Open Access Article
Open Access ArticleTuning the immune system by nanoparticle–biomolecular corona
Valentina
Palmieri
 *a and
Giulio
Caracciolo
*a and
Giulio
Caracciolo
 *b
*b
aInstitute for Complex Systems, National Research Council of Italy, Via dei Taurini 19, 00185, Rome, Italy. E-mail: valentina.palmieri@cnr.it
bNanoDelivery Lab, Department of Molecular Medicine, Sapienza University of Rome, Viale Regina Elena 291, 00161, Rome, Italy. E-mail: giulio.caracciolo@uniroma1.it
First published on 18th July 2022
Abstract
Nanotechnology has a great potential to revolutionize the landscape of medicine, but an inadequate understanding of the nanomaterial-biological (nano–bio) interface hampers its ultimate clinical translation. Surface attachment of biomolecules provides a new biological identity of nanoparticles that plays a crucial role in vivo as it can activate the immune system triggering inflammatory responses, clearance from the body, and cellular toxicity. In this review, we summarize and critically analyze progress in understanding the relationship between the biological identity of nanoparticles and immune system activation. Accordingly, we discuss the implications of biomolecular corona on nanotoxicity, immune safety, and biocompatibility. We also highlight a perspective on engineering the biological identity of nanoparticles for modulating immunological responses.
The role of the immune system in the safety and efficacy of nanomedicines
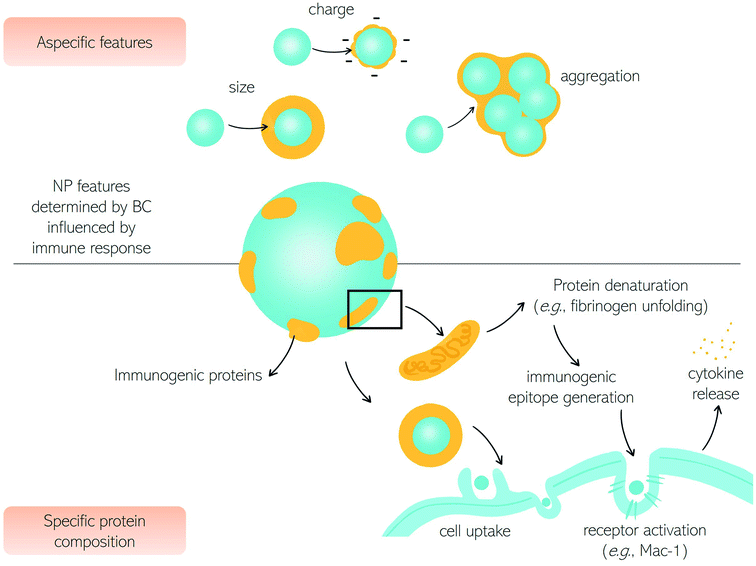
Among various types of interactions of nanomaterials within the human body, contact with immune cells is of remarkable interest due to the significant role of the immune system in the clearance of foreign substances from blood circulation.1 Unintended detection of nanoparticles (NPs) as harmful substances by the immune cells may cause an immune response which in turn may cause toxic effects in the patient and/or impede NPs' therapeutic efficacy.2,3 There are however therapies, like cancer immunotherapy or vaccination (e.g., the recent COVID-19 vaccination), in which there is an intended NP-mediated activation of immune response.4,5 Conversely, NPs-mediated suppression of immune reactions is currently studied for anti-rejection therapies after transplants and autoimmune disorders.6 Therefore, for safe nanomedicine translation, the interaction with the immune system needs to be fully understood.7–10Here we discuss the exchanges between NPs and the immune system in terms of fundamental questions about the correct targeting in vivo, i.e., biodistribution and biosafety for clinical applications.11,12 The first interaction occurs with the innate immune system immediately when NPs enter the human body, depending on their physicochemical characteristics and administration route.6,13,14 In Fig. 1, we summarize the principal cells involved in the immune response. The first mechanism of defense (i.e., the innate immune system) consists of a series of biological responses to remove foreign nanomaterials from blood circulation. Innate immune system cellular components interact with NPs, which have dimensions comparable to viruses, and therefore are competently recognized by immune cells. Recognition of NPs may lead to opsonization by proteins of the complement system that can trigger inflammation and engulfment by the mononuclear phagocyte system (MPS). The complement system consists of a network of proteins activated by proteolytic cascades that start with the identification of a foreign substance. The ways to activate the complement system are threefold, the most common pathway activated by NPs is the so-called “alternative pathway”, started by a variety of surfaces.6,15 Cells that form the MPS are monocytes of the blood, splenic and liver macrophages, and dendritic cells (DCs), all capable of engulfing pathogens or more in general foreign material circulating in the bloodstream.16 If phagocytosis is evaded, NPs half-life is prolonged with higher chances of reaching the target. In general, the size, surface charge, hydrophilicity, and chemical composition of NPs are critical factors for MPS recognition. Consequently, the interaction with the innate immune system has been explored as a function of these particle features.
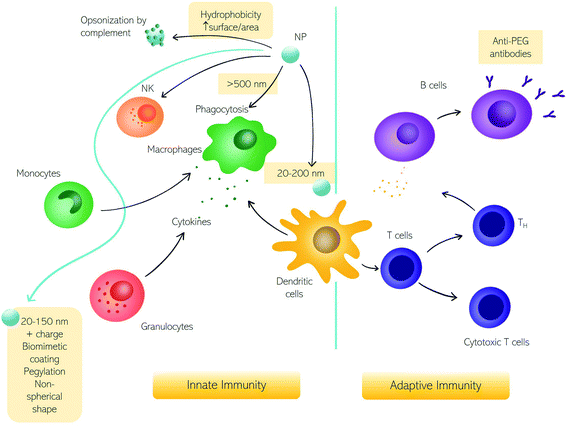 | ||
| Fig. 1 The impact of nanoparticles (NPs) on the immune system. Schematic representation of NPs interacting with the immune system and the corresponding innate and adaptive immune responses. | ||
The size of NPs is usually easily modulated to influence targeting, cellular uptake in vivo, and intracellular distribution as well as permeation of biological barriers. NPs that easily cross biological barriers have a size around 10–500 nm, while the exploitation of the enhanced permeation and retention (EPR) effect,17i.e., the enhanced penetrability of defenestrated tumor vessels, occurs with NPs having a size between 10 and 200 nm.6 The effect of size on the clearance rates of NPs has been known for decades,18 with 5–10 nm NPs eliminated by renal clearance and particles with a size around 200 nm or larger eliminated by the MPS systems. Immune MPS cells recognize different sizes with DCs able to phagocytize particles with a size between 20 and 200 nm and macrophages specialized for larger particles (>500 nm).19,20 It seems that larger particles induce a humoral adaptative response compared to smaller ones.5,21,22 An NPs size between 20 and 150 nm can be suitable to reduce elimination by the liver or kidneys, to prolong the in vivo circulation time.23
Besides size, a high surface area to volume ratio increases the chances of particle toxicity due to an increased probability of opsonization, therefore smaller NPs suffer this problem and can trigger adverse reactions.24 Several materials and surface compositions are available for modern nanotechnology systems production. Hydrophobic materials promote opsonization, act as a danger signal, and induce inflammation.25,26 A positive low absolute charge of the surface, in turn, reduces opsonization and phagocytosis.23 It is known that charged cationic NPs are better phagocytized due to the nonspecific electric attraction with macrophages' negative surface charge.25,27 Therefore, using negatively charged NPs reduces interaction with macrophages due to repulsion. However, these repulsive interactions with cell membranes also reduce the chances of uptake by target cells. Interestingly a class of “charge reversing materials” is currently studied to avoid this problem: the charge reversal NPs have a negative charge and are not phagocytized by the MPS at physiological pH. When the NPs are exposed to the acidic environment of the tumor, their charge becomes positive, thus promoting charge-mediated cellular internalization.23 A relevant example is provided by ionizable cationic lipids that are the main component of marketed mRNA lipid NPs (LNPs) (i.e., BNT162b2/Comirnaty by BioNTech/Pfizer and mRNA-1273/SpikeVax by Moderna). Ionizable cationic lipids possess a cationic charge that consents to high loading efficiencies of mRNA during the synthesis step (pH < 6) while keeping a neutral charge at physiological pH. This allows for reducing toxicity associated with the cationic charge and allows a prolonged circulation time of the LNPs in the bloodstream. Acidification occurring in the endosome's environment promotes interaction and fusion with anionic cellular lipids with mRNA release.
To impede the recognition by the immune system, another strategy consists of surface coating with natural membranes such as membranes of extracellular vesicles (e.g., exosomes), or red and white blood cells.28,29 Alternatively, the most common coating strategy envisages the use of polyethylene glycol (PEG) that neutralizes surface charge and increases the hydrophilicity to prolong NP circulation.30,31 However, some studies evidenced the activation of adaptative immune response with the production of anti-PEG antibodies31,32 leading to accelerated blood clearance of NPs referred to as the “ABC phenomenon”.33
Besides the size, the shape of NPs also has a crucial effect on the response of the immune system.34 Some classes of microorganisms are initially recognized by their shape, though non-spherical bacteria or viruses evade the identification by the immune system. It seems that non-spherical particles, like rods, disks, and unidentified flying object or UFO-like particles are capable of evading MPS as well. The hypothesis explaining this “invisibility” of elongated nano-objects is (i) a faster flow rate due to fluid dynamics or (ii) the local shape impeding internalization.23 Thanks to the possibility of developing materials with any shape, it is now possible to systematically investigate the role of shape in the interactions between NPs and biological systems. Dawson, Yan, and coworkers have provided researchers with a robust discovery framework to rigorously investigate the biological effects caused by the shape of nanomaterials.35 As a proof of concept, distinct shape-dependent immunological regimes have been identified. The exploration of shape biology at the nanoscale may open new avenues in promoting collective cellular processes and controlling the immunogenicity of nanomaterials.
After drug delivery to the intended target, NPs should be cleared from circulation to avoid long-term inflammation. Some degradable NPs or small-sized NPs are eliminated by biliary or renal excretion. Reports indicate that after 2 weeks, a relevant dose of administered NPs is accumulated in clearance organs (spleen, liver, kidneys) with possible chronic toxicity.36 Several in vivo studies show correlations between macrophage uptake and accumulation, such that the exploitation of macrophages as disease-homing carriers has been proposed.37,38 It is important to understand that, even if physicochemical properties of NPs have been discussed separately, there is a strong interplay between size, charge, hydrophilicity, and so on when interactions occur in biological fluids. Upon exposure to physiological environments, NPs evolve and transform due to adsorption of biomolecules with sophisticated functional patterns and dynamics leading to significant alteration of in vivo pharmacokinetics, toxicity, and biodistribution.39 Consequently, the complement activation is strictly dependent on the biomolecules that immediately adsorb on the NP's surface, i.e., the biomolecular corona (BC), as will be further discussed in the following sections. As a fundamental step towards tuning the immune system by BC, the nanotechnologist will carefully understand factors regulating the bio-nano interactions, i.e., the interaction between NPs and biomolecules leading to BC formation.40
The biomolecular corona of nanoparticles
Upon exposure to biological media abundant and high exchange rate proteins rapidly cover the particle surface which results in the formation of a fast-evolving “soft” BC made of loosely bound proteins.41 Over time high-affinity proteins with a very slow exchange rate undermine the soft BC and form a long-standing “hard” BC around NPs.42 Typically, most studies focused on the characterization of the hard corona as the characterization of the soft corona is affected by experimental biases arising from the lack of precise protocols for its isolation. For this reason, many open-ended questions have long remained elusive including if the soft corona is different in composition from its hard counterpart, and whether proteins enriching the soft BC trigger nanoparticle–cellular interactions. Recently, it has been demonstrated that only a minor fraction of proteins is unique to soft BC, while the largest fraction is also present in the hard BC, showing that the same proteins can interact strongly or weakly with nanoparticles or pre-adsorbed neighboring proteins.Over the last decade, we have learned that hard BC equilibrium composition and structure is due to the interplay of shaping factors belonging to three main categories: (i) the physicochemical properties of NPs (e.g., size, curvature radius, the chemistry of surface, shape, charge, aggregation after synthesis, etc., reviewed in ref. 43), (ii) environmental factors (e.g., incubation time,44 temperature,45 local heating,46 and shear stress47); and (iii) the protein source and concentration.48–50 Among them, the protein source (e.g., human vs. mouse serum) is surely the factor contributing most to the enrichment of the biomolecular corona with immunogenic proteins (e.g., immunoglobulins, complement proteins). For instance, Caracciolo et al. demonstrated that two different formulations of cationic liposomes were more enriched in immunogenic proteins following exposure to human plasma (HP) than they were after exposure to mouse plasma (MP).51 Subsequently, Müller and coworkers demonstrated that the biomolecular corona of polystyrene and magnetite NPs was markedly different in rabbit, sheep, and mouse plasma, especially in terms of Ig content.52 Among other implications, different profiles in immunogenic proteins may significantly affect the pharmacokinetic profile of NPs in the bloodstream thus questioning the real significance of animal testing for predicting physiological response in humans.
Even different protein sources from the same animal species can cause different responses by the immune system to the administration of NPs. For instance, Mirshafiee et al. showed that incubating NPs in HP produces a greater enrichment of immunogenic proteins than incubation in human serum (HS) and, consequently, a more adverse immune reaction.53 Subsequent investigations clarified that alterations in the human proteome as those caused by life-threatening diseases (e.g., cancer) lead to personalized disease-specific BC.54,55
A turning point was reached when it became clear that protein binding cannot be prevented by grafting stealth components (e.g., PEG) to the NP surface.56 We now know with certainty that the synthetic identity of any nanomaterial, be it stealth or not, is changed into a biological identity by the inevitable formation of a BC on its surface. As the formation of BC seems to be inevitable, it is now believed that the clinical translation of the results of basic research will necessarily have to pass from the control and exploitation of the biological identity of nanomaterials. A decisive contribution to achieving this goal will come from increasingly advanced computational methods that correlate the biological identity of nanomaterials and their biological effects in a physiological environment. Among the emerging approaches, QSAR (quantitative structure–activity relation) is based on the assumption that nanomaterials with a similar biological identity elicit a similar physiological response.57 In a QSAR investigation, Bigdeli et al. explored the correlation between the biological identity of a library of 17 liposome-protein coronas with typical biological effects such as cellular uptake and cytotoxicity in human cervical cancer cells (HeLa) and prostate cancer cells (PC3). The authors demonstrated that a small subset of the more than 400 plasma proteins that made up the 17 BCs was able to rationalize the results of biological experiments.58 A paradigmatic example is represented by nanoimmunotherapy, a combination of nanomedicine and immunotherapy that could significantly increase patient response.59 There are several approaches to nanoimmunotherapy in which NPs target tumor cells, the tumor's immune microenvironment, or the peripheral immune system. A promising prospect is the development of artificial BCs that can selectively activate the immune system forcing it to transform cold non-immunoresponsive metastases into warm immunoreactive lesions.58 In the following section, we will review the most significant literature that links BC of NPs and activation of the immune response.
The biomolecular corona directs nanomedicine interaction with the immune system
Being located on the surface of the NPs, the BC is the interface that controls the interaction between the NPs themselves and the immune system. This aspect has significant consequences on the use of NPs in nanomedicine, nanopharmacology, and nanotoxicology. At a first level, BC changes the physicochemical properties of nanoparticles that regulate the interaction with the immune system (e.g., surface charge, size, hydrophobicity, etc.) as summarized in Table 1. This is a kind of nonspecific activation of the immune system as it is not directly related to protein composition and structure, but only to the physicochemical properties of nanoparticle–protein complexes. As most plasma proteins are anionic at physiological pH, BC provides nanomaterials with a negative surface charge. Despite its complexity, and large variability across different nanomaterial types, the BC provides the NPs with a negative zeta-potential, between −10 and −20 mV regardless of their chemical–physical properties. This attenuates not specific electrostatic interactions between nanomaterials and negatively charged plasma membranes thus reducing cellular association and internalization (Fig. 2).| BC feature | Effect |
|---|---|
| Negative surface charge/artificial BC | Reduced immune system recognition due to limited interaction with plasma membrane60 |
| Size increase/aggregation | Immune clearance42,60 |
| Opsonins enrichment | Phagocytosis61,62 |
| Protein conformational changes | Inflammatory response triggering,63 amyloid fibrils formation, autoimmunity64,65 |
Many studies have shown that pre-coating nanoparticles with an artificial BC reduces capture by immune cells in vitro, ex vivo, and in vivo. Simon et al. exploited an artificial BC that reduces cellular uptake by immune cells in vitro.60 Giulimondi et al. demonstrated that an artificial BC renders the zeta-potential of cationic liposomes negative thus contributing to reducing the capture by circulating leukocytes charge in patients' whole blood ex vivo.66 More recently, the negative charge of DNA was exploited to produce a multilayered biomimetic NP type called “proteoDNAsome” whose negatively charged BC allows to elude the immune system more efficiently than PEGylated liposomes in vivo.67
A second effect of the BC is the enlargement of the size of NPs. Such enlargement can vary by about one order of magnitude depending on the NP type. The smallest increase of about 20 nm was reported for 30–50 nm citrate-stabilized gold NPs, while the largest of roughly 200 nm was reported for PSOSO3 nanoparticles.42 The increase in the size of the NPS is not only due to the steric hindrance of the protein coating but also to the formation of BC-induced aggregates. This happens when the adsorbed proteins neutralize the surface charge of the NPs reducing the electrostatic repulsion between particles and promoting the formation of short-range van der Waals bonds.60 Large aggregates are immediately recognized by the cells of the immune system which remove them from the bloodstream.
In addition to activating the immune system through charge- and size-dependent mechanisms, the BC surrounding NPs can activate the immune system through molecular recognition mechanisms. This occurs when BC is enriched with opsonins which bind to macrophage and phagocyte receptors, promote cellular internalization of NPs and, consequently, their removal from the bloodstream. The notion that host proteins bind to NPs by making them recognized as “non-self” by immune system cells and labeling them with an “eat me” signal on the surface has been known for over forty years so far.61,62 The liposomal formulations, which can be considered a model system of NPs, are coated with significantly different protein coronas depending on their physical–chemical properties. Enrichment with immunogenic proteins results in phagocytes' activation, reducing the circulation time in the blood and preventing accumulation in the target tissue.68 The uptake of liposomes by phagocytes is a process mediated by a set of receptors expressed on the phagocytes themselves (FcR and C3) that recognize NPs decorated by complement derived C3d/C3bi opsonins. Other membrane proteins expressed on the surface of phagocytes can bind to innate collectins (e.g., Ficoline, C1q). Phagocytes can also eliminate NPs coated with antibodies (e.g., IgG).
If, on the one hand, the success of NPs-mediated drug delivery requires the ability of the NPs themselves to evade the immune system, on the other hand, the activation of immune cells is emerging as a promising approach in advanced medical applications. For example, NPs are capable of targeting tumor-associated macrophages (TAMs) that have been identified as an essential component of the tumor microenvironment.69 Although this field of research is still in its infancy and the mechanisms that regulate the interaction between TAMs and cancer cells are not yet fully understood, there is general agreement that TAMs may be associated with both tumor progression. On other hand, the antitumor cytotoxicity of TAMs can be stimulated by NPs loaded with immunomodulators leading to the elimination of cancer cells and metastases.70
However, the BC maybe not be able, per se, to lead to immune system activation. Previous studies have shown that certain BCs trigger neither innate nor adaptive immune responses and do not give rise to the production of reactive oxygen species (ROS) or pro-inflammatory cytokines.71 On the other side, an inflammatory response can be triggered by conformational changes in proteins bound to nanomaterials. Variations in the tertiary structure of proteins are driven by interactions with the surface of the NPs. Such conformational changes can lead to the exposure of functional groups that are folded into the hydrophobic core of the naïve protein inducing protein aggregation and formation of amyloid fibers. The formation of amyloids triggers inflammatory fiber responses via the activation of the immune cascade through receptor-mediated recognition. The concept of “amyloid protein corona” has recently been introduced to indicate a protein corona enriched with amyloid fibers.64 In most cases, the interaction between a naïve protein and NPs promotes the conversion of α-helices to β sheets and the NP surface acts as a nucleation center for the growth of amyloid fibers.65 Although the number of studies that have explored the formation of amyloid fibers within the protein corona is still limited, some conclusions already seem general. First, for each specific protein, the extent of the transformation from α-helices to β sheets transition is strictly related to the type of NP on which it is adsorbed. For example, the native HSA shows a different variation of the α-helices fraction depending on the NP type on which it is adsorbed. On the other hand, some types of NPs inhibit α-helices to β-sheets transition and thus may be exploited to inhibit amyloidosis and immune reactions.72,73 Another possibility is to use NP inhibitors capable of selectively sequestering amyloid proteins from biological fluids (e.g., blood, cerebrospinal fluid, etc.) instead of functional proteins so as not to evoke immune responses. The same concepts underlying the formation of the nanoparticle-protein corona are currently being exploited to develop novel sensing and mitigation strategies64 against amyloid-associated diseases, such as Parkinson's disease,74 Alzheimer's disease,75 type 2 diabetes76 and familial amyloidosis.77
Upon NP–protein interaction immunogenic epitope generation can also occur.78–80 This is intended as the change in the protein tertiary structure that makes proteins immunogenic and triggers autoimmune reactions. Moreover, the denaturation of corona proteins eventually promotes the activation of signaling pathways and stimulates autoimmune responses. Changes in the secondary structure of proteins bound to nanoparticles can be explored in situ by Circular Dichroism (CD) spectroscopy eventually implemented with synchrotron radiation (SR) as a light source that allows accessing the extreme ultraviolet (UV) wavelength region and, in turn, more precise structural determination.81 Deng et al. demonstrated that fibrinogen undergoes denaturation when it binds to negatively charged poly (acrylic acid) conjugated gold NPs. The denatured protein binds to the Mac-1 integrin receptor on the surface of macrophages and stimulates the NF-κB signaling pathway leading to the release of inflammatory cytokines.63 Caruso and coworkers82 have demonstrated that denaturation of corona proteins results in a significant reduction of cellular internalization of nanoporous polymer NPs in human monocytic THP-1 cells with respect to pristine NPs. On the other side, protein denaturation was found to promote class A scavenger receptor-mediated phagocytosis in differentiated THP-1 cells.
Protein denaturation within the biomolecular corona is also dependent on the pH of the medium. Shang et al. demonstrated that protein denaturation is more pronounced at basic pH.65 Exposing spherical and flat gold NPs with covalently bound yeast cytochrome C (Cyt C) to different pH values, it was shown that Cyt C undergoes reversible denaturation at acidic pH and refolds at basic pH and that this transition does not depend on the particle morphology. However, these pH denaturation experiments were done on single proteins so probably should not be generalised. Park et al. demonstrated protein denaturation occurring on carbon nanotubes (CNTs) produced high ROS levels and induced major proinflammatory cytokine release both in human and murine macrophages.71 Furthermore, activation of innate and adaptive immunity was only stimulated by “unfolded BC” while immune responses were not activated by a normal BC.
Moreover, BC considerably influences interactions between NPs and cells through different activation of pathways along with internalization. Furthermore, BC considerably influences the interactions between NP and cells as it can inhibit or promote different endocytosis pathways than pristine NP. For example, it has been shown that cationic liposomes mainly use macropinocytosis as a mechanism of cellular internalization and that BC can significantly reduce this mechanism by promoting cellular uptake through clathrin-dependent endocytosis.83 Furthermore, since the toxicity of NPs also derives from direct contact with membrane cells, BC can mitigate the cytotoxicity of NPs.
For example, Mbeh et al. showed that the cytotoxicity of GO nanosheets results from direct interactions that cause physical damage to the cell membrane.84 This effect is largely attenuated by the biomolecular corona by a reduction of the interaction between graphene and membrane lipids.85 Cells exposed to silica NP have a degree of adhesion to the cell membrane and permeability depends on the presence of BC.86 Yin et al. showed that coating zinc oxide NP with corona proteins decreases their cytotoxicity in skin fibroblasts and human hepatocarcinoma cells.87 The change in the mechanism of interaction with cell membranes and cell internalization can have a cascade effect on numerous cellular events such as transcription, proliferation, signal transduction, cell cycle regulation, metabolism, apoptosis, and so on. However, studies on the role of BC in the internalization of NPs in immune cells are currently limited.
Conclusions and future roadmaps
There is a large debate in the literature regarding the safety and efficacy of nanomaterials in nanomedicine, despite their potential advancements. When nanomaterials enter a biological fluid, several biomolecules adsorb on their surface and form a biomolecular corona. In this review, we focused on the ways the immune system is triggered by the biomolecular corona. However, despite several general principles that have been established, most aspects need to be elucidated. Among hidden factors, the role of sex as a biological variable on the activation of the immune system has been only marginally explored so far. It is now being well documented that genetics, hormones, and microbiome variances are sex-related biological factors that stimulate the immune response, control sensitivity to infections, disease onset,88 and ultimately mortality.89 Despite this, gender is a largely overlooked factor in research related to immune response and infectious diseases and has generally been ignored in most clinical trials. Across species, females tend to develop a greater innate and adaptive immune response to infections. On the other side, the augmented immune function exhibited by females is associated with a roughly 10-fold amplified risk of developing inflammatory and autoimmune diseases (AD) in men. In addition, it is shown that such sex differences play a critical role in the development, progress, and treatment of diseases. Surprisingly, very recent studies revealed the critical role of sex differences in the clinical outcomes of the novel coronavirus disease 2019 (COVID-19).90 A major portion of the published reports revealed that male individuals have a higher mortality rate than females. These outcomes along with the observed substantial differences in the response of male and female immune cells to nanomaterials demand greater attention and consideration of sex and gender in research about COVID-19.Finally, there are other metabolites besides protein in the biomolecular corona to be considered. This class includes lipid metabolites, blood and dietary metabolites as well as exogenous metabolites in the environment, also known as xenometabolites or xenobiotics as recently reviewed by Chetwynd and Lynch.91 These compounds intervene in cell signalling and growth thus understanding their contribution in the biomolecular corona can offer important research hints in the field of NP uptake, distribution and immunotoxicity.
With a substantially improved understanding of NP–BC interactions in the immune system, the adverse aspects of NPs will be anticipated in advance and inhibited through rational design.
Conflicts of interest
There are no conflicts to declare.References
- C. Farrera and B. Fadeel, Eur. J. Pharm. Biopharm., 2015, 95, 3–12 CrossRef CAS PubMed.
- G. La Barbera, A. L. Capriotti, G. Caracciolo, C. Cavaliere, A. Cerrato, C. M. Montone, S. Piovesana, D. Pozzi, E. Quagliarini and A. Laganà, Talanta, 2020, 209, 120487 CrossRef CAS PubMed.
- Y. Liu, J. Hardie, X. Zhang and V. M. Rotello, in Seminars in immunology, Elsevier, 2017, vol. 34, pp. 25–32 Search PubMed.
- M. S. Goldberg, Nat. Rev. Cancer, 2019, 19, 587–602 CrossRef CAS PubMed.
- M. O. Oyewumi, A. Kumar and Z. Cui, Expert Rev. Vaccines, 2010, 9, 1095–1107 CrossRef CAS PubMed.
- A. L. Silva, C. Peres, J. Conniot, A. I. Matos, L. Moura, B. Carreira, V. Sainz, A. Scomparin, R. Satchi-Fainaro and V. Préat, in Seminars in immunology, Elsevier, 2017, vol. 34, pp. 3–24 Search PubMed.
- H. He, L. Liu, E. E. Morin, M. Liu and A. Schwendeman, Acc. Chem. Res., 2019, 52, 2445–2461 CrossRef CAS PubMed.
- J. Mo, Q. Xie, W. Wei and J. Zhao, Nat. Commun., 2018, 9, 1–11 CrossRef CAS PubMed.
- B. Sun, Z. Ji, Y.-P. Liao, M. Wang, X. Wang, J. Dong, C. H. Chang, R. Li, H. Zhang and A. E. Nel, ACS Nano, 2013, 7, 10834–10849 CrossRef CAS PubMed.
- C. Corbo, R. Molinaro, A. Parodi, N. E. Toledano Furman, F. Salvatore and E. Tasciotti, Nanomedicine, 2016, 11, 81–100 CrossRef CAS PubMed.
- S. Surana, A. R. Shenoy and Y. Krishnan, Nat. Nanotechnol., 2015, 10, 741–747 CrossRef CAS PubMed.
- R. H. Fang and L. Zhang, Annu. Rev. Chem. Biomol. Eng., 2016, 7, 305–326 CrossRef PubMed.
- D. Chenthamara, S. Subramaniam, S. G. Ramakrishnan, S. Krishnaswamy, M. M. Essa, F.-H. Lin and M. W. Qoronfleh, Biomater. Res., 2019, 23, 1–29 CrossRef PubMed.
- Y. Min, J. M. Caster, M. J. Eblan and A. Z. Wang, Chem. Rev., 2015, 115, 11147–11190 CrossRef CAS PubMed.
- C. Giannakou, M. V. D. Z. Park, W. H. de Jong, H. van Loveren, R. J. Vandebriel and R. E. Geertsma, Int. J. Nanomed., 2016, 11, 2935 CrossRef CAS PubMed.
- K. M. Tsoi, S. A. MacParland, X.-Z. Ma, V. N. Spetzler, J. Echeverri, B. Ouyang, S. M. Fadel, E. A. Sykes, N. Goldaracena and J. M. Kaths, Nat. Mater., 2016, 15, 1212–1221 CrossRef CAS PubMed.
- H. Maeda, J. Wu, T. Sawa, Y. Matsumura and K. Hori, J. Contr. Release, 2000, 65, 271–284 CrossRef CAS.
- R. áL Juliano and D. Stamp, Biochem. Biophys. Res. Commun., 1975, 63, 651–658 CrossRef.
- S. Hirn, M. Semmler-Behnke, C. Schleh, A. Wenk, J. Lipka, M. Schäffler, S. Takenaka, W. Möller, G. Schmid and U. Simon, Eur. J. Pharm. Biopharm., 2011, 77, 407–416 CrossRef CAS PubMed.
- G. Sonavane, K. Tomoda and K. Makino, Colloids Surf., B, 2008, 66, 274–280 CrossRef CAS PubMed.
- F. A. Sharp, D. Ruane, B. Claass, E. Creagh, J. Harris, P. Malyala, M. Singh, D. T. O'Hagan, V. Pétrilli and J. Tschopp, Proc. Natl. Acad. Sci., 2009, 106, 870–875 CrossRef CAS PubMed.
- V. B. Joshi, S. M. Geary and A. K. Salem, AAPS J., 2013, 15, 85–94 CrossRef CAS PubMed.
- J. Di, X. Gao, Y. Du, H. Zhang, J. Gao and A. Zheng, Asian J. Pharm. Sci., 2021, 16, 444–458 CrossRef PubMed.
- L. Shang, K. Nienhaus and G. U. Nienhaus, J. Nanobiotechnol., 2014, 12, 1–11 CrossRef PubMed.
- A. Verma and F. Stellacci, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- A. Badiee, V. H. Shargh, A. Khamesipour and M. R. Jaafari, Vaccine, 2013, 31, 735–749 CrossRef CAS PubMed.
- M. Lucarelli, A. M. Gatti, G. Savarino, P. Quattroni, L. Martinelli, E. Monari and D. Boraschi, Eur. Cytokine Netw., 2004, 15, 339–346 CAS.
- R. Molinaro, C. Corbo, J. O. Martinez, F. Taraballi, M. Evangelopoulos, S. Minardi, I. K. Yazdi, P. Zhao, E. De Rosa and M. B. Sherman, Nat. Mater., 2016, 15, 1037–1046 CrossRef CAS PubMed.
- E. Ben-Akiva, R. A. Meyer, H. Yu, J. T. Smith, D. M. Pardoll and J. J. Green, Sci. Adv., 2020, 6, eaay9035 CrossRef CAS PubMed.
- J. Xie, C. Xu, N. Kohler, Y. Hou and S. Sun, Adv. Mater., 2007, 19, 3163–3166 CrossRef CAS.
- Y. Sheng, Y. Yuan, C. Liu, X. Tao, X. Shan and F. Xu, J. Mater. Sci.: Mater. Med., 2009, 20, 1881–1891 CrossRef CAS PubMed.
- E. T. M. Dams, P. Laverman, W. J. G. Oyen, G. Storm, G. L. Scherphof, J. W. M. Van der Meer, F. H. M. Corstens and O. C. Boerman, J. Pharmacol. Exp. Ther., 2000, 292, 1071–1079 CAS.
- A. S. A. Lila, H. Kiwada and T. Ishida, J. Contr. Release, 2013, 172, 38–47 CrossRef PubMed.
- D. R. Getts, L. D. Shea, S. D. Miller and N. J. C. King, Trends Immunol., 2015, 36, 419–427 CrossRef CAS PubMed.
- W. Zhang, H. Lopez, L. Boselli, P. Bigini, A. Perez-Potti, Z. Xie, V. Castagnola, Q. Cai, C. P. Silveira, J. M. De Araujo, L. Talamini, N. Panini, G. Ristagno, M. B. Violatto, S. Devineau, M. P. Monopoli, M. Salmona, V. A. Giannone, S. Lara, K. A. Dawson and Y Yan, ACS Nano, 2022, 16, 1547–1559 CrossRef CAS PubMed.
- H. H. Gustafson, D. Holt-Casper, D. W. Grainger and H. Ghandehari, Nano Today, 2015, 10, 487–510 CrossRef CAS PubMed.
- G. Escobar, B. Gentner, L. Naldini and R. Mazzieri, Oncoimmunology, 2014, 3, e28696 CrossRef PubMed.
- E. V. Batrakova, H. E. Gendelman and A. V. Kabanov, Expert Opin. Drug Delivery, 2011, 8, 415–433 CrossRef CAS PubMed.
- T. L. Moore, L. Rodriguez-Lorenzo, V. Hirsch, S. Balog, D. Urban, C. Jud, B. Rothen-Rutishauser, M. Lattuada and A. Petri-Fink, Chem. Soc. Rev., 2015, 44, 6287–6305 RSC.
- L. M. Ernst, E. Casals, P. Italiani, D. Boraschi and V. Puntes, Nanomaterials, 2021, 11, 2991 Search PubMed.
- H. Mohammad-Beigi, Y. Hayashi, C. M. Zeuthen, H. Eskandari, C. Scavenius, K. Juul-Madsen, T. Vorup-Jensen, J. J. Enghild and D. S. Sutherland, Nat. Commun., 2020, 11, 1–16 CrossRef PubMed.
- C. D. Walkey and W. C. W. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- C. Corbo, R. Molinaro, M. Tabatabaei, O. C. Farokhzad and M. Mahmoudi, Biomater. Sci., 2017, 5, 378–387 RSC.
- A. L. Barrán-Berdón, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso, C. Cavaliere, A. Riccioli, S. Palchetti and A. Laganà, Langmuir, 2013, 29, 6485–6494 CrossRef PubMed.
- M. Mahmoudi, A. M. Abdelmonem, S. Behzadi, J. H. Clement, S. Dutz, M. R. Ejtehadi, R. Hartmann, K. Kantner, U. Linne and P. Maffre, ACS Nano, 2013, 7, 6555–6562 CrossRef CAS PubMed.
- M. Mahmoudi, S. E. Lohse, C. J. Murphy, A. Fathizadeh, A. Montazeri and K. S. Suslick, Nano Lett., 2014, 14, 6–12 CrossRef CAS PubMed.
- L. Digiacomo, S. Palchetti, F. Giulimondi, D. Pozzi, R. Z. Chiozzi, A. L. Capriotti, A. Laganà and G. Caracciolo, Lab Chip, 2019, 19, 2557–2567 RSC.
- G. Caracciolo, D. Pozzi, A. L. Capriotti, C. Cavaliere, P. Foglia, H. Amenitsch and A. Lagana, Langmuir, 2011, 27, 15048–15053 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 Search PubMed.
- V. Palmieri, M. De Spirito and M. Papi, Nanomedicine, 2021, 16, 20 CrossRef PubMed.
- G. Caracciolo, D. Pozzi, A. L. Capriotti, C. Cavaliere, S. Piovesana, G. La Barbera, A. Amici and A. Laganà, J. Mater. Chem. B, 2014, 2, 7419–7428 RSC.
- L. K. Müller, J. Simon, C. Rosenauer, V. Mailänder, S. Morsbach and K. Landfester, Biomacromolecules, 2018, 19, 374–385 CrossRef PubMed.
- V. Mirshafiee, R. Kim, M. Mahmoudi and M. L. Kraft, Int. J. Biochem. Cell Biol., 2016, 75, 188–195 CrossRef CAS PubMed.
- G. Caracciolo, D. Caputo, D. Pozzi, V. Colapicchioni and R. Coppola, Colloids Surf., B, 2014, 123, 673–678 CrossRef CAS PubMed.
- M. J. Hajipour, S. Laurent, A. Aghaie, F. Rezaee and M. Mahmoudi, Biomater. Sci., 2014, 2, 1210–1221 RSC.
- D. Pozzi, V. Colapicchioni, G. Caracciolo, S. Piovesana, A. L. Capriotti, S. Palchetti, S. De Grossi, A. Riccioli, H. Amenitsch and A. Laganà, Nanoscale, 2014, 6, 2782–2792 RSC.
- C. D. Walkey, J. B. Olsen, F. Song, R. Liu, H. Guo, D. W. H. Olsen, Y. Cohen, A. Emili and W. C. W. Chan, ACS Nano, 2014, 8, 2439–2455 CrossRef CAS PubMed.
- A. Bigdeli, S. Palchetti, D. Pozzi, M. R. Hormozi-Nezhad, F. Baldelli Bombelli, G. Caracciolo and M. Mahmoudi, ACS Nano, 2016, 10, 3723–3737 CrossRef CAS PubMed.
- Y. Shi and T. Lammers, Acc. Chem. Res., 2019, 52, 1543–1554 CrossRef CAS PubMed.
- J. Simon, L. K. Müller, M. Kokkinopoulou, I. Lieberwirth, S. Morsbach, K. Landfester and V. Mailänder, Nanoscale, 2018, 10, 10731–10739 RSC.
- Y. J. Kao and R. L. Juliano, Biochim. Biophys. Acta, Gen. Subj., 1981, 677, 453–461 CrossRef CAS.
- S. M. Moghimi, D. Simberg, T. Skotland, A. Yaghmur and A. C. Hunter, J. Pharmacol. Exp. Ther., 2019, 370, 581–592 CrossRef CAS PubMed.
- Z. Deng, Nat. Nanotechnol., 2011, 6(1), 39–44 CrossRef CAS PubMed.
- P. Chen, F. Ding, R. Cai, I. Javed, W. Yang, Z. Zhang, Y. Li, T. P. Davis, P. C. Ke and C. Chen, Nano Today, 2020, 35, 100937 CrossRef CAS PubMed.
- L. Shang, Y. Wang, J. Jiang and S. Dong, Langmuir, 2007, 23, 2714–2721 CrossRef CAS PubMed.
- F. Giulimondi, L. Digiacomo, D. Pozzi, S. Palchetti, E. Vulpis, A. L. Capriotti, R. Z. Chiozzi, A. Laganà, H. Amenitsch and L. Masuelli, Nat. Commun., 2019, 10, 1–11 CrossRef PubMed.
- F. Giulimondi, E. Vulpis, L. Digiacomo, M. V. Giuli, A. Mancusi, A. L. Capriotti, A. Laganà, A. Cerrato, R. Zenezini Chiozzi and C. Nicoletti, ACS Nano, 2022, 16(2), 2088–2100 CrossRef CAS PubMed.
- S. M. Moghimi and H. M. Patel, FEBS Lett., 1988, 233, 143–147 CrossRef CAS PubMed.
- A. R. Poh and M. Ernst, Front. Oncol., 2018, 12(8), 49 CrossRef PubMed.
- S.-Y. Kim, S. Kim, J.-E. Kim, S. N. Lee, I. W. Shin, H. S. Shin, S. M. Jin, Y.-W. Noh, Y. J. Kang and Y. S. Kim, ACS Nano, 2019, 13, 12671–12686 CrossRef CAS PubMed.
- J. Park, S. J. Park, J. Y. Park, S. Kim, S. Kwon, Y. Jung and D. Khang, Adv. Sci., 2021, 8, 2004979 CrossRef CAS PubMed.
- R. Capomaccio, I. O. Jimenez, P. Colpo, D. Gilliland, G. Ceccone, F. Rossi and L. Calzolai, Nanoscale, 2015, 7, 17653–17657 RSC.
- F. Ramezani and H. Rafii-Tabar, Mol. BioSyst., 2015, 11, 454–462 RSC.
- D. Grassi, S. Howard, M. Zhou, N. Diaz-Perez, N. T. Urban, D. Guerrero-Given, N. Kamasawa, L. A. Volpicelli-Daley, P. LoGrasso and C. I. Lasmézas, Proc. Natl. Acad. Sci., 2018, 115, E2634–E2643 CrossRef CAS PubMed.
- U. Dettmer, A. J. Newman, F. Soldner, E. S. Luth, N. C. Kim, V. E. Von Saucken, J. B. Sanderson, R. Jaenisch, T. Bartels and D. Selkoe, Nat. Commun., 2015, 6, 1–16 Search PubMed.
- X. Ge, Y. Yang, Y. Sun, W. Cao and F. Ding, ACS Chem. Neurosci., 2018, 9, 967–975 CrossRef CAS PubMed.
- M. Schmidt, S. Wiese, V. Adak, J. Engler, S. Agarwal, G. Fritz, P. Westermark, M. Zacharias and M. Fändrich, Nat. Commun., 2019, 10, 1–9 CrossRef PubMed.
- L. K. Bogart, G. Pourroy, C. J. Murphy, V. Puntes, T. Pellegrino, D. Rosenblum, D. Peer and R. Lévy, ACS Nano, 2014, 8, 3107–3122 CrossRef CAS PubMed.
- S. Goy-López, J. Juárez, M. Alatorre-Meda, E. Casals, V. F. Puntes, P. Taboada and V. Mosquera, Langmuir, 2012, 28(24), 9113–9126 CrossRef PubMed.
- D. Zhang, O. Neumann, H. Wang, V. M. Yuwono, A. Barhoumi, M. Perham, J. D. Hartgerink, P. Wittung-Stafshede and N. J. Halas, Nano Lett., 2009, 9, 666–671 CrossRef CAS PubMed.
- D. Baimanov, R. Cai and C. Chen, Bioconjugate Chem., 2019, 30, 1923–1937 CrossRef CAS PubMed.
- Y. Yan, K. T. Gause, M. M. J. Kamphuis, C.-S. Ang, N. M. O'Brien-Simpson, J. C. Lenzo, E. C. Reynolds, E. C. Nice and F. Caruso, ACS Nano, 2013, 7, 10960–10970 CrossRef CAS PubMed.
- L. Digiacomo, F. Cardarelli, D. Pozzi, S. Palchetti, M. A. Digman, E. Gratton, A. L. Capriotti, M. Mahmoudi and G. Caracciolo, Nanoscale, 2017, 9, 17254–17262 RSC.
- D. A. Mbeh, O. Akhavan, T. Javanbakht and M. Mahmoudi, Appl. Surf. Sci., 2014, 320, 596–601 CrossRef CAS.
- V. Palmieri, G. Perini, M. De Spirito and M. Papi, Nanoscale Horiz., 2018, 273–290 Search PubMed.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CrossRef CAS PubMed.
- H. Yin, R. Chen, P. S. Casey, P. C. Ke, T. P. Davis and C. Chen, RSC Adv., 2015, 5, 73963–73973 RSC.
- E. P. Scully, J. Haverfield, R. L. Ursin, C. Tannenbaum and S. L. Klein, Nat. Rev. Immunol., 2020, 1–6 Search PubMed.
- N. Gadi, S. C. Wu, A. P. Spihlman and V. R. Moulton, Front. Immunol., 2020, 11, 2147 CrossRef CAS PubMed.
- M. D. Park, Nat. Rev. Immunol., 2020, 20, 461 CrossRef CAS PubMed.
- A. J. Chetwynd and I. Lynch, Environ. Sci.: Nano, 2020, 7, 1041–1060 RSC.
| This journal is © The Royal Society of Chemistry 2022 |