Open Access Article
Open Access ArticleCell morphology-guided de novo hit design by conditioning GANs on phenotypic image features†
Paula A.
Marin Zapata‡
 *a,
Oscar
Méndez-Lucio‡
*a,
Oscar
Méndez-Lucio‡
 bc,
Tuan
Le
bc,
Tuan
Le
 a,
Carsten Jörn
Beese
a,
Carsten Jörn
Beese
 d,
Jörg
Wichard
e,
David
Rouquié
b and
Djork-Arné
Clevert
d,
Jörg
Wichard
e,
David
Rouquié
b and
Djork-Arné
Clevert
 a
a
aBayer AG, Machine Learning Research, Pharmaceuticals, Berlin, Germany. E-mail: paula.marinzapata@bayer.com
bBayer SAS, Early Toxicology, Crop Science, Sophia Antipolis, France
cBloomoom, 13 Avenue Albert Einstein 69100, Villeurbane, France
dLeibniz-Forschungsinstitut für Molekulare Pharmakologie, Berlin, Germany
eBayer AG, Genetic Toxicology, Pharmaceuticals, Berlin, Germany
First published on 25th November 2022
Abstract
Developing novel bioactive molecules is time-consuming, costly and rarely successful. As a mitigation strategy, we utilize, for the first time, cellular morphology to directly guide the de novo design of small molecules. We trained a conditional generative adversarial network on a set of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds using their cell painting morphological profiles as conditioning. Our model was able to learn chemistry-morphology relationships and influence the generated chemical space according to the morphological profile. We provide evidence for the targeted generation of known agonists when conditioning on gene overexpression profiles, even though no information on biological targets was used during training. Based on a target-agnostic readout, our approach facilitates knowledge transfer between biological pathways and can be used to design bioactives for many targets under one unified framework. Prospective application of this proof-of-concept to larger chemical spaces promises great potential for hit generation in drug and phytopharmaceutical discovery and chemical safety.
000 compounds using their cell painting morphological profiles as conditioning. Our model was able to learn chemistry-morphology relationships and influence the generated chemical space according to the morphological profile. We provide evidence for the targeted generation of known agonists when conditioning on gene overexpression profiles, even though no information on biological targets was used during training. Based on a target-agnostic readout, our approach facilitates knowledge transfer between biological pathways and can be used to design bioactives for many targets under one unified framework. Prospective application of this proof-of-concept to larger chemical spaces promises great potential for hit generation in drug and phytopharmaceutical discovery and chemical safety.
Introduction
Recent studies have categorized pharmaceutical research and development (R&D) as a very inefficient process in terms of the high cost and small number of approved molecules.1,2 Given that the cost of bringing a new drug to market is doubling approximately every 9 years, the current R&D model might not be sustainable in a few more decades.1 This concern raises the question of how to improve the current pharmaceutical R&D efficiency. Some authors propose to focus on desired biological responses, such as in systems-based approaches, moving away from the currently popular target-based approach.3–5 The latter is characterized for focusing only on a well-characterized target or mode of action. On the other hand, systems-based drug discovery aims to identify or optimize a molecule with little knowledge of the biological target or mode of action, relying more on phenotypic changes. Recent studies have compared the efficiency of these approaches based on the number of first-in-class drugs approved by the FDA but it is too soon to draw a conclusion due to the long time frame of drug discovery projects.6,7 The same principles apply to the phytopharmaceutical sector where innovative active substances are needed to find new mode of action in target species (plant, insect, fungi) while having an optimized safety profiles for humans and the environment.8Despite the many advantages of the systems-based approach,3 only a few experimental settings have been used in large-scale screens. One of these is transcriptomic analysis where the change in gene expression levels caused by perturbation (either chemical or biological) is used as a readout to select active compounds. This approach has been used in the Connectivity Map project9,10 to connect disease, genes and drugs and has been successfully applied to identify new active molecules,11–13 drug repurposing,14,15 and mode of action identification.16,17 An alternative is to observe the effect of the perturbation at the morphological level rather than at the transcriptomic level. In this regard, the cell painting assay, a new technique based on High-Content Imaging (HCI), has been extremely useful. Cell painting uses six stains to label eight cellular components, thus capturing a good overview of the cellular state.18 In the context of profiling, images are usually processed by computational pipelines to extract feature representations called morphological profiles19–24 (reviewed in ref. 25), which serve as phenotype descriptors in further tasks. This technique has been used for drug repurposing,26 to cluster small molecules by similar phenotypic effect,19 map cellular morphology to gene function,27 predict biochemical assays,28,29 infer modes of action,30 and characterize toxicity31 among other applications (reviewed in ref. 32 and 33).
Interestingly, most cell painting applications in small molecule research have focused on clustering or classification tasks. To our best knowledge, no work has used this information-rich assay to guide de novo molecular design in drug discovery.34,35De novo molecular design aims to propose novel chemical structures with desired profiles, for example, bioactivity on a target protein36 or optimized molecular properties.37,38 A popular method in the field is the use of generative adversarial neural networks (GANs),36 having numerous applications to drug discovery39–43 and materials sciences.44 These techniques promise to play a crucial role in pharmaceutical and phytopharmaceutical research by allowing a cost-effective exploration of the vast drug-like chemical space (estimated to be in the range of 1023 to 1060).45
In our previous work,46 we proposed to guide the de novo generation of active compounds using gene expression signatures. While transcriptional profiling is costly, high content imaging provides a less expensive profiling technology, which is broadly accessible and offers higher throughput. Therefore, in this work, we combine cell painting with generative adversarial networks to design compounds inducing a specific morphological effect. We show that by conditioning a GAN on morphological profiles we can influence the generated chemical space and suggest compounds with high molecular similarity to known agonists without using biological activity annotations in the training set.
Results
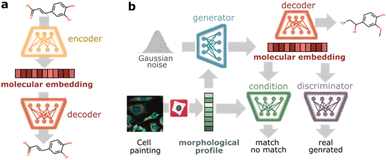
Model architecture
To design phenotype tailored compounds, we propose a modular architecture with two main components: a molecular autoencoder and a conditional Wasserstein generative adversarial network (cGAN)47 (Fig. 1). The goal of the molecular autoencoder is to provide a representation of compounds (molecular embedding) and decode those representations back to compounds. Inspired by recent work encoding molecules into continuous spaces,38,48,49 we trained a variational autoencoder50 to reconstruct discrete representations of compounds and used the bottleneck layer as continuous molecular embedding. As input, we chose SELFIES51 (self-referencing embedded strings) since they offer 100% validity to describe molecular graphs. The autoencoder architecture follows the implementation proposed by Winter et al.48 based on stacked Gated Recurrent Unit (GRU) cells, and it was trained on ChEMBL22 (ref. 52) to cover a large chemical space. The goal of the cGAN is to produce phenotype-tailored compounds using morphological profiles from cell painting images as conditioning input. The model is composed of three neural networks: a generator, a discriminator, and a condition network. The generator takes a random noise vector and a morphological profile to generate a vector in the molecular embedding space targeted to the input profile, the discriminator evaluates if the generated molecular embedding corresponds to a real or generated molecule, and the condition network evaluates whether the molecular embedding matches the morphological profile. The cGAN was trained in an adversarial manner on the compound profiling dataset BBBC036v1,53 which contains 126![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 779 morphological profiles induced by 30
779 morphological profiles induced by 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 616 compounds in U2OS cells (see Methods for more details).
616 compounds in U2OS cells (see Methods for more details).
Generating molecules from compound-induced morphological profiles
First, we examined the overall generative performance of our model. To this end, we generated 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules conditioned on 30
000 molecules conditioned on 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 morphological profiles randomly selected from the training set (i.e., 1 molecule per profile) and computed several statistics on generated compounds (Table 1, column Cpds). Although only ∼50% of the molecules were valid, most of them were unique (99%), displayed low similarity to the training set (90%), and showed relatively high scaffold diversity (83% Murcko scaffold uniqueness vs. 70% in the training set). Furthermore, a Fréchet ChemNet Distance (FCD)54 in the range of 12–15 shows a modest difference between the distribution of generated molecules and the training set. These results indicate that the generative adversarial network was able to generate novel and diverse sets and did not suffer from the mode-collapse problem commonly observed in this type of model.
000 morphological profiles randomly selected from the training set (i.e., 1 molecule per profile) and computed several statistics on generated compounds (Table 1, column Cpds). Although only ∼50% of the molecules were valid, most of them were unique (99%), displayed low similarity to the training set (90%), and showed relatively high scaffold diversity (83% Murcko scaffold uniqueness vs. 70% in the training set). Furthermore, a Fréchet ChemNet Distance (FCD)54 in the range of 12–15 shows a modest difference between the distribution of generated molecules and the training set. These results indicate that the generative adversarial network was able to generate novel and diverse sets and did not suffer from the mode-collapse problem commonly observed in this type of model.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 randomly selected training set profiles per condition group and repetition. Cpds: compounds, DMSO: neutral controls, %: percentage of valid unique molecules satisfying specified constraints. Values report mean ± standard deviation among 3 repetitions, except for Physchem properties and ChEMBL FCD, which report mean and std among all generated molecules. NN: nearest neighbor, scaff: scaffold, dice sim: dice similarity of Morgan fingerprints, CFD: Fréchet ChemNet distance
000 randomly selected training set profiles per condition group and repetition. Cpds: compounds, DMSO: neutral controls, %: percentage of valid unique molecules satisfying specified constraints. Values report mean ± standard deviation among 3 repetitions, except for Physchem properties and ChEMBL FCD, which report mean and std among all generated molecules. NN: nearest neighbor, scaff: scaffold, dice sim: dice similarity of Morgan fingerprints, CFD: Fréchet ChemNet distance
| Property | Details | Generated | Train | |
|---|---|---|---|---|
| Cpds | DMSO | |||
| Validity | % Valid | 49.8 ± 0.4 | 56.6 ± 0.3 | — |
| Uniqueness | % Unique | 99.9 ± 0.1 | 99.7 ± 0.1 | — |
| Novelty | % Dice sim to train NN < 0.5 | 90.8 ± 0.5 | 81.5 ± 0.4 | — |
| Train set FCD | 15.2 ± 0.025 | 12.3 ± 0.028 | ||
| Diversity | % Unique Murcko scaff | 82.8 ± 0.4 | 83.0 ± 0.4 | 69.6 |
| Condition match | % Class prob > 0.75 | 99.3 ± 0.0 | 97.6 ± 0.2 | 73.9 |
| Shuffled samples | 9.4 ± 0.6 | 10.9 ± 0.3 | 13.5 | |
| Shuffled features | 10.3 ± 0.1 | 8.1 ± 0.3 | 5.3 | |
| Synthesizability | % SA score < 4.5 | 81.6 ± 0.6 | 92.3 ± 0.1 | 96.2 |
| % RSA score > 0.5 | 54.0 ± 0.5 | 74.6 ± 0.4 | 76.1 | |
| % (RSA > 0.5) & (SA < 4.5) | 50.9 ± 0.7 | 72.6 ± 0.6 | 75.2 | |
| Drug-likeness | ChEMBL FCD | 8.5 ± 0.07 | 4.9 ± 0.05 | 8.4 |
| % QED > 0.5 | 41.5 ± 1.1 | 63.9 ± 0.2 | 75.8 | |
| Toxicity | % cpds with toxicophores | 24.8 ± 0.2 | 21.9 ± 0.4 | 12.5 |
| Physchem | Molecular weight | 443.6 ± 124.5 | 391.3 ± 101.8 | 381.0 ± 96.4 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
2.5 ± 1.9 | 3.0 ± 1.6 | 3.2 ± 1.3 | |
| HBA + HBD | 9.0 ± 3.2 | 7.0 ± 2.7 | 6.5 ± 2.3 | |
| NRB | 7.6 ± 4.4 | 5.9 ± 3.3 | 4.9 ± 2.4 | |
To provide a comprehensive characterization of generated molecules, Table 1 also lists physicochemical and drug-like properties, although they were not part of our optimization objective. On average, generated molecules present drug-like physicochemical properties, most of them fulfilling the ‘Rule of 5’ proposed by Lipinski,55 which is considered a standard of drug-likeness (molecular weight < 500D, Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 5, H-bond donors < 5, H-bond acceptors < 10, and number of rotatable bonds < 10). Accordingly, more than 40% of the generated molecules have a high Quantitative Estimate of Drug-likeness56 (QED > 0.5), and the similarity between ChEMBL and generated compounds is comparable to that between ChEMBL and the training set in terms of FCD score. In addition, more than 50% of the generated molecules are expected to be synthesizable since they display an acceptable Retrosynthetic Accessibility Score57 (RSA > 0.5) and a low Synthetic Accessibility score58 (SA < 4.5). Finally, only less than 25% of the generated molecules contained one or more toxicophores, i.e., substructures that are highly correlated with adverse effects.59 Together, these findings demonstrate that our model can generate molecules with drug-like characteristics.
P < 5, H-bond donors < 5, H-bond acceptors < 10, and number of rotatable bonds < 10). Accordingly, more than 40% of the generated molecules have a high Quantitative Estimate of Drug-likeness56 (QED > 0.5), and the similarity between ChEMBL and generated compounds is comparable to that between ChEMBL and the training set in terms of FCD score. In addition, more than 50% of the generated molecules are expected to be synthesizable since they display an acceptable Retrosynthetic Accessibility Score57 (RSA > 0.5) and a low Synthetic Accessibility score58 (SA < 4.5). Finally, only less than 25% of the generated molecules contained one or more toxicophores, i.e., substructures that are highly correlated with adverse effects.59 Together, these findings demonstrate that our model can generate molecules with drug-like characteristics.
As a preliminary assessment of the effect of morphological conditioning, we compared conditioning on training set compounds vs. conditioning on Dimethyl Sulfoxide (DMSO) negative controls, finding subtle differences between both groups. DMSO-conditioned molecules (Table 1, column DMSO) display higher validity, synthesizability and slightly closer physicochemical properties to the training set than compound-conditioned molecules (Table 1, column Cpds). However, both groups display similar diversity, uniqueness, and toxicophore content. The high similarity of physicochemical properties between DMSO-conditioned molecules and the training set might be due to the large proportion of compounds without distinguishable morphological signature in the training set. This is evident from the overlap between morphological profiles from compounds and DMSO (ESI Fig. 1†), leading to an increased density of training samples near the DMSO phenotypic neighborhood. Such overlap also advocates for a more fine-grained stratification of morphological profiles to judge the effect of morphological conditioning.
Translating morphological similarity into chemical similarity
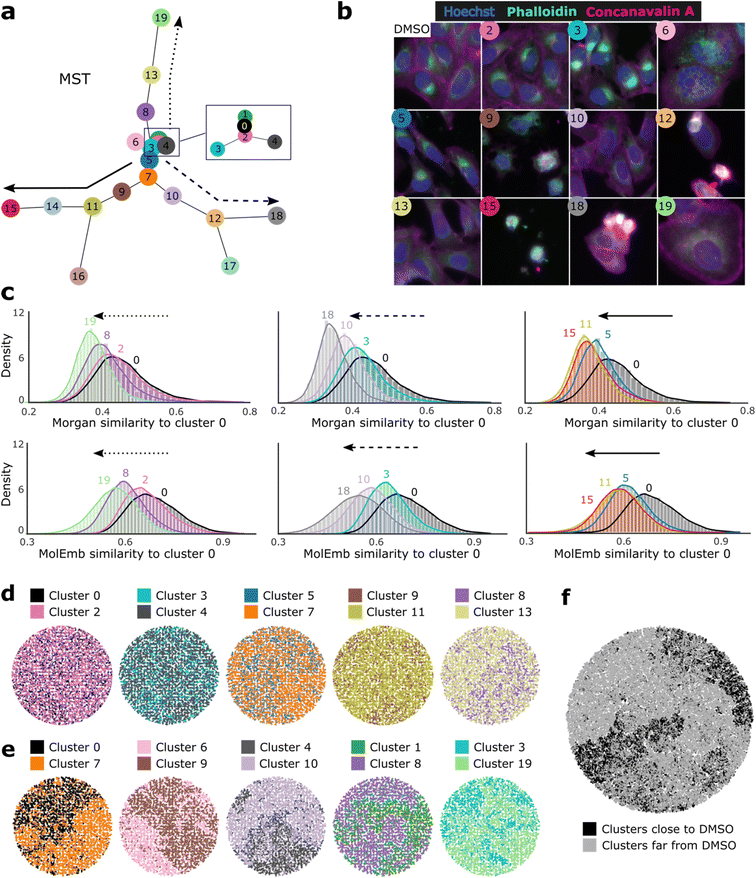
Next, we asked whether morphological conditioning could influence the chemical space of the generated compounds in a sensible manner. To answer this, we clustered morphological features and compared the chemical similarity of the generated molecules conditioned on profiles from different clusters (more details in Methods). To get an impression of the cluster phenotypes and their proximity in the feature space, we created a minimum spanning tree (MST) based on the cluster centroids (Fig. 2a) and displayed representative images of the closest neighbors to selected centroids (Fig. 2b). For easy reference, clusters were enumerated by increasing distance to the cluster with most DMSO control samples (cluster 0).Comparison of the generated chemical space between cluster pairs showed that our model effectively translates morphological similarity into chemical similarity and even preserves distance relationships between both levels. Taking cluster 0 as a reference and following the hierarchical paths proposed by the spanning tree, we observe a reduced chemical similarity to cluster 0 for increasing distances in the morphological space (Fig. 2c). This trend holds for most pairwise comparisons (except clusters 11 and 15) and is observable based on molecular fingerprints, as well as molecular embeddings. The latter indicates that distances in the chemical embedding space learned by the model are in accordance with established metrics of chemical similarity. Additionally, we used t-map60 projections of Morgan fingerprints to visualize differences in the generated chemical space between pairs of nearby (Fig. 2d) and distant (Fig. 2e) clusters judging from the MST. While proximal clusters do not segregate well, pairs of distant clusters display a clear differentiation of their chemical space. Moreover, visualization of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 generated molecules from all clusters depicts a good distinction between the 10 closest and 10 farthest clusters to the DMSO control reference (Fig. 2f). Similar segregation patterns were observed among the training set molecules (ESI Fig. 2a†), although no obvious morphology-chemistry associations could be observed from bulk, un-clustered profiles (ESI Fig. 2b and c†). Together, these results indicate that our model learned morphology-chemistry relationships from the training set and can manipulate the generated chemical space through its morphological conditioning in a specific and logical manner.
000 generated molecules from all clusters depicts a good distinction between the 10 closest and 10 farthest clusters to the DMSO control reference (Fig. 2f). Similar segregation patterns were observed among the training set molecules (ESI Fig. 2a†), although no obvious morphology-chemistry associations could be observed from bulk, un-clustered profiles (ESI Fig. 2b and c†). Together, these results indicate that our model learned morphology-chemistry relationships from the training set and can manipulate the generated chemical space through its morphological conditioning in a specific and logical manner.
Generating molecules from genetic perturbations
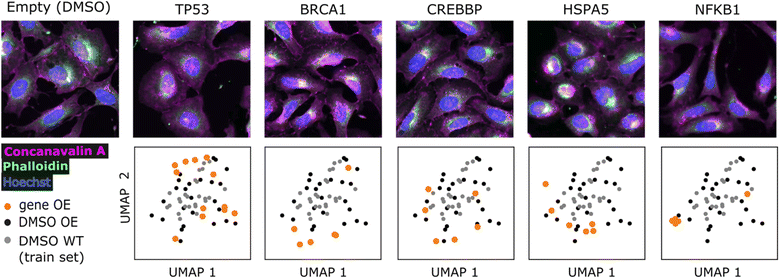
We sought to find evidence for the generalizability of the model and biological relevance of generated molecules by conditioning on profiles outside the training set and comparing them to known bioactive molecules. We used profiles from the gene overexpression profiling dataset BBBC037v1 (ref. 27) and compared generated molecules to known agonists retrieved from the ExCAPE database,61 following the reasoning that overexpression should resemble agonism. We could retrieve known agonists for 9 genes out of 220 overexpressed genes (summarized in ESI Table 1†), making sure to exclude those compounds present in the training set.First, we visualized the overexpression phenotypes to inspect the strength of the morphological signal (Fig. 3 and ESI Fig. 3†). Profile projections indicate that only NFKB1, BRCA1 and HSPA5 strongly to mildly differ from DMSO controls, while other genes show low differentiation (TP53, CREBBP, STAT1, STAT3, HIF1A, NFKBIA). Nevertheless, generated molecules conditioned on most genes showed some differentiation compared to conditioning on DMSO (ESI Fig. 4†). We include all genes in subsequent analyses, later confirming that only differentiable genes lead to conclusive results. Good overlap between DMSO controls from the overexpression (test set) and training set is also evident, corroborating the comparability of both datasets after applying quantile transform (see Methods).
Scaffold enrichment analysis identifies morphology-tailored molecular substructures
Next, we generated molecules conditioned on profiles from overexpressed genes (see methods) and examined if the generated molecules were similar to known agonists. A key question is whether resemblance to a known agonist happens due to the conditioning or by chance, since the larger the generated sample size, the higher the probability to find an active-like molecule. To address this, we first performed an enrichment analysis to select scaffolds that are significantly overrepresented for each gene compared to DMSO, thus ensuring, to a certain extent, the causality of the morphological conditioning (see Methods). Subsequently, we checked whether known agonists contain enriched scaffolds from the corresponding gene. Highlighting the fairness of our scaffold selection process is the fact that it does not guarantee a given number of scaffolds per target (if any).On average, generated molecules conditioned on overexpressed genes displayed 96% and 79% compound and scaffold uniqueness, respectively (ESI Table 1†), closely following the results from the training set. Scaffold enrichment analysis relative to DMSO showed that overrepresented scaffolds were consistent and slightly discriminative. Within a given gene conditioning, we observed groups of scaffolds sharing a good proportion of molecular substructures (ESI Fig. 5†), reassuring that the model learned chemical patterns. Between different genes, we observed some diversity in enriched scaffolds, although there was a frequent pattern of two aromatic rings linked by an aliphatic chain of variable length.
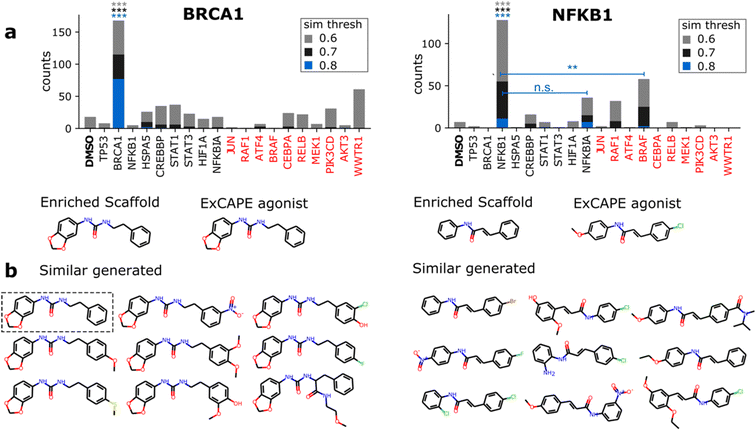
Among the 9 tested genes, we could identify ExCAPE agonists with enriched scaffolds for only two genes (BRCA1 and NFKB1) which interestingly, displayed the strongest phenotypic signal (Fig. 3). For BRAC1, we proposed 8 active scaffolds, among which, 1 was found in a potent BRCA1 agonist (hit rate of 12.5%). For NFKB1, 2 out of 25 proposed scaffolds were found in an agonist (hit rate of 8%). The training set provides a comparison baseline, where 7.6% and 0.23% of 9750 unique scaffolds were found in BRCA1 and NFKB1 agonists, respectively. Thus, we provide a respective increase in hit rate of 1.6 and 34-fold, but most importantly, we dramatically reduce the search possibilities (from 9750 to just 8 or 25). The small number of proposed active scaffolds would make it feasible to prospectively conduct experimental validation. However, since chemical synthesis of de novo molecules is time and resource-consuming, we consider it is out of scope for this proof-of-concept work.
Comparative analyses reassure attribution to the morphological conditioning
Having identified 3 active scaffolds suggested by the model, we further scrutinize the contribution of the morphological conditioning by checking the specificity with which active-like molecules are generated. For a given agonist, we compared the number of similar generated molecules conditioned on the respective gene vs. the remaining 8 overexpressed genes and DMSO. To provide a robust comparison, we also included a set of 10 highly differentiable genes (see Methods) and report counts for 3 Morgan fingerprint similarity thresholds (0.6, 0.7 and 0.8) to designate active-like compounds (Fig. 4). Results are only reported for 2 of the ExCAPE agonists since no similar molecules were found for one of the NFKB1 compounds (ESI Fig. 6†).Significant differences in the frequency of active-like molecules among the 20 tested genes reveal good specificity of the generation process towards the corresponding gene targets (Fig. 4a). Both BRCA1 and NFKB1 agonists displayed significantly increased active-like counts when conditioning on their respective gene compared to any other gene, with only NFKB1 lacking significance vs. NFKBIA for the lowest similarity threshold (0.6). Besides this, active-like generated compounds showed different substituents and side chains at several locations, suggesting that the model can mimic diverse medicinal chemistry decorations (Fig. 4b). Interestingly, one of the generated compounds for BRCA1 was a perfect match to the known agonist (highlighted in Fig. 4b).
As a second check, we used the condition network to quantify changes in the matching probability towards both ExCAPE agonists as a function of the phenotypic distance to their respective gene overexpression profile (ESI Fig. 7†). Following our expectations, the classification probability towards the NFKB1 agonist strongly decreased for training set and overexpression profiles with increasing cosine distance to the NFKB1 overexpression profile. A similar but milder trend was observed for BRCA1, which as formerly pointed out, has a less distinctive morphological signature. Overall, these results encourage the idea that the generation of active-like molecules was attributed to the morphological conditioning.
Interpolation of chemical and morphological spaces
A benefit of using continuous chemical descriptors as well as continuous biological readouts is that it allows us to smoothly navigate both spaces. We leverage this to further assess the behavior of our model with respect to the two selected ExCAPE agonists, performing linear interpolation and monitoring the predictions of the condition network. Concretely, we study how the matching probability towards a given gene changes as we navigate the chemical space (Fig. 5a), and conversely, how the matching probability towards a given compound changes as we navigate the phenotypic space (Fig. 5b).For interpolation of the chemical space, we pick the NFKB1 and BRCA1 ExCAPE agonists with enriched scaffolds as starting points and evaluate the matching probability against NFKB1 and BRCA1 overexpression profiles, respectively (see Methods). To cover increased and decreased activity directions, we choose interpolation endpoints from other ExCAPE agonists outside the training set with higher matching probability towards the corresponding gene (Fig. 5c) as well as random generated molecules with lower matching probability (Fig. 5d). Further examples are provided in Fig. 8.† Chemical interpolation experiments show that incremental shifts in the molecular embedding correspond to smooth transitions in the discrete chemical space and induce sensible changes in the output from the condition network. The matching probability towards NFKB1 and BRCA1 overexpression profiles increased progressively along interpolation trajectories towards increased activity directions, reinforcing the belief that our model learned meaningful trends instead of isolated phenotype-compound matching occurrences. Interpolation towards decreased activity directions showed more irregular responses, possibly due to the randomness of the selected direction and increased distance to the start molecule.
For interpolation of the morphological space, we follow the matching probability against the compounds used in Fig. 5c and d and pick the DMSO profile from the overexpression dataset as starting point (Fig. 5e) and NFKB1 and BRCA1 profiles as endpoints. As comparison baselines, we also include trajectories towards random directions or along the axes of highest variation in the training set (see Methods). Phenotypic interpolation revealed a dose–response towards the distance to DMSO and confirmed boosted matching in the direction of overexpression profiles. The matching probability against the 3 NFKB1 agonists (start, end1, end2 in Fig. 5e) increased from DMSO towards the NFKB1 phenotype, while decreasing for the NFKB1 inactive molecule (end3). Interestingly, the DMSO probability was zero for the 3 agonists, indicating that they were recognized as bioactives even though neither bioactivity labels nor DMSO profiles were used during training. Notably, there was a general trend towards higher matching probabilities for larger distances to DMSO (random curve). However, the trend was more pronounced along axes of highest variation (top 10 PCA) and even more towards the NFKB1 phenotype, with similar results for BRCA1 (Fig. 9†). This suggests that the model does not simply rely on overall phenotypic strength but displays some specificity towards different regions in the morphological space.
Together, interpolation experiments highlight the quality of our molecular descriptors and the soundness of the morphology-chemistry associations learned by the model. Moreover, they demonstrate the potential of our framework for molecular optimization.
Discussion
We propose an approach to generate phenotype-customized molecules by conditioning a generative adversarial network on cell painting morphological profiles and demonstrate how our model can influence the chemical space and propose tailored bioactives without prior information of biological targets. By using a target-agnostic readout, we learn general relationships between cell morphology and molecular structure and intrinsically provide a framework for knowledge transfer between diverse biological pathways. Our work provides a first step towards the systematic use of high content imaging for molecular design.To our knowledge, we are the first to use cell painting data to guide the de novo design of small molecules. A recent work tackled the inverse problem i.e., synthesizing cell painting images conditioned on molecular structures.62 We believe that our approach is more applicable to small molecule discovery since it directly proposes compounds given a desired biological response and allows straightforward integration of genetic perturbations. Other related works have used gene expression profiles to guide de novo molecular design.46,63 However, given the low cost of high content imaging, the large-scale data generation needed to bring these methods to their full potential will favor the morphological over the transcriptomic domain. For instance, joint initiatives, such as the JUMP-CP consortium, promise to release morphological profiling data for over 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 chemical and genetic perturbations.64
000 chemical and genetic perturbations.64
Evaluating the significance of model outputs in generative chemistry is not straightforward.65 In this proof-of-concept, we focused on demonstrating a sensible influence of morphological conditioning on the generated chemical space and evaluating the relative merit of the conditioning. Using gene overexpression profiles, we provide evidence of the targeted generation of agonist compounds, where we could attribute the overrepresentation of active-like substructures to morphological conditioning. However, assessment of the potentiality of our approach to generate truly novel compounds inducing a desired phenotype will require larger datasets or experimental testing. Our evaluation strategy relied on public datasets and was based on the premise that gene overexpression should resemble agonism. We were therefore limited by the low number of agonists we could retrieve from public resources, which reduced our evaluation set to only 9 genes, many of them with rather weak phenotypes. Although gene knock-down perturbations would have certainly provided more reference compounds (in this case antagonists), we avoided using public cell painting RNAi data due to the reported seeding effects reflected in poor specificity.66
Agreeing with our expectations, we only found enriched scaffolds present in ExCAPE agonists for a subset of two genes displaying good morphological differentiation from DMSO controls (NFKB1 and BRCA1). A deeper look into the ExCAPE compounds shows that both agonists have strong activity towards their target. The NFKB1 agonist was identified as a potent activator of the NFKB1 pathway in NFkB-bla cells (PubChem AID: 928, PubChem CID: 1628214), with half-maximal efficacy (EC50) of 1.4 μM. The BRCA1 agonist showed high efficacy on a BRCA1 expression activation assay in MCF7 cells (EC50 = 3.4 μM), although the activity value was calculated from only a partial Hill curve reaching ∼80% efficacy at the highest concentration (PubChem AID: 624![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 202, PubChem CID: 6465485). Interestingly, this assay directly measured BRCA1 expression levels, matching the overexpression perturbation on which we based our analysis. The fact that both agonists showed micromolar potencies in cell-based assays reassures their functionality activating their respective target in a relevant biological context.
202, PubChem CID: 6465485). Interestingly, this assay directly measured BRCA1 expression levels, matching the overexpression perturbation on which we based our analysis. The fact that both agonists showed micromolar potencies in cell-based assays reassures their functionality activating their respective target in a relevant biological context.
A notable aspect of our approach is the continuous nature of our chemical and biological descriptors, which enables efficient exploration and interpolation of both spaces. We exploited this to confirm the consistency of our model predictions regarding the two selected ExCAPE agonists, revealing logical dose-like effects and smooth, meaningful changes in chemical structure. These observations reinforce the notion that the observed phenotype-chemistry matches resulted from relevant associations learned by the model. Furthermore, they underline the quality of our molecular representations and provide useful evidence about the remarkable performance of SELFIES, as it was recently reported in neural translation tasks for molecular representation.67
Conditioning on morphological profiles opens many possibilities inherent to profiling itself. The multiparametric, target-agnostic nature of the readout allows for capturing complex biological responses, being particularly helpful when experimental endpoints are not easily defined. Moreover, morphological profiles can implicitly capture toxicity or off-target effects, thereby providing an alternative to multi-objective optimization for de novo molecular design and chemical de-risking. Finally, having a standardized cell painting protocol will potentially ameliorate the lack of comparability between experimental set-ups and dependence of labels on external context, two common difficulties in modeling biological systems.68 It is however worth noting that we observed differences between the distributions of morphological profiles from chemical and genetic perturbations, even though both datasets were produced in the same research group. Although distributions became more comparable after applying a quantile transformation, this emphasizes the need for profile harmonization methods to enable the collaborative assembly of large morphological datasets.
Conclusions
We were able to combine generative models with cellular morphology information to design compounds that potentially induce a desired biological response. Our work bridges the fields of de novo molecular design and morphological profiling, taking the first steps into an exciting wave of system-based approaches which promise to overcome reductionist, single-target views of biological systems. By using larger datasets, we expect that we will be able to push our approach beyond proof-of-concept and ultimately, accelerate small molecule discovery by providing fast access to innovative chemistry influencing diverse biological pathways.Methods
Model architecture and training
where x and c are a molecule representation and a morphology profile, respectively, sampled from the real data distribution preal, z is a vector with random noise sampled from a Gaussian distribution (pz) and f0 is a function (in this case, a neural network) that measures the probability of a morphology profile to correspond to a molecular representation. The λ and α terms are regularization parameters, both empirically set to 10. λ weights the influence of the gradient penalty in the discriminator loss. α term weights the influence of f0 in the generator loss.
The model was trained for 500 epochs with a batch size of 256 (496 steps per epoch). The discriminator was updated after each step, whereas the generator every 10 steps. The network was trained using the RMSprop optimizer with a learning rate of 1 × 10−4 for both the generator and discriminator. During training, we monitored the similarity between real and generated molecular representations using Fréchet distance49 (ESI Fig. 11†). The weights of the condition network were pre-trained on a binary cross-entropy loss and frozen during the GAN training process. All neural networks were built and trained using Tensorflow 1.14.72
Morphological profiles
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 616 compounds at 10 μM, 24 h and 4 replicates, and imaged with the cell painting protocol. Per-well profiles were downloaded from Bray et al.53 and normalized relative to negative controls per plate using robust z-scoring. i.e., subtraction of DMSO median and division by DMSO median absolute deviation (mad). 334 Features with zero mad were removed, reducing the number of features to 1449. Profiles were preprocessed with the scikit-learn73 implementation of a quantile transformer fitted on the training set to improve the comparability between training and overexpression datasets. In total, the training set consisted of 126
616 compounds at 10 μM, 24 h and 4 replicates, and imaged with the cell painting protocol. Per-well profiles were downloaded from Bray et al.53 and normalized relative to negative controls per plate using robust z-scoring. i.e., subtraction of DMSO median and division by DMSO median absolute deviation (mad). 334 Features with zero mad were removed, reducing the number of features to 1449. Profiles were preprocessed with the scikit-learn73 implementation of a quantile transformer fitted on the training set to improve the comparability between training and overexpression datasets. In total, the training set consisted of 126![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 779 morphological profiles which were treated independently.
779 morphological profiles which were treated independently.
Molecule generation and post inference analyses
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 valid molecules were generated based on a random sample of 500 profiles per cluster (or less for clusters with less than 500 samples).
000 valid molecules were generated based on a random sample of 500 profiles per cluster (or less for clusters with less than 500 samples).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, valid molecules passing custom physicochemical filters were generated per gene using per-well profiles. Physicochemical filters were applied to enhance similarity to drug-like molecules and included: −2 < log
000, valid molecules passing custom physicochemical filters were generated per gene using per-well profiles. Physicochemical filters were applied to enhance similarity to drug-like molecules and included: −2 < log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 7, −2 < Mol weight < 7, H acceptors + H donors < 10, TPSA < 150, rotatable bonds < 150, and Sure ChEMBL alerts = 0. The generation experiment was repeated 3 times using different random seeds.
P < 7, −2 < Mol weight < 7, H acceptors + H donors < 10, TPSA < 150, rotatable bonds < 150, and Sure ChEMBL alerts = 0. The generation experiment was repeated 3 times using different random seeds.
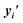 ) and reran the embedding to SELFIES decoding step. This was repeated until a valid molecule was found or up to maximum of 20 iterations.
) and reran the embedding to SELFIES decoding step. This was repeated until a valid molecule was found or up to maximum of 20 iterations. 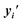 was sampled from a uniform distribution between 0, and 2% of the per-dimension values. Morphological interpolation was performed on quantile transformed profiles. To increase comparability between all trajectories starting from DMSO towards different directions, we subtract the median DMSO profile from the overexpression dataset, normalize the end profiles to their unit vector and scale this vector with the magnitude of the overexpression profile (BRCA1 or NFKB1) being compared against. Thus, morphological endpoints point to different directions but preserve the same distance to DMSO as the compared overexpression profile. In this setting, all trajectories have a constant cosine distance to DMSO, and therefore, we report the Euclidean distance. The random baseline directions were chosen from a uniform distribution. The highest variation directions were obtained from the coefficients of a PCA model with 10 components fitted on the per-compound median profiles of the training set (after subtracting the DMSO profile).
was sampled from a uniform distribution between 0, and 2% of the per-dimension values. Morphological interpolation was performed on quantile transformed profiles. To increase comparability between all trajectories starting from DMSO towards different directions, we subtract the median DMSO profile from the overexpression dataset, normalize the end profiles to their unit vector and scale this vector with the magnitude of the overexpression profile (BRCA1 or NFKB1) being compared against. Thus, morphological endpoints point to different directions but preserve the same distance to DMSO as the compared overexpression profile. In this setting, all trajectories have a constant cosine distance to DMSO, and therefore, we report the Euclidean distance. The random baseline directions were chosen from a uniform distribution. The highest variation directions were obtained from the coefficients of a PCA model with 10 components fitted on the per-compound median profiles of the training set (after subtracting the DMSO profile).
Data availability
The data that support the findings of this study are available on https://github.com/Bayer-Group/CPMolGAN.Code availability
The code used to generate results shown in this study will be available from the corresponding author upon request after the final publication of this manuscript.Author contributions
P. A. M. Z. processed images and profiles, ran inference experiments, prepared figures and wrote the manuscript. O. M. L. trained and designed the deep learning architecture and helped writing the manuscript. T. L. helped with inference experiments and manuscript preparation. C. J. B. made image compositions and helped with manuscript and figures preparation. D. R. and J. W. provided guidance and helped with the manuscript preparation. D. A. C. conceived the study and supervised the work. O. M. L., P. A. M. Z., T. L., C. J. B., J. W., D. R. and D. A. C. read and approved the manuscript.Conflicts of interest
During the development of this work, D. A. C., P. A. M. Z. and J. W. were employees of Bayer AG, D. R. and O. M. L. were employees of Bayer SAS, and C. B. was an employee of the Leibniz-Forschungsinstitut für Molekulare Pharmakologie.Acknowledgements
The authors thank Joerg Tiebes for providing chemistry expert feedback and for his useful comments. We are also grateful to Arwa Al-Dilaimi and Angela Becker for supporting the project and for insightful discussions.References
- W. J. Scannell, A. Blanckley, H. Boldon and B. Warrington, Nat. Rev. Drug Discovery, 2012, 11, 191–200 CrossRef PubMed.
- F. Pammolli, L. Magazzini and M. Riccaboni, Nat. Rev. Drug Discovery, 2011, 10, 428–438 CrossRef CAS PubMed.
- E. C. Butcher, Nat. Rev. Drug Discovery, 2005, 5, 7 CrossRef.
- W. Zheng, N. Thorne and J. C McKew, Drug Discovery Today, 2013, 18, 1067–1073 CrossRef CAS PubMed.
- J. G. Moffat, F. Vincent, J. A. Lee, J. Eder and M. Prunotto, Nat. Rev. Drug Discovery, 2017, 16, 531–543 CrossRef CAS PubMed.
- D. C. Swinney and J. Anthony, Nat. Rev. Drug Discovery, 2011, 10, 507–519 CrossRef CAS PubMed.
- J. Eder, R. Sedrani and C. Wiesmann, Nat. Rev. Drug Discovery, 2014, 13, 577–587 CrossRef CAS PubMed.
- C. Lamberth, S. Jeanmart, T. Luksch and A. Plant, Science, 2013, 341, 742–746 CrossRef PubMed.
- J. Lamb, E. D. Crawford and D. Peck, et al. , Science, 2006, 313, 1929–1935 CrossRef CAS PubMed.
- A. Subramanian, R. Narayan and S. M. Corsello, et al. , Cell, 2017, 171, 1437–1452 CrossRef CAS PubMed.
- H. Hieronymus, J. Lamb and K. N. Ross, et al. , Cancer Cell, 2006, 10, 321–330 CrossRef CAS PubMed.
- G. Wei, D. Twomey and J. Lamb, et al. , Cancer Cell, 2006, 10, 331–342 CrossRef CAS PubMed.
- H. De Wolf, L. Cougnaud and K. Van Hoorde, et al. , Assay Drug Dev. Technol., 2018, 16, 162–176 CrossRef CAS PubMed.
- A. Aliper, S. Plis, A. Artemov, A. Ulloa, P. Mamoshina and A. Zhavoronkov, Mol. Pharm., 2016, 13, 2524–2530 CrossRef CAS PubMed.
- F. Iorio, T. Rittman, H. Ge, M. Menden and J. Saez-Rodriguez, Drug Discovery Today, 2013, 18, 350–357 CrossRef CAS PubMed.
- M. Iwata, R. Sawada, H. Iwata, M. Kotera and Y. Yamanishi, Sci. Rep., 2017, 7, 40164 CrossRef CAS PubMed.
- S. A. Wacker, B. R. Houghtaling, O. Elemento and T. M. Kapoor, Nat. Chem. Biol., 2012, 8, 235–237 CrossRef PubMed.
- M. A. Bray, S. Singh and H. Han, et al. , Nat. Protoc., 2016, 11, 1757–1774 CrossRef CAS PubMed.
- S. M. Gustafsdottir, V. Ljosa and K. L. Sokolnicki, et al. , PLoS One, 2013, 8, e80999 CrossRef PubMed.
- C. Scheeder, F. Heigwer and M. Boutros, Curr. Opin. Syst. Biol., 2018, 10, 43–52 CrossRef.
- E. Moen, D. Bannon, T. Kudo, W. Graf, M. Covert and D. Van Valen, Nat. Methods, 2019, 16, 1233–1246 CrossRef CAS.
- P. T. Jackson, Y. Wang, S. Knight, et al., 16th International Conference on Machine Vision Applications (MVA) 1–4, IEEE, 2019, DOI:10.23919/MVA.2019.8757871.
- A. X. Lu, O. Z. Kraus, S. Cooper and A. M. Moses, PLoS Comput. Biol., 2019, 15, e1007348 CrossRef CAS PubMed.
- A. E. Carpenter, T. R. Jones and M. R. Lamprecht, et al. , Genome Biol., 2006, 7, R100 CrossRef PubMed.
- J. C. Caicedo, S. Cooper and F. Heigwer, et al. , Nat. Methods, 2017, 14, 849–863 CrossRef CAS.
- C. C. Gibso, W. Zhu and C. T. Davis, et al. , Circulation, 2015, 131, 289–299 CrossRef PubMed.
- M. H. Rohban, S. Singh and X. Wu, et al. , eLife, 2017, 6, e24060 CrossRef PubMed.
- J. Simm, G. Klambauer and A. Arany, et al. , Cell Chem. Biol., 2018, 25, 611–618 CrossRef CAS PubMed .e3..
- M. Hofmarcher, E. Rumetshofer, D.-A. Clevert, S. Hochreiter and G. Klambauer, J. Chem. Inf. Model., 2019, 59, 1163–1171 CrossRef CAS PubMed.
- M. J. Cox, S. Jaensch and J. Van de Waeter, et al. , Sci. Rep., 2020, 10, 13262 CrossRef CAS PubMed.
- J. Nyffeler, C. Willis, R. Lougee, A. Richard, K. Paul-Friedman and J. A. Harrill, Toxicol. Appl. Pharmacol., 2020, 389, 114876 CrossRef CAS PubMed.
- J. C. Caicedo, S. Singh and A. E. Carpenter, Curr. Opin. Biotechnol., 2016, 39, 134–142 CrossRef CAS PubMed.
- S. N. Chandrasekaran, H. Ceulemans, J. D. Boyd and A. E. Carpenter, Nat. Rev. Drug Discovery, 2021, 20, 145–159 CrossRef CAS PubMed.
- J. Meyers, B. Fabian and N. Brown, Drug Discovery Today, 2021, 26, 2707–2715 CrossRef CAS PubMed.
- D. C. Elton, Z. Boukouvalas, M. D. Fuge and P. W. Chung, Mol. Syst. Des. Eng., 2019, 4, 828–849 RSC.
- M. H. S. Segler, T. Kogej, C. Tyrchan and M. P. Waller, ACS Cent. Sci., 2018, 4, 120–131 CrossRef CAS PubMed.
- P.-C. Kotsias, J. Arús-Pous, H. Chen, O. Engkvist, C. Tyrchan and E. J. Bjerrum, Nat. Mach. Intell., 2020, 2, 254–265 CrossRef.
- R. Gómez-Bombarelli, J. N. Wei and D. Duvenaud, et al. , ACS Cent. Sci., 2018, 4, 268–276 CrossRef PubMed.
- G. L. Guimaraes, B. Sanchez-Lengeling, C. Outeiral, P. L. C. Farias and A. Aspuru-Guzik, arXiv, 2018, preprint, arXiv:1705.10843, DOI:10.48550/arXiv.1705.10843.
- A. Kadurin, S. Nikolenko, K. Khrabrov, A. Aliper and A. Zhavoronkov, Mol. Pharm., 2017, 14, 3098–3104 CrossRef CAS PubMed.
- B. Sanchez-Lengeling, C. Outeiral, G. L. Guimaraes, ChemRxiv Camb. Camb. Open Engage, 2017, preprint, DOI:10.26434/chemrxiv.5309668.v3.
- N. De Cao and T. Kipf, arXiv, 2018, preprint, arXiv:1805.11973, DOI:10.48550/arXiv.1805.11973.
- E. Putin, A. Asadulaev, Y. Ivanenkov and V. Aladinskiy, et al. , J. Chem. Inf. Model., 2018, 58, 1194–1204 CrossRef CAS PubMed.
- B. Sanchez-Lengeling and A. Aspuru-Guzik, Science, 2018, 361, 360–365 CrossRef CAS PubMed.
- W. P. Walters, J. Med. Chem., 2019, 62, 1116–1124 CrossRef CAS PubMed.
- O. Méndez-Lucio, B. Baillif, D.-A. Clevert, D. Rouquié and J. Wichard, Nat. Commun., 2020, 11, 10 CrossRef PubMed.
- M. Mirza and S. Osindero, arXiv, 2014, preprint, arXiv1411.1784v1, DOI:10.48550/arXiv.1411.1784.
- R. Winter, F. Montanari, F. Noé and D.-A. Clevert, Chem. Sci., 2019, 10, 1692–1701 RSC.
- R. Winter, F. Montanari, A. Steffen, H. Briem, F. Noé and D.-J. Clevert, Chem. Sci., 2019, 10, 8016–8024 RSC.
- D. P. Kingma and M. Welling, arXiv, 2014, preprint, arXiv:1312.6114v10, DOI:10.48550/arXiv.1312.6114.
- M. Krenn, F. Häse, A. Nigam, P. Friederichand and A. Aspuru-Guzik, 33rd Conference on Neural Information Processing Systems, NeurIPS 2019, 2019 Search PubMed.
- A. Gaulton, A. Hersey and M. Nowotka, et al. , Nucleic Acids Res., 2017, 45, D945–D954 CrossRef CAS PubMed.
- M.-A. Bray 1, S. M. Gustafsdottir and M. H. Rohban, et al. , GigaScience, 2017, 6, 1–5 CrossRef PubMed.
- K. Preuer, P. Renz, T. Unterthiner, S. Hochreiter and G. Klambauer, J. Chem. Inf. Model., 2018, 58, 1736–1741 CrossRef CAS PubMed.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 1997, 23, 3–25 CrossRef CAS.
- G. R. Bickerton, G. V. Paolini, J. Besnard, S. Muresan and A. L. Hopkins, Nat. Chem., 2012, 4, 90–98 CrossRef CAS PubMed.
- A. Thakkar, V. Chadimova, E. J. Bjerrum, O. Engkvist and J.-L. Reymond, Chem. Sci., 2021, 12, 3339–3349 RSC.
- P. Ertl and A. Schuffenhauer, J. Cheminf., 2009, 1, 8 Search PubMed.
- I. Sushko, E. Salmina, V. A. Potemkin, G. Poda and I. V. Tetko, J. Chem. Inf. Model., 2012, 52, 2310–2316 CrossRef CAS PubMed.
- D. Probs and J. L. Reymond, J. Cheminf., 2020, 12, 12 Search PubMed.
- J. Sun, N. Jeliazkova and V. Chupakhin, et al. , J. Cheminf., 2017, 9, 17 Search PubMed.
- K. Yang, S. Goldman, W. Jin, et al., arXiv, 2020, preprint, arXiv:200608532, DOI:10.48550/arXiv.2006.08532.
- J. Born, M. Manica, A. Oskooei, J. Cadow, G. Markert and M. Rodríguez Martínez, iScience, 2021, 24, 102269 CrossRef CAS PubMed.
- JUMP-Cell Painting Consortium. https://jump-cellpainting.broadinstitute.org/.
- W. P. Walters and M. Murcko, Nat. Biotechnol., 2020, 38, 143–145 CrossRef CAS PubMed.
- S. Singh, X. Wu and V. Ljosa, et al. , PLoS One, 2015, 10, e0131370 CrossRef PubMed.
- K. Rajan, A. Zielesny and C. Steinbeck, J. Cheminf., 2021, 13, 34 CAS.
- A. Bender and I. Cortes-Ciriano, Drug Discovery Today, 2021, 26, 1040–1052 CrossRef CAS PubMed.
- G. A. Landrum, RDKit-Open-source cheminformatics, https://www.rdkit.org Search PubMed.
- R. J. Williams and D. Zipser, Neural Comput., 1989, 1, 270–280 CrossRef.
- I. Gulrajani, F. Ahmed, M. Arjovsky, V. Dumoulin and A. Courville, arXiv, 2017, preprint, arXiv:1704.00028, DOI:10.48550/arXiv.1704.00028.
- M. Abadi, A. Agarwal, P. Barham, et al., arXiv, 2016, preprint, arXiv:160304467, DOI:10.48550/arXiv.1603.04467.
- F. Pedregosa, G. Varoquaux and A. Gramfort, et al. , J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- N. Brown, M. Fiscato, M. H. S. Segler and A. C. Vaucher, J. Chem. Inf. Model., 2019, 59, 1096–1108 CrossRef CAS PubMed.
- S. Genheden, A. Thakkar and V. Chadimová, et al. , J. Cheminf., 2020, 12, 70 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2dd00081d |
| ‡ Equal contribution: these authors contributed equally and the order was decided alphabetically. |
| This journal is © The Royal Society of Chemistry 2023 |