to gain valuable insights into the mechanisms of biological events, it is a significant challenge. Recently, researchers worldwide have tried to integrate diverse machine learning methods into metal-detection based assay platforms, thereby facilitating improved elucidation of the multi-dimensional data.The utilization of machine learning has revolutionized the analysis of multi-omics data acquired from individual patients. Both supervised algorithms, such as decision trees, and unsupervised algorithms, like PCA, have been employed in ICP-MS based metallome and proteome profiling data analysis to classify or predict diseases. For instance, tSNE has been utilized to successfully classify COVID-19 patients based on distinct metallome patterns.24 A disease severity prediction model was also further developed using a decision tree algorithm, which relied on specific antibody patterns derived from multiplexed proteome data.24 In another study, Wang et al. employed a random forest algorithm to develop a bladder cancer diagnosis model using two-dimensional copper signatures, namely the copper isotopic composition and concentration in plasma and red blood cells.62 These examples highlight the successful application of machine learning algorithms to the analysis of metal-detection based data for disease diagnosis and prediction.
Moreover, machine learning is exceptionally valuable for analyzing single-cell omics data, particularly due to the presence of high-dimensional datasets containing millions of cells obtained from this type of assay. Over the past few years, there have been notable breakthroughs in analyzing multiplexed single-cell data, such as single-cell proteomics data obtained from mass cytometry, as well as spatially resolved data collected from IMC and MIBI. A variety of well-established machine learning tools have been developed specifically for mass cytometry data analysis, primarily focusing on dimensionality reduction and clustering analysis. For instance, the t-Stochastic Neighbor Embedding (tSNE) algorithm is commonly used to analyze mass cytometry data by comparing the similarity of high-dimensional expression profiles.98 Additionally, artificial neural networks can approximate the tSNE embedding function,99 allowing for the projection and comparison of additional data that were not initially included in the embedding process. Recently, the introduction of uniform manifold approximation and projection (UMAP)100 has further improved the preservation of the global structure in high-dimensional datasets, while also reducing the time required for calculations. Amodio et al. have developed a deep neural network,101i.e., SAUCIE, that leverages the parallelization and scalability offered by neural networks. This novel approach integrates the powerful deep representation learning capabilities of neural networks to successfully accomplish a range of tasks in single-cell data analysis. These tasks include imputation and denoising, batch effect removal, clustering, and visualization. In a subsequent study, SAUCIE was validated as the best overall performer among a group of top-performing methods, including t-SNE,102 UMAP,100 scvis103 and PHATE.104 Recently, Wang et al.102 have created CytofDR, a user-friendly package that integrates several dimensionality reduction methods. This package streamlines the process of applying multiple techniques to a single dataset and enables researchers to efficiently explore different approaches for analyzing single-cell data.
In addition to achieving dimensionality reduction, various machine learning methods have also been developed specifically for data clustering purposes. Spanning-tree progression analysis of density normalized events (SPADE)105 is a powerful method that offers an overview of all populations present in a sample by representing them as clusters on a minimum spanning tree. This tree can be color-coded based on any marker of interest, and differential expression can be analyzed when one or more reference samples are included. SPADE is particularly well-suited for comparing the characteristics of multiple populations simultaneously across a set of samples. FlowSOM is a robust machine learning tool used for unsupervised clustering and visualization of high-dimensional data.106 It is based on the self-organizing map (SOM) approach, which allows for the identification of distinct cell populations within the data. FlowSOM divides the data into a predefined number of clusters, assigning each cell to a cluster based on its similarity to other cells. This method enables researchers to explore the phenotypic characteristics of these populations and gain insights into their functional states.
With the emergence of single-cell multiplex imaging techniques like MIBI-TOF and IMC, there is an increasing demand for machine learning tools that can effectively analyze spatial information in imaging data. Unlike mass cytometry data, imaging data necessitates several preprocessing steps, including imaging compensation, normalization, and cell segmentation, to ensure accurate analysis and interpretation. These steps are crucial in preparing the data for subsequent machine learning algorithms. Therefore, the development of machine learning tools that can seamlessly integrate these preprocessing steps and facilitate the analysis of spatial information in imaging data is of paramount importance. In recent years, several machine learning methods have been developed for cell segmentation purposes.107,108 For example, a deep learning-based segmentation method has been employed for MIBI analysis.109 This method involves training a classifier to distinguish between nuclear and non-nuclear pixels, thereby facilitating the automatic identification of nuclei and subsequent cells from extensive image datasets. Once the imaging data from MIBI and IMC have undergone the segmentation process, subsequent analysis can be conducted in a similar manner to single-cell data analysis from mass cytometry.
5. Conclusions and prospects
This perspective presents a comprehensive overview of the latest advancements in metal-detection based techniques, with a specific emphasis on single-cell approaches, and highlights their significant applications in enhancing our comprehension of the roles of metals in different medical contexts. In addition to the technique discussed above, synchrotron radiation-based methods, such as synchrotron radiation X-ray fluorescence, have also emerged as crucial tools for high-resolution elemental mapping in tissues, cellular compartments, or even individual cells.110 The insights gained from these techniques offer invaluable perspectives into the diverse functions of metals, paving the way for the development of innovative metallo-pharmacophores. As the utilization and recognition of metal-detection based techniques continue to expand, their applications in various disciplines will further expand our understanding of the roles of metals in complex biological systems. There are several emerging directions that hold promise for the future development of metal-detection based techniques.One area of focus is the development of more sensitive and stable metal tags, as these tags serve as crucial components in various metal-detection based technologies such as ICP-MS multi-omics assay, mass cytometry, IMC, and MIBI. Improving the sensitivity and stability of these metal tags would enable greater accuracy and precision in detecting and quantifying biomolecules (e.g., protein markers) in complex biological samples. While nanoparticles have shown promise in improving sensitivity, addressing the issue of non-specific binding is crucial. In addition, the development of novel metal tags with unique properties and characteristics has the potential to broaden the application of metal-detection based techniques. One example is the development of metal-containing probes that specifically target enzyme activity.111 With such a probe, metal-detection based techniques can be extended to monitor cellular metabolism and investigate cell-specific responses. Moreover, the development of multi-functional metal tags that are compatible with both fluorescence and metal-detection based detection techniques shows promise as a method to achieve integrative analysis. This approach has the potential to overcome the technical limitations of each individual technique and offer a more comprehensive and accurate analysis.
Another area of focus will be the exploration of metal-detection based techniques, specifically single-cell techniques in metallo-pharmaceutical related research. While these techniques are still in their infancy in this field, they hold great potential for exciting discoveries, particularly in the emerging field of metallo-immunology and precision medicine.
Moreover, as artificial intelligence (AI) continues to advance rapidly, there is growing interest in exploring its applications to facilitate data analysis for metal-detection based techniques, which holds great promise in biomarker discovery, target identification, disease diagnosis, and biomarker screening when integrated into metal-detection based assay platforms.
Last but not least, the integration of multi-omics approaches represents a promising strategy for gaining an understanding of multifaceted metal-involved biological events. By combining multiple layers of data, such as genomics, proteomics, and metabolomics, we can obtain a comprehensive view of the molecular and cellular changes that occur in response to metal-based interactions. The integration of multi-omics approaches has the potential to uncover new insights into the mechanisms of action, toxicity, and therapeutic potential of metal-based compounds, and is expected to drive significant advancements in metallobiology.
Author contributions
Y. Z., and H. L. wrote the manuscript, and all authors reviewed and edited the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Our research on metallomics and metallobiology has been supported by the Research Grants Council of Hong Kong SAR (C7034-20E, R7070-18, 17308921, SRFS2122-7S04, and 17306323) and the University of Hong Kong (URC (202107185074) and Norman & Cecilia Yip Foundation).References
- P. K. Bhattacharya and P. B. Samnani, Metal ions in biochemistry, CRC press, 2020 Search PubMed.
- A. K. Goswami and I. Kostova, Medicinal and Biological Inorganic Chemistry, Walter de Gruyter GmbH & Co KG, 2022 Search PubMed.
- P. Kroneck and M. S. Torres, Metals, Microbes, and Minerals-The Biogeochemical Side of Life, Walter de Gruyter GmbH & Co KG, 2021 Search PubMed.
- Y. Zhou, H. Li and H. Sun, Annu. Rev. Biochem., 2022, 91, 449–473 CrossRef CAS PubMed.
- E. J. Anthony, E. M. Bolitho, H. E. Bridgewater, O. W. L. Carter and J. M. Donnelly, Chem. Sci., 2020, 11, 12888–12917 RSC.
- Y. Zhou, K. I. T. Ip, Q. Zhang, H. Li and H. Sun, Comprehensive Coordination Chemistry III. Elsevier, 2021, 680–705 Search PubMed.
- I. Yousuf, M. Bashir, F. Arjmand and S. Tabassum, Coord. Chem. Rev., 2021, 445, 214104 CrossRef CAS.
- L. Riccardi, V. Genna and M. De Vivo, Nat. Rev. Chem, 2018, 2, 100–112 CrossRef CAS.
- A. Frei, A. D. Verderosa, A. G. Elliott, J. Zuegg and M. A. T. Blaskovich, Nat. Rev. Chem, 2023, 7, 202–224 CrossRef CAS PubMed.
- C. Weng, Y. L. K. Tan, W. G. Koh and W. H. Ang, Angew. Chem., Int. Ed., 2023, 62, e202310040 CrossRef CAS PubMed.
- J. Li, H. Ren and Y. Zhang, Coord. Chem. Rev., 2022, 455, 214345 CrossRef CAS.
- R. Yang, L. Chen, Y. L. Wang, L. J. Zhang, X. Zheng, Y. Yang and Y. X. Zhu, et al. , Front. Immunol., 2023, 14, 1237361 CrossRef CAS PubMed.
- Z. Y. Li, Q. H. Shen, Z. W. Mao and C. P. Tan, Chem.–Asian J., 2022, 17, e202200270 CrossRef CAS PubMed.
- H. Li, R. Wang and H. Sun, Acc. Chem. Res., 2018, 52, 216–227 CrossRef PubMed.
- T. R. Steel and C. G. Hartinger, Metallomics, 2020, 12, 1627–1636 CrossRef CAS PubMed.
- X. Sun, C.-N. Tsang and H. Sun, Metallomics, 2009, 1, 25–31 CrossRef CAS.
- Y.-T. Lai, Y.-Y. Chang, L. G. Hu, Y. Yang and A. L. Chao, et al. , Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2948–2953 CrossRef CAS PubMed.
- H. Wang, L. G. Hu, H. Y. Li, Y.-T. Lai and X. Y. Wei, et al. , Nat. Commun., 2023, 14, 1738 CrossRef CAS PubMed.
- A. Magnusdottir, I. Johansson, L.-G. Dahlgren, P. Nordlund and H. Berglund, Nat. Methods, 2009, 6, 477–478 CrossRef CAS PubMed.
- L. Hu, L. G. Hu, T. F. Cheng, B. He, L. Li, Y. C. Wang, Y.-T. Lai, G. B. Jiang and H. Z. Sun, Angew. Chem., Int. Ed., 2013, 18, 4916–4920 CrossRef PubMed.
- Y. Zhou, S. F. Yuan, F. Xiao, H. Li and Z. W. Ye, et al. , Chem. Sci., 2023, 14, 10570–10579 RSC.
- Y. Zhou, T. F. Cheng, K. M. Tang, H. Y. Li and C. T. Luo, et al. , Clin. Immunol., 2024, 263, 110205 CrossRef CAS PubMed.
- R. S. Amais, G. L. Donati and M. A. Z. Arruda, Trends Anal. Chem., 2020, 133, 116094 CrossRef CAS.
- Y. Zhou, S. F. Yuan, K. K.-W. To, X. H. Xu and H. Li, et al. , Chem. Sci., 2022, 13, 3216–3226 RSC.
- X. Xu, H. Wang, H. Li and H. Sun, Chem. Lett., 2020, 49, 697–704 CrossRef CAS.
- R. Liu, S. X. Zhang, C. Wei, Z. Xing, S. C. Zhang and X. R. Zhang, Acc. Chem. Res., 2016, 49, 775–783 CrossRef CAS PubMed.
- L. P. Arnett, R. Rana, W. W.-Y. Chung, X. C. Li and M. Abtahi, et al. , Chem. Rev., 2023, 123, 1166–1205 CrossRef CAS PubMed.
- Y. Zhou, H. B. Wang, E. Tse, H. Li and H. Sun, Anal. Chem., 2018, 90, 10465–10471 CrossRef CAS PubMed.
- Y. Zhou, H. Li and H. Sun, Chem. Commun., 2017, 53, 2970–2973 RSC.
- M. Corte Rodríguez, A.-F. García, E. B. J. Bettmer and M. Montes-Bayón, Anal. Chem., 2017, 89, 11491–11497 CrossRef PubMed.
- C.-N. Tsang, K.-S. Ho, H. Sun and W.-T. Chan, JACS, 2011, 133, 7355–7357 CrossRef CAS PubMed.
- H. Pan, L. X. Feng, Y. L. Lu, Y. C. Han, J. P. Xiong and H. M. Li, Trends Anal. Chem., 2022, 156, 116710 CrossRef CAS.
- M. Schaier, S. Theiner, D. Baier, G. Braun, W. Berger and G. Koellensperger, JACS Au, 2023, 3, 419–428 CrossRef CAS PubMed.
- X. Xu, H. B. Wang, H. Li, X. Q. Hu, Y. Zhang, X. Y. Guan, P. H. Toy and H. Sun, Chem. Commun., 2019, 55, 13120–13123 RSC.
- H. Wang, A. X. Yan, Z. G. Liu, X. M. Yang and Z. L. Xu, et al. , PLoS Biol., 2019, 17, e3000292 CrossRef PubMed.
- H. Wang, M. J. Wang, X. H. Xu, P. Gao and Z. L. Xu, et al. , Nat. Commun., 2021, 12, 3331 CrossRef CAS PubMed.
- S. C. Bendall, E. F. Simonds, P. Qiu, E. D. Amir and P. O. Krutzik, et al. , Science, 2011, 332, 687–696 CrossRef CAS PubMed.
- M. H. Spitzer and G. P. Nolan, Cell, 2016, 165, 780–791 CrossRef CAS PubMed.
- D. R. Bandura, V. I. Baranov, O. I. Ornatsky, A. Antonov and R. Kinach, et al. , Anal. Chem., 2009, 81, 6813–6822 CrossRef CAS.
- F. J. Hartmann and S. C. Bendall, Nat. Rev. Rheumatol., 2020, 16, 87–99 CrossRef.
- T. Selvanantham, S. K. H. Li, N. Zabinyakov, A. Bouzekri, R. Jong, M. Sullivan, A. Laboda, D. Majonis and C. Loh, Cancer Res., 2022, 82, 3908 CrossRef.
- C. Giesen, H. A. O. Wang, D. Schapiro, N. Zivanovic and A. Jacobs, et al. , Nat. Methods, 2014, 11, 417–422 CrossRef CAS PubMed.
- B. Bodenmiller, E. R. Zunder, R. Finck, T. J. Chen and E. S. Savig, et al. , Nat. Biotechnol., 2012, 30, 858–867 CrossRef CAS PubMed.
- A. P. Frei, F.-A. Bava, E. R. Zunder, E. W. Y. Hsieh, S.-Y. Chen, G. P. Nolan and P. F. Gherardini, Nat. Methods, 2016, 13, 269–275 CrossRef CAS PubMed.
- X. Lou, G. H. Zhang, I. Herrera, R. Kinach, O. Ornatsky, V. Baranov, M. Nitz and M. A. Winnik, Angew. Chem., Int. Ed., 2007, 46, 6111–6114 CrossRef CAS PubMed.
- W. Lin, Y. Hou, Y. J. Lu, A. I. Abdelrahman, P. P. Cao and G. Y. Zhao, et al. , Langmuir, 2014, 30, 3142–3153 CrossRef CAS PubMed.
- X. Wu, Q. DeGottardi, I. C. Wu, J. B. Yu, L. Wu and F. M. Ye, et al. , Angew. Chem., Int. Ed., 2017, 56, 14908–14912 CrossRef CAS PubMed.
- J. Dang, H. X. Li, L. L. Zhang, S. J. Li and S. Y. Huang, et al. , Adv. Mater., 2021, 33, 2008297 CrossRef CAS PubMed.
- Y. Yu, Y. Y. Yu, X. Wang, X. L. Jia, Z. J. Feng, L. L. Zhang, H. X. Li, J. He, G. X. Shen and X. T. Ding, Adv. Sci., 2021, 8, 2102812 CrossRef CAS PubMed.
- K. Li, J. L. Liu, C. R. M. Grovenor and K. L. Moore, Annu. Rev. Anal. Chem., 2020, 13, 273–292 CrossRef CAS PubMed.
- Q. Li, J. J. Chang, L. F. Li, X. Y. Lin and Y. C. Li, Sci. Total Environ., 2023, 905, 167257 CrossRef CAS PubMed.
- A. A. Legin, A. Schintlmeisterc, M. A. Jakupecab, M. S. Galanskiab, I. Lichtscheidle, M. Wagnercd and B. K. Keppler, Chem. Sci., 2014, 5, 3135–3143 RSC.
- B. Neuditschko, A. A. Legin, D. Baier, A. Schintlmeister, S. Reipert and M. Wagner, et al. , Angew. Chem., Int. Ed., 2021, 60, 5063–5068 CrossRef CAS PubMed.
- M. Angelo, S. C. Bendall, R. Finck, M. B. Hale and C. Hitzman, et al. , Nat. Med., 2014, 20, 436–442 CrossRef CAS PubMed.
- L. Keren, M. Bosse, S. Thompson, T. Risom and K. Vijayaragavan, et al. , Sci. Adv., 2019, 5, eaax5851 CrossRef CAS PubMed.
- S. Harmsen, H. S. Zeng, H. L. Jin and H. B. Wang, Adv. Mater. Technol., 2020, 5, 2000098 CrossRef CAS PubMed.
- X. Rovira-Clave, S. Z. Jiang, Y. H. Bai, B. K. Zhu and G. Barlow, et al. , Nat. Commun., 2021, 12, 4628 CrossRef CAS PubMed.
- L. Wang, Y. L. Yin, X. Z. Liu, P. Shen, Y. G. Zheng, X. R. Lan, C. B. Lu and J. Z. Wang, Transl. Neurodegener., 2020, 9, 1–13 CrossRef PubMed.
- M. T. Maung, A. Carlson, M. Olea-Flores, L. Elkhadragy, K. M. Schachtschneider, N. Navarro-Tito and T. Padilla-Benavides, FASEB J., 2021, 35, e21810 CrossRef CAS PubMed.
- S. A. Saleh, H. M. Adly, A. A. Abdelkhaliq and A. M. Nassir, Curr. Urol., 2020, 14, 44–49 CrossRef CAS PubMed.
- E. C. S. Cruz, K. C. Madrid, M. A. Z. Arruda, A. Sussulini and J. Trace, Elem. Med. Biol., 2020, 59, 126467 CrossRef PubMed.
- W. Wang, X. Liu, C. W. Zhang, F. Sheng and S. J. Song, et al. , Chem. Sci., 2022, 13, 1648–1656 RSC.
- D. M. Griffith, H. Y. Li, M. V. Werrett, P. C. Andrews and H. Sun, Chem. Soc. Rev., 2021, 50, 12037–12069 RSC.
- H. Wang, Y. Zhou, X. H. Xu, H. Y. Li and H. Sun, Curr. Opin. Chem. Biol., 2020, 55, 171–179 CrossRef CAS PubMed.
- X. Yang, M. Koohi-Moghadam, R. M. Wang, Y. Y. Chang, P. C. Y. Woo, J. W. Wang, H. Li and H. Sun, PLoS Biol., 2018, 16, e2003887 CrossRef PubMed.
- Y. Wang, Z. Zhang, Y. X. Xie, X. Q. Hu, H. B. Wang, W. Xia, Y. L. Wang, H. Y. Li, Y. C. Wang and H. Sun, Chem. Sci., 2017, 8, 4626–4633 RSC.
- H. Wang, M. J. Wang, X. M. Yang, X. H. Xu, Q. Hao, A. X. Yan, M. L. Hu, R. Lobinski, H. Y. Li and H. Sun, Chem. Sci., 2019, 10, 7193–7199 RSC.
- H. Wang, X. M. Yang, M. J. Wang, M. L. Hu, X. H. Xu, A. X. Yan, Q. Hao, H. Li and H. Sun, Chem. Sci., 2020, 11, 11714–11719 RSC.
- Q. Chang, O. I. Ornatsky, C. J. Koch, N. Chaudary, D. T. Marie-Egyptienne, R. P. Hill, S. D. Tanner and D. W. Hedley, Int. J. Cancer, 2015, 136, 1202–1209 CrossRef CAS PubMed.
- L. Corte-Real, R. G. Teixeira, P. Gírio, E. Comsa, A. Moreno and R. Nasr, et al. , Inorg. Chem., 2018, 57, 4629–4639 CrossRef CAS PubMed.
- E. Comsa, K.-A. Nguyen, F. Loghin, A. Boumendjel, M. Peuchmaur, T. Andrieu and P. Falson, Future Med. Chem., 2018, 10, 1349–1360 CrossRef CAS PubMed.
- Y. Cao, Q. Chang, W. J. Zhang, O. Ornatsky, D. Hedley and E. X. Chen, Cancer Chemother. Pharmacol., 2019, 84, 1195–1200 CrossRef CAS PubMed.
- Q. Chang, O. I. Ornatsky, I. Siddiqui, R. Straus, V. I. Baranov and D. W. Hedley, Sci. Rep., 2016, 6, 36641 CrossRef CAS PubMed.
- A. Legin, S. Theinera, A. Schintlmeisterb, S. Reipertc and P. Heffeterd, et al. , Chem. Sci., 2016, 7, 3052–3061 RSC.
- D. Hu, C. Yang, C.-N. Lok, F. R. Xing, P.-Y. Lee, Y. M. E. Fung, H. B. Jiang and C.-M. Che, Angew. Chem., Int. Ed., 2019, 131, 11030–11034 CrossRef.
- K.-C. Tong, K.-C. Tong, C.-N. Lok, P.-K. Wan and C.-M. Che, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 1321–1329 CrossRef CAS PubMed.
- B. Neuditschko, A. A. Legin, D. Baier, A. Schintlmeister and S. Reipert, et al. , Angew. Chem., Int. Ed., 2021, 60, 5063–5068 CrossRef CAS PubMed.
- Y. Lin, K. Wu, F. F. Jia, L. Chen, Z. Y. Wang and Y. Y. Zhang, Chem. Sci., 2021, 12, 5419–5429 RSC.
- J. J. Rennick, A. P. Johnston and R. G. Parton, Nat. Nanotechnol., 2021, 16(3), 266–276 CrossRef CAS PubMed.
- Y. S. Yang, P. U. Atukorale, K. D. Moynihan, A. Bekdemir, K. Rakhra, L. Tang, F. Stellacci and D. J. Irvine, Nat. Commun., 2017, 8, 14069 CrossRef CAS PubMed.
- S. Park, M. K. Ha, Y. Lee, J. Song and T. H. Yoon, ACS Nanosci. Au, 2023, 3, 323–334 CrossRef CAS PubMed.
- M. K. Ha, S. J. Kwon, J.-S. Choi, N. T. Nguyen, J. Song, Y. Lee, Y.-E. Kim, I. Shin, J.-W. Nam and T. H. Yoon, Small, 2020, 16, e1907674 CrossRef PubMed.
- X. Wang, X. Cui, J. Wu, L. Bao, Z. Tan and C. Chen, Sci. Adv., 2023, 9, 2252 CrossRef PubMed.
- A. Malysheva, A. Ivask, C. L. Doolette, N. H. Voelcker and E. Lombi, Nat. Nanotechnol., 2021, 16(8), 926–932 CrossRef CAS PubMed.
- C. C. Murdoch and E. P. Skaar, Nat. Rev. Microbiol., 2022, 20, 657–670 CrossRef CAS PubMed.
- D. Skrajnowska and B. Bobrowska-Korczak, Nutrients, 2019, 11, 2273 CrossRef CAS PubMed.
- T. R. Kramer and W. T. Johnson, Copper and Immunity, CRC Press, 2020, pp. 239–254 Search PubMed.
- Y. Jiang, C. F. Li, Q. Wu, P. An and L. Q. Huang, Nat. Commun., 2019, 10, 2935 CrossRef PubMed.
- Y. Yao, Z. Chen, H. Zhang, C. L. Chen and M. Zeng, et al. , Nat. Immunol., 2021, 22, 1127–1139 CrossRef CAS PubMed.
- Z. Wei, Y. Yi, Z. Luo, X. Y. Gong and Y. X. Jiang, et al. , Adv. Mater., 2022, 34, 2108167 CrossRef CAS PubMed.
- S. Hua, F. H. Chen, G. Xu and S. H. Gou, Eur. J. Med. Chem., 2019, 169, 29–41 CrossRef CAS PubMed.
- S. Wan, S. Pestka, R. G. Jubin, Y. L. Lyu, Y.-C. Tsai and L. F. Liu, PLoS One, 2012, 7, e32542 CrossRef CAS PubMed.
- Q. Chen, L. Z. He, X. Y. Li, L. G. Xu and T. F. Chen, Biomaterials, 2022, 281, 121371 CrossRef CAS PubMed.
- S. Jin, E. Yin, C. Y. Feng, Y. W. Sun, T. Yang, H. Yuan, Z. J. Guo and X. Y. Wang, Chem. Sci., 2023, 14, 8327–8337 RSC.
- L. Porter, J. J. Zhu, N. L. Lister, S. G. Harrison, S. Keerthikumar and D. L. Goode, et al. , Nat. Commun., 2023, 14(1), 5346 CrossRef CAS PubMed.
- X. W. Wu, Y. Zheng, F.-X. Wang, J.-J. Cao and H. Zhang, et al. , Chem. Eur J., 2019, 25, 7012–7022 CrossRef CAS PubMed.
- X. Su, W.-J. Wang, Q. Cao, H. Zhang, B. Liu, Y. Y. Ling, X. T. Zhou and Z. W. Mao, Angew. Chem., Int. Ed., 2022, 61, e202115800 CrossRef CAS PubMed.
- D. Kobak and G. C. Linderman, Nat. Biotechnol., 2021, 39, 156–157 CrossRef CAS PubMed.
- H. Cho, B. Berger and J. Peng, Cell Syst., 2018, 7, 185–191 CrossRef CAS PubMed.
- E. Becht, L. McInnes, J. Healy, C.-A. Dutertre, I. W. H. Kwok, L. G. Ng, F. Ginhoux and E. W. Newell, Nat. Biotechnol., 2019, 37, 38–44 CrossRef CAS PubMed.
- M. Amodio, D. V. Dijk, K. Srinivasan, W. S. Chen and H. Mohsen, et al. , Nat. Methods, 2019, 16, 1139–1145 CrossRef CAS PubMed.
- K. Wang, Y. Yang, F. Wu, B. Song, X. Wang and T. Wang, Nat. Commun., 2023, 14, 1836 CrossRef CAS PubMed.
- J. Ding, A. Condon and S. P. Shah, Nat. Commun., 2018, 9, 2002 CrossRef PubMed.
- K. R. Moon, D. van Dijk, Z. Wang, S. Gigante and D. B. Burkhardt, et al. , Nat. Biotechnol., 2019, 37, 1482–1492 CrossRef CAS PubMed.
- P. Qiu, E. F. Simonds, S. C Bendall, K. D. G. Jr, R. V. Bruggner, M. D. Linderman, K. Sachs, G. P. Nolan and S. K. Plevritis, Nat. Biotechnol., 2011, 29, 886–891 CrossRef CAS PubMed.
- S. Van Gassen, B. Callebaut, M. J. Van Helden, B. N. Lambrecht, P. Demeester, T. Dhaene and Y. Saeys, Cytometry, Part A, 2015, 87, 636–645 CrossRef PubMed.
- D. Schapiro, H. W. Jackson, S. Raghuraman, J. R. Fischer and V. R. T. Zanotelli, et al. , Nat. Methods, 2017, 14, 873–876 CrossRef CAS PubMed.
- M. G. Haberl, C. Churas, L. Tindall, D. Boassa and S. Phan, et al. , Nat. Methods, 2018, 15, 677–680 CrossRef CAS PubMed.
- L. Keren, M. Bosse, S. Thompson, T. Risom, K. Vijayaragavan and E. McCaffrey, et al. , Sci. Adv., 2019, 5, 1–16 Search PubMed.
- J. H. Lovett and H. H. Harris, Curr. Opin. Chem. Biol., 2021, 61, 135–142 CrossRef CAS PubMed.
- L. J. Edgar, R. N. Vellanki, A. Halupa, D. Hedley, B. G. Wouters and M. Nitz, Angew. Chem., Int. Ed., 2014, 53, 11473–11477 CrossRef CAS PubMed.
Footnote |
| † This paper is dedicated to Professor Benli Huang on the occasion of his 100th birthday in 2024 to honor his contribution to atomic spectrometry. |
| This journal is © The Royal Society of Chemistry 2024 |