Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Indicator displacement assays (IDAs): the past, present and future
Adam C.
Sedgwick
 a,
James T.
Brewster
II
a,
James T.
Brewster
II
 a,
Tianhong
Wu
b,
Xing
Feng
b,
Steven D.
Bull
a,
Tianhong
Wu
b,
Xing
Feng
b,
Steven D.
Bull
 c,
Xuhong
Qian
de,
Jonathan L.
Sessler
c,
Xuhong
Qian
de,
Jonathan L.
Sessler
 *a,
Tony D.
James
*a,
Tony D.
James
 *c,
Eric V.
Anslyn
*c,
Eric V.
Anslyn
 *a and
Xiaolong
Sun
*a and
Xiaolong
Sun
 *b
*b
aDepartment of Chemistry, The University of Texas at Austin, 105 East 24th Street A5300, Austin, Texas 78712-1224, USA. E-mail: sessler@cm.utexas.edu; anslyn@austin.utexas.edu
bThe Key Laboratory of Biomedical Information Engineering of Ministry of Education, School of Life Science and Technology, Xi’an Jiaotong University, Xi’an, 710049, P. R. China. E-mail: x.l.sun86@xjtu.edu.cn
cDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
dState Key Laboratory of Bioreactor Engineering, Shanghai Key Laboratory of Chemical Biology, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China
eSchool of Chemistry and Molecular Engineering, East China Normal University, 3663 Zhongshan Road, Shanghai 200062, China
First published on 10th November 2020
Abstract
Indicator displacement assays (IDAs) offer a unique and innovative approach to molecular sensing. IDAs can facilitate the detection of a range of biologically/environmentally important species, provide a method for the detection of complex analytes or for the determination and discrimination of unknown sample mixtures. These attributes often cannot be achieved by traditional molecular sensors i.e. reaction-based sensors/chemosensors. The IDA pioneers Inouye, Shinkai, and Anslyn inspired researchers worldwide to develop various extensions of this idea. Since their early work, the field of indicator displacement assays has expanded to include: enantioselective indicator displacement assays (eIDAs), fluorescent indicator displacement assays (FIDAs), reaction-based indicator displacement assays (RIAs), DimerDye disassembly assays (DDAs), intramolecular indicator displacement assays (IIDAs), allosteric indicator displacement assay (AIDAs), mechanically controlled indicator displacement assays (MC-IDAs), and quencher displacement assays (QDAs). The simplicity of these IDAs, coupled with low cost, high sensitivity, and ability to carry out high-throughput automation analysis (i.e., sensing arrays) has led to their ubiquitous use in molecular sensing, alongside the other common approaches such as reaction-based sensors and chemosensors. In this review, we highlight the various design strategies that have been used to develop an IDA, including the design strategies for the newly reported extensions to these systems. To achieve this, we have divided this review into sections based on the target analyte, the importance of each analyte and then the reported IDA system is discussed. In addition, each section includes details on the benefit of the IDAs and perceived limitations for each system. We conclude this Tutorial Review by highlighting the current challenges associated with the development of new IDAs and suggest potential future avenues of research.
 Tianhong Wu | Tianhong Wu, obtained BSc from Shandong Normal University in 2015. He is now a 1st year PhD student under the supervision of Xiaolong Sun in Xi’an Jiaotong University. |
Key learning points(1) Various aspects of chemistry involved in the design of IDAs.(2) Use of IDAs for sensing applications and “sensing arrays”. (3) Recent examples of IDA-based systems. (4) Current limitations of each IDA strategy. (5) Challenges and future perspectives related to IDAs (discussed within the review). |
Introduction
Molecular sensors have become a key component to the advancement of biological, environmental, agricultural, and industrial sciences. Since as early as 1867, optical-based sensors have been developed and used to detect biologically and environmentally important species.1 Despite the significant progress made in this field, a number of limitations continue to exist, which include the complex, multi-step syntheses required to obtain the desired sensor. Researchers have attempted to solve such problems using the knowledge acquired from several decades of progress in supramolecular chemistry.2 Inspired by the high specificity found in nature, supramolecular chemists have attempted to mimic various host–guest interactions with the development of synthetic molecular-based receptors. This has led to the identification of a number of host–guest interactions that are capable of forming unique supramolecular ensembles. With this acquired knowledge, various host–guest interactions have been exploited to design supramolecular-based molecular sensors, known as indicator displacement assays (IDAs). In this Tutorial Review, we aim to educate the reader regarding the design principles of indicator displacement assays (IDAs) and highlight the importance of IDAs in the area of molecular sensing, as demonstrated by select literature examples.The beginnings of IDAs: What is an indicator displacement assay (IDA)? Traditional IDAs are supramolecular ensembles comprised of an optical guest (indicator) that is reversibly bound to a synthetic receptor (host). In the presence of an analyte, the indicator is displaced from the host resulting in a change in its optical signal (detection) (Scheme 1). What is required? The design of a suitable IDA requires knowledge of supramolecular chemistry to identify the appropriate size, charge, and hydrophobicity of the chosen indicator to achieve complementary binding within the synthetic host. The target analyte must also have a higher affinity at a particular concentration. We use affinity here to mean the extent that the target analyte is bound relative to the indicator, and thus the affinity is related to the binding constant and concentration of the host, guest, and indicator, in order to achieve an effective displacement of the indicator and provide an optical response. In this Tutorial Review, we have aimed to take a step back and provide an overview of this area, starting from basic concepts to new advancements in the form of historical and recent literature examples. For an extensive explanation on the various criteria required for the development of IDAs, the reader is directed to an excellent review by Kim et al.3
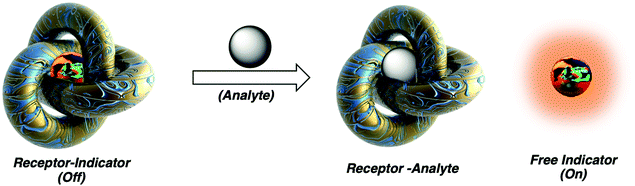 | ||
| Scheme 1 Basic schematic of a traditional indicator displacement assay (IDA). Analyte mediated displacement of an indicator from a supramolecular ensemble. | ||
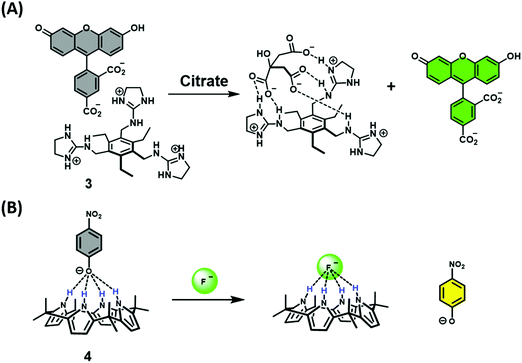
Early examples of traditional IDAs were first reported by the groups of Inouye, Shinkai, and Anslyn.4 As seen in these seminal demonstrations, IDAs were formed using the synthetic receptors resorcin[4]arene (1), calix[4]arene (2), and a tri-guanidinium pinwheel (3) host with a fluorescent guest (Fig. 1). These receptors provided the appropriate non-covalent interactions (i.e., ionic, cation–π, or hydrogen bonding) to yield suitable complexation and, when exposed to the appropriate analyte, chromophore release was observed by UV-Vis and fluorescence spectroscopy. More specifically, tri-guanidinium “pinwheel” 3 was intentionally designed to be pre-organized to afford a receptor with several hydrogen bond acceptors and three sets of positively charged species on the same face of the benzene spacer. This pinwheel system (3) provided the appropriate hydrogen bonding and charge pairing to form an IDA with the commercially available 5-carboxyfluorescein fluorophore (Scheme 2A). The three negatively charged carboxylate anions on citrate led to its higher binding affinity for 3 and the successful displacement of 5-carboxyfluorescein to facilitate the detection of citrate by UV-Vis and fluorescence spectroscopy in aqueous solution. This IDA proved effective in determining the concentration levels of citrate in beverages and displayed no interference from competing analytes, such as fructose and sucrose.4 It is important to note that an IDA and fluorescent indicator displacement assay (FIDA) are often used in the same regard due to the close relationship between a colorimetric and fluorometric response. In general, a FIDA is defined when the displaced indicator provides a change in fluorescence output.
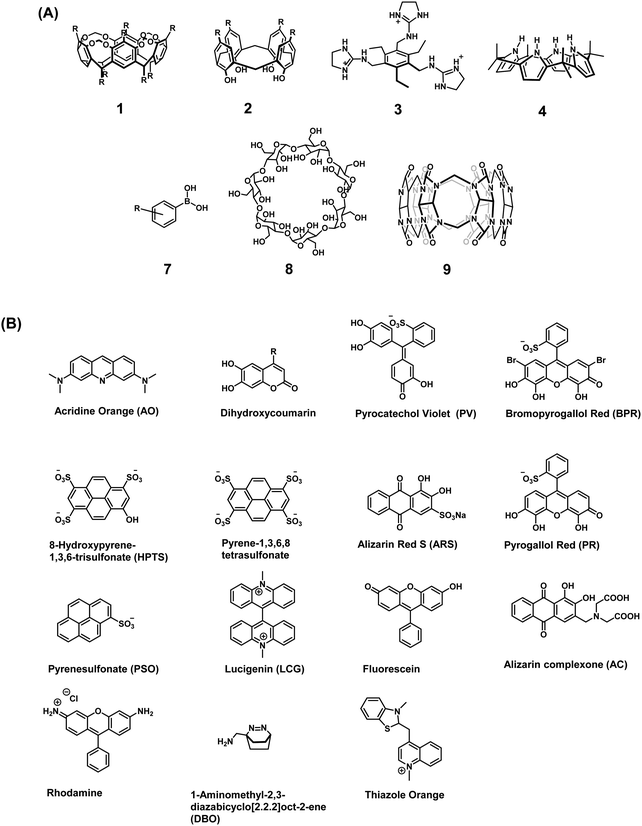 | ||
| Fig. 1 (A) Commonly used receptor/host systems. (B) Commonly used optical indicators/reporters used for the design of IDAs. | ||
The identification and application of new synthetic receptors can be seen from the early work of Sessler and co-workers, in which the anionic receptor known as calixpyrrole (4) was developed (Fig. 1). Exploiting the propensity of this receptor to bind to anions through hydrogen bonding, the colorimetric indicator 4-nitrophenolate anion was used to form the corresponding chemosensor ensemble.4 Due to the high affinity of the fluoride anion (F−) for calixpyrrole, this IDA was able to detect F− as a result of the displacement of the 4-nitrophenolate indicator, which led to a visual colour change (Scheme 2B). Following this seminal example, calixpyrrole has been exploited in numerous IDA strategies.
In recent years, the field has witnessed substantial growth with the emergence of new IDA strategies complementary to traditional IDAs. To avoid confusion, all of the systems in this review will be referred to as IDAs, however, when discussed individually, the correct abbreviation for that particular system will be used. As detailed below, extensions to the IDA concept include enantioselective indicator displacement assays (eIDAs), fluorescent indicator displacement assays (FIDAs), reaction-based indicator displacement assays (RIAs), allosteric indicator displacement assays (AIDAs), DimerDye Disassembly assays (DDAs), intramolecular indicator displacement assays (IIDAs), mechanically controlled indicator displacement assays (MC-IDAs), and a new extension that we term quencher displacement assays (QDAs). The basic design concepts for each IDA system are shown in Scheme 3. Note – the real-life examples of each IDA system are then referenced for a specific target analyte.
 | ||
| Scheme 3 Design strategies used for the indicator displacement assays (IDAs) included in this review. | ||
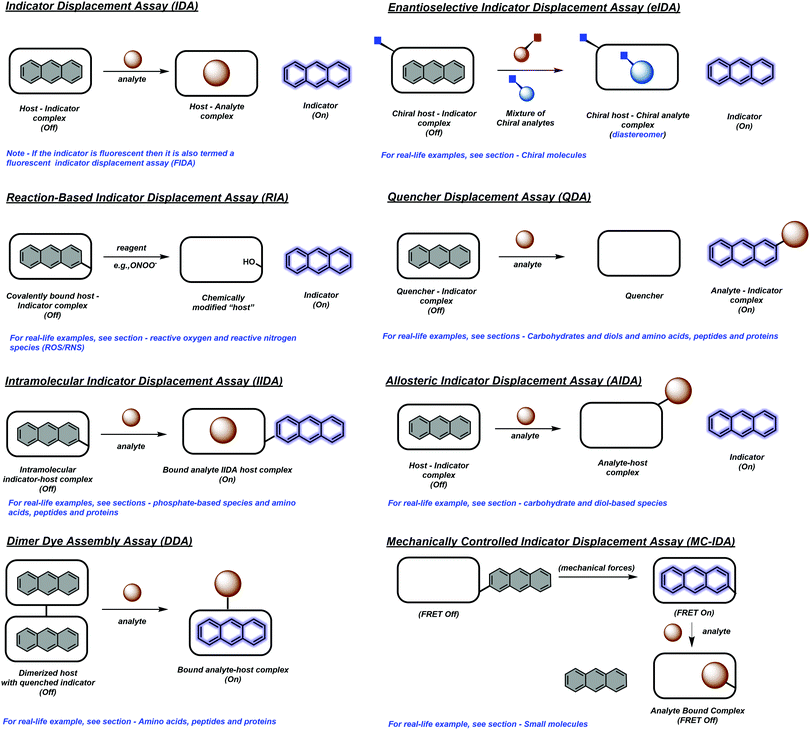
The continued development of synthetically accessible receptor scaffolds and commercially available dyes has led to their use as sensing arrays that allow the pattern-based recognition of analytes. The characteristic pattern for each analyte provides the ability to identify and discriminate unknown sample mixtures using chemometric methods, and is generally referred to as “differential sensing” (DS).5 This latter strategy provides an attractive alternative to the use of reaction-based molecular sensors (irreversible)/chemosensors (reversible) (Scheme 4). In addition, IDAs and their extensions, often provide a method to detect complex analytes that cannot be detected by traditional molecular sensors. For an extensive review of reaction-based molecular sensors and chemosensors, the reader can be directed to two excellent reviews by Chang and James, respectively.1,6
 | ||
| Scheme 4 Reaction-based molecule sensor (irreversible) and chemosensor (reversible). Compound 5 is an example of a reaction based molecular sensor for the detection of hydrogen peroxide (H2O2), which is highlighted in the review by Chang and co-workers.6 Receptor 6 is an example of a chemosensor for the detection of calcium ions (Ca2+), which is highlighted in the review by James and co-workers.1 | ||
Commonly used indicators and synthetic receptors for the design of IDAs
Traditionally, the indicators used in IDAs as reporter groups have been either colorimetric or fluorescent species with specific excitation and emission profiles. It is common to find the choice of indicator being dependent on its specific application and, in the context of general use, a turn-on approach or a clear colour change is favourable. For biological applications, colorimetric indicators in some cases are unsuitable and near-infrared (NIR) emitting fluorophores are highly desired as this allows deeper penetration and a higher sensitivity with minimal background auto-fluorescence and photodamage.1 Unfortunately, there are a lack of NIR fluorophores available that complement the currently reported synthetic receptors that allow the development of NIR-based IDAs. A full list of indicators that are discussed in this review are shown in Fig. 1.3The synthetic receptor unit is also a key element in designing an IDAs. Synthetic modifications of common recognition motifs coupled with fine-tuning of the inherent binding properties can afford constructs with highly selective sensing properties. Common receptor systems include calix[n]arenes (2), calixpyrroles (4), boronic acid derivatives (7), cyclodextrins (8), and cucurbit[n]urils (9) (Fig. 1). It is important to note these are only select examples and new synthetic receptors and reporters continue to emerge in the literature. While a comprehensive summary of all work in this fast-evolving area is not possible here, a number of illustrative indicators and hosts will be highlighted throughout this Tutorial Review.
As implied above, our goal is to outline the requirements of an effective IDA and illustrate the various design strategies that have been used to develop various specialised IDAs. In this context, we have used recently reported examples, however, earlier examples have also been used to provide a historical framework and provide a description of the basic concepts. In particular, we cover the application of IDAs for determining enantiomeric excess (ee) of chiral molecules and detecting biologically and environmentally important species, such as anions, cations, small neutral molecules, reactive oxygen species/reactive nitrogen species (ROS/RNS), saccharides, amino acid, peptides, proteins, nucleic acids, gases, and mechanical force effects. It is hoped that this treatment will serve to educate young scientists and seasoned researchers alike on the importance of IDAs and the underlying design principles, rather than providing an extensive catalogue of examples. For extensive listings of IDAs, the reader is directed to previous reviews by Kim et al. and Anslyn et al.3,4
IDA-based sensors for chiral molecules
Asymmetric synthetic methods allow for the controlled preparation of well-defined, three-dimensional architectures, in an enantioselective manner. Such chemistry is desired in the area of biomedical sciences as stereochemical differences can afford molecules with either beneficial or deleterious therapeutic properties. Most advances within the area of enantioselective synthesis have relied on empirical testing as a guide to iterative experimental design. The ability to rapidly determine enantioselectivity is essential. The majority of methods used to determine enantiomeric and diastereomeric excess rely on chiral chromatography and NMR spectroscopy, respectively. However, over the past decade we have witnessed an acceleration in the discovery of asymmetric reactions through the use of high-throughput experimentation (HTE).7 Unfortunately, the serial nature of current analytical methods is not readily amenable to high-throughput screening (HTS) protocols. This in turn has led to the development of enantioselective indicator displacement assays (eIDAs) for the analysis of the enantiomeric excess (ee) of a synthesized chiral molecule. For the uses of sensors for chirality determination in HTE, we point the reader to the review by Anslyn and Wolf.7In contrast to traditional IDAs, eIDAs incorporate chirality into the host to form a chiral host–indicator complex. Thus, in the presence of a chiral analyte, the indicator is displaced from the chiral receptor to form a diastereomeric complex, which allows the direct quantification of the enantiomeric excess (ee) of the putative chiral analyte using UV or fluorescence spectroscopic methods. The energetic differences between each resulting diastereomer provides a unique response that allows the ee of the sample to be determined. An attractive feature of eIDAs is that ee can be determined without the need for reaction work-up or product isolation. This approach also permits high-throughput analysis using 96-well plate readers without the need for expensive and time-consuming chiral HPLC.
A milestone in the development of eIDAs was the use of chiral boronic acid receptors for the simultaneous determination of the concentration and ee of chiral α-hydroxycarboxylates and vicinal diols.8 This seminal study took advantage of the fact that boronic acids were known to be excellent receptors for diols and α-hydroxycarboxylates (Scheme 5).9
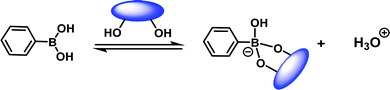 | ||
| Scheme 5 The reversible interaction between boronic acid derivatives and diols-containing molecules. | ||
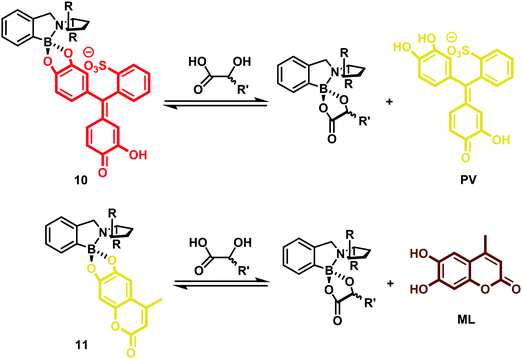
In this work, several colorimetric and/or fluorescent catechol (1,2-diol)-containing indicators were used to form the corresponding chiral boronate–dye complexes (10) and (11). This study revealed that steric differences between each indicator–receptor complex allowed the enantioselectivity and sensitivity of the assay to be tuned. Specifically, dihydroxycoumarin derivative 4-methylesculetin (ML), pyrocatechol violet (PV), and alizarin complexone (AC) proved effective as indicators and allowed for the development of eIDAs that could analyse chiral α-hydroxycarboxylates and diol samples over a broad sample range (Scheme 6). For optimization of the high throughput analysis of chiral samples, mathematical principles were established that permitted the concurrent determination of concentration and enantiomeric excess (ee) for a given chiral analyte.8
 | ||
| Scheme 6 eIDA for determining the concentration and ee of α-hydroxycarboxylates using either PV (colorimetric) or ML (fluorescent) as the indicators and a boronic acid as the chiral host. | ||
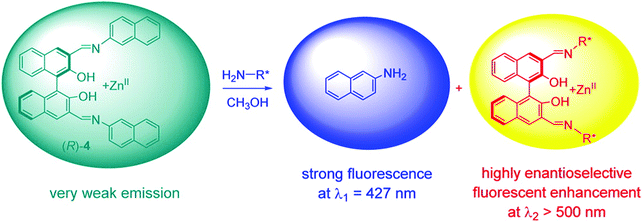
Pu and co-workers recently reported a rationally designed chiral 1,1′-bi-2-naphthol (BINOL)-based bis(naphthylimine) zinc complex ((Zn(II))(R)-4) for use as an eIDA. Remarkably, this strategy enabled the simultaneous determination of concentration and ee of chiral amines (i.e., diamines, amino alcohols, and amino acids) by analysing two separate emission wavelengths (Scheme 7).10 Receptor (R)-4 was developed through the simple imine formation between a chiral BINOL aldehyde and a fluorescent aromatic amine (2-naphthylamine). In the presence of Zn(OAc)2, the addition of a chiral amine to (R)-4 resulted in imine metathesis and displacement of the 2-naphthylamine units, which led to a concentration-dependent enhancement in the fluorescence emission intensity at λ1 = 427 nm (highlighted in blue). The enhancement in the fluorescence emission intensity at λ2 > 500 nm was shown to be dependent on the enantioselectivity of the chiral species (highlighted in yellow). As a result, this system enabled both the concentration and the enantiomeric composition to be determined using one fluorescence measurement.
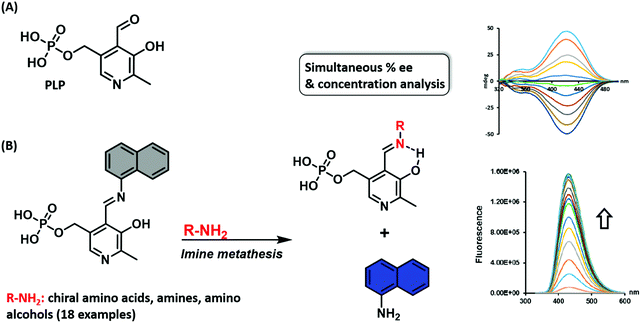
Wolf and co-workers reported a similar imine metathesis approach in creating a biomimetic eIDA that relies on the ubiquitous enzyme co-factor pyridoxal phosphate (PLP) for the simultaneous determination of the absolute configuration, enantiomeric composition, and concentration of unprotected amino acids, amino alcohols, and amines.11 This strategy exploited the condensation of an aromatic fluorescent amine with PLP. In the presence of a chiral amine, imine metathesis yielded a CD spectrum with characteristic Cotton effects thereby allowing the determination of the major enantiomer and ee (Scheme 8). In addition, displacement of the fluorescent amine led to a non-enantioselective dependent increase in fluorescence intensity, a feature that allowed the total analyte concentration to be determined. This example highlights a multimodal IDA strategy that takes advantage of two separate but complementary analytical techniques, namely circular dichroism and fluorescence, to determine analyte ee and concentrations.
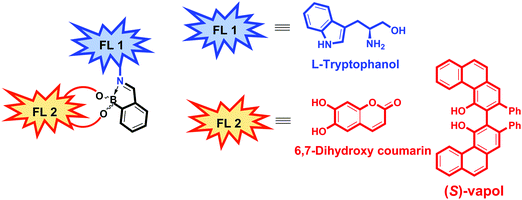
In 2019, Anzenbacher and co-workers reported a unique dual fluorophore (FL1 and FL2) eIDA platform that enabled the determination of the ee of more than one class of compound, including amines, amino alcohols, diols, and hydroxyacids.12 Their approach exploits both imine metathesis and diol displacement strategies by combining tryptophanol (FL1) and the diol fluorophores, dihydroxycoumarin or (S)-Vapol (FL2) with 2-formylphenylboronic acid (2-FPBA) (Fig. 2). For determining the ee of chiral diols or hydroxy acids, a supramolecular ensemble consisting of tryptophanol (FL1), dihydroxycoumarin (FL2) and 2-FPBA was prepared. This construct is characterised by FRET between the FL1 (donor) and FL2 (acceptor) moieties. In the presence of a chiral diol or hydroxy acid, the boronic-acid bound FL2 is displaced resulting in a concomitant increase in the fluorescence emission of FL1 and a decrease in the fluorescence emission of FL2. The specific fluorescence response was dependent on the binding affinity of each analyte, enabling the determination of the ee of diols (strong binding guests) and hydroxy acids (weak binding guests). To achieve the chiral recognition of amines and amino alcohols, (S)-Vapol was used as FL2. In the resulting ensemble, the fluorescence emission of both FL1 and FL2 were quenched via a photoinduced electron transfer (PeT) process. Upon addition of either an amine or amino alcohol, the imine metathesis-based displacement of FL1 resulted in an increase in the fluorescence emission intensity. Interestingly, the addition of a chiral amine quenched the fluorescence of FL2, whereas, the addition of a chiral amino alcohol resulted in an increase in the fluorescence signature ascribed FL2. This difference allowed the species in question to be distinguished. This dual fluorophore ensemble was utilised in a high throughput assay to classify 13 pairs of enantiomers with additional qualitative and quantitative LDA-based determination of ee of various classes of chiral compounds with high accuracy being further demonstrated.
Despite a number of eIDAs being reported for chiral carboxylates, simple strategies for determining the ee of chiral aliphatic carboxylates are limited. In a continuation of their efforts, Anzenbacher and co-workers developed a simple eIDA that exploits ligand–copper (Cu) coordination chemistry to determine the enantiomeric composition of chiral carboxylates.13 Two chiral copper(II)-based receptors—[CuRRL]2+ and [CuSSL]2+ were developed and shown to bind and quench the fluorescence of the carboxylate-based indicator coumarin 343 (Scheme 9). The addition of chiral carboxylates resulted in the release of the fluorescent dye coumarin 343 with a concomitant increase in fluorescence emission. These sensors were able to discriminate the enantiomers of ibuprofen and naproxen, two commonly used anti-inflammatory drugs, as well as atorvastatin, a cholesterol reducing medication.
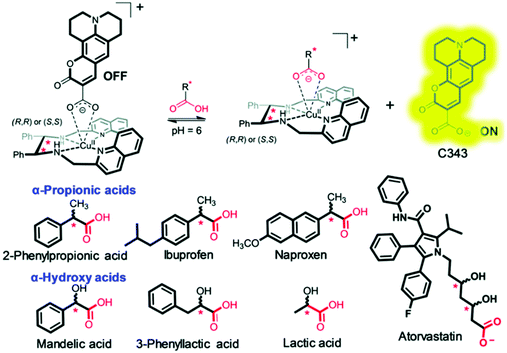 | ||
| Scheme 9 Chiral copper(II)-based receptors—[CuRRL]2+ and [CuSSL]2+ that allow the enantiomeric composition of chiral carboxylates, such as ibuprofen, to be determined. Reproduced from ref. 13 with permission from [The Royal Society of Chemistry], copyright [2019]. | ||
As seen by these examples, eIDAs offer an attractive high throughput method that can be used to determine the enantioselectivity of a given asymmetric transformation, as well as reaction yields. Unfortunately, the development of eIDAs for both enantiomers and diastereomers (diastereomeric excess, de) remains challenging. In fact, the current eIDAs are limited to determining the ee, not de, of a chiral species. To overcome this limitation, efforts have increasingly focused on a different strategy, one involving dynamic covalent self-assembly. This approach has allowed both ee and de values to be determined. A first example of this emerging paradigm was recently reported by Herrera and co-workers, in which the stereoisomers of 2-aminocyclohexanol, and their concentrations were differentiated.14 Thus, the use or IDAs to determine both ee and de is an open playing-field for research.
IDA-based sensors for carbohydrates and diol-based species
Carbohydrates
Carbohydrates act as the main source of energy for most living systems and are used as key building blocks for polysaccharides, DNA, RNA, glycolipids, glycoproteins, and ATP. In addition, measuring glucose concentration finds commercial relevance with diabetes mellitus patient care, where the inability to control the levels of blood glucose can lead to serious health complications. The development of strategies to detect blood glucose concentrations selectively at physiologically relevant levels, or visualise other biologically important carbohydrates are thus desirable. As noted in Scheme 5, boronic acids are excellent receptors for 1,2 and 1,3-diols. James, Kubo, Czarnik, and Shinkai have exploited this motif for the development of chemosensors for monosaccharide detection. Continued progress has yielded new systems able to discriminate between fructose and glucose monosaccharides.9 There are, however, few examples of IDA based platforms useful for the selective detection of a specific monosaccharides.15 This section will highlight the various IDA based strategies that have been reported to date. Unfortunately, due to the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 boronate complexes with monosaccharides, the IDA detection systems reported in this review are selective for fructose and not applicable for glucose sensing. To understand the various design modifications that are needed to achieve glucose selectivity, the reader is directed to a review by Sun et al.9 The hope is that this brief synopsis will inspire the development of new and effective monosaccharide (i.e. glucose) selective IDA based strategies.
1 boronate complexes with monosaccharides, the IDA detection systems reported in this review are selective for fructose and not applicable for glucose sensing. To understand the various design modifications that are needed to achieve glucose selectivity, the reader is directed to a review by Sun et al.9 The hope is that this brief synopsis will inspire the development of new and effective monosaccharide (i.e. glucose) selective IDA based strategies.
Fossey and co-workers reported a hydrogel-based IDA used for the colorimetric detection of monosaccharides.16 In this work, a series of meta-substituted phenylboronate methylacrylamides (12) were used to develop boronate-containing polyacrylamide hydrogels, known as borogels. Subsequent doping with Alizarin Red S (ARS) and washing steps, yielded orange coloured borogels. A visual colour change from red to orange was shown to be indicative of a boronate–ARS complexation, as shown in Scheme 10. The sensing properties were demonstrated by exposing the orange ARS–borogels to an aqueous solution containing specific monosaccharides. This led to the effective displacement of ARS from the borogel and result in an increase in the UV absorption of the surrounding solution at 513 nm, which corresponded to free ARS. This enabled the detection of monosaccharides.
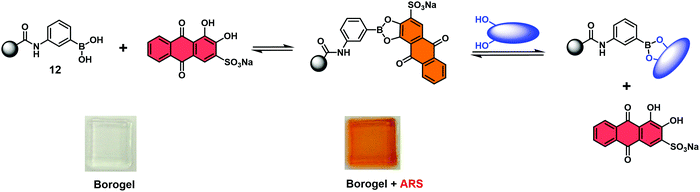 | ||
| Scheme 10 Binding and monosaccharide-induced release of ARS from hydrogel-bound boronic acid/ARS complexes (meta-substituted phenylboronate 12). Monosaccharide highlighted in Blue. Reproduced from ref. 16 with permission from [The Royal Society of Chemistry], copyright [2009]. | ||
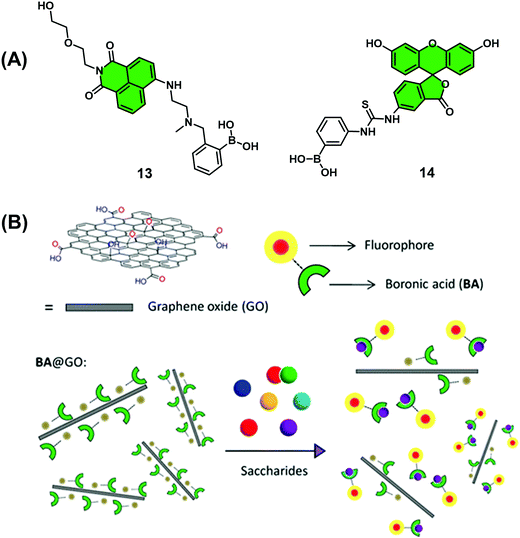
Graphene oxide (GO) has become a material of choice for the development of chemical sensors since it displays near-ideal electrical properties, good water solubility, and low toxicity in human cell lines. GO also displays the ability to complex most fluorescent dyes through aromatic interactions while acting as a universal quencher. In a continuation of their efforts to develop boronic acid-based monosaccharide sensors, James, He and co-workers prepared two mono-boronic acid-functionalised fluorescent probes, 13 and 14 (Scheme 11), which were used for development of a quencher displacement assay (QDA), wherein the host (quencher) is displaced rather than the indicator.17 What is required for the development of a QDA? As briefly shown in Scheme 3 and seen in this example, the indicators are designed to incorporate the receptor unit on its scaffold for analyte binding. Significant interaction between quencher (often material-based) and indicator is then required to induce a change to the optical properties of the system. Then, in the presence of the target analyte, the indicator binds to the analyte, which weakens the quencher–indicator interaction resulting in the displacement of the quencher resulting in a detectable change in optical signal.
Analyses showed these dyes readily complexed GO (quencher), which led to the quenching of their fluorescence emission intensity. Interestingly, studies with 14@GO showed no recovery in fluorescence intensity in the presence of excess fructose (1 M). This was presumed to be the result of strong aromatic interactions between 14 and GO. However, in the presence of fructose (0–2 M), 13@GO complex showed fluorescence recovery to enable the detection of fructose. The marked difference between 13@GO and 14@GO complexes was ascribed to the weaker non-covalent interactions of 13 with GO to allow dissociation and fluorescence increase upon exposure to fructose. In spite of showing an incomplete fluorescence recovery and encountering difficulties associated with accuracy, the simplicity of this QDA system represents an attractive sensing approach for the detection of biologically important saccharides. Further developments and optimisation will be needed, however, before widespread application.
Singaram and co-workers reported the quenching of the fluorescence emission of the anionic dye, 8-hydroxypyrene-1,3,6-trisulfonic acid (HPTS) in the presence of the cationic boronic acid-based viologen derivatives (o-BBV, m-BBV, p-BBV and BV – Scheme 12A).18 Electrostatic interactions between HPTS and viologen hosts facilitated the formation of each ensemble in aqueous solution and the associated fluorescence quenching of HPTS. The subsequent addition of glucose to each ensemble resulted in the formation of the corresponding glucoboronic esters and displacement of HPTS, resulting in an increase in fluorescence intensity. Gratifyingly, each BBV provided a fluorescence response within the clinically relevant glucose concentration range of 2.5–20 mM. This glucose-mediated displacement of HPTS was confirmed by non-boronic acid functionalised control viologen, BV, which was shown to be unaffected by the addition of glucose. The o-BBV derived construct demonstrated the greatest enhancement in fluorescence intensity, which was rationalised in terms of effective charge neutralization of the cationic nitrogen on viologen by the sp3 glucoboronate anion (Schemes 5 and 12B). In contrast to the QDA system highlighted in Scheme 11, the indicator is displaced by an analyte binding to an “allosteric site” on the host, which induces changes in the extent to which the indicator is bound to the receptor. This IDA-based system is an example of an allosteric indicator displacement assay (AIDA).19 To summarise, an AIDA requires the host to interact with both analyte and indicator using different interactions and at separate locations. Finally, analyte binding must influence the host–indicator interaction in order to facilitate displacement of the indicator and provide an optical response.
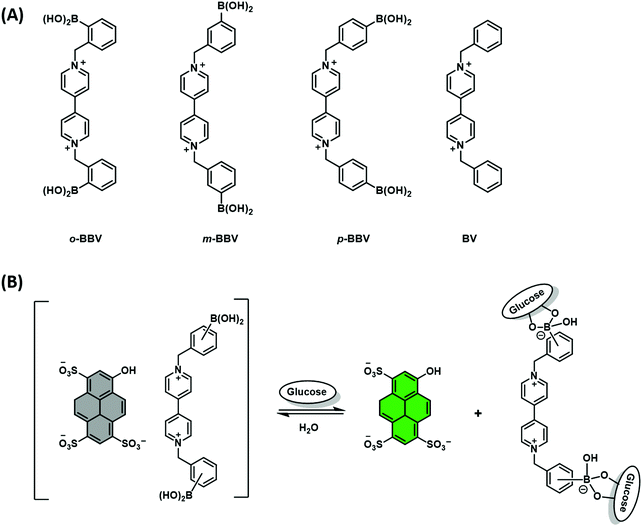 | ||
| Scheme 12 (A) Chemical structure of viologen-based hosts. (B) Proposed AIDA mechanism for the detection of glucose through charge neutralisation. | ||
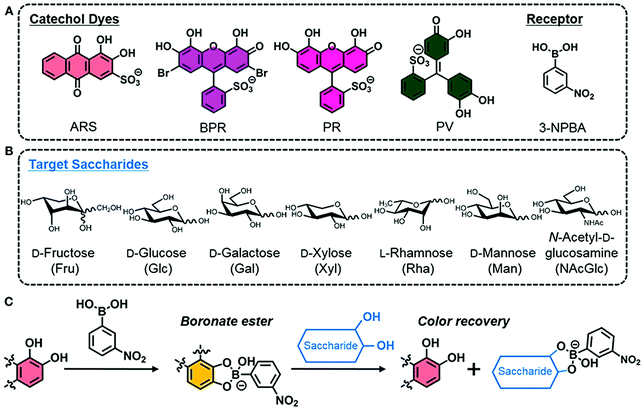
More recently, Minami and co-workers reported a simple colorimetric IDA sensing array for the detection and discrimination of monosaccharides. Notably, the authors developed a system that utilised all commercially available reagents; 3-nitrophenylboronic acid (3-NPBA) as the diol receptor and four catechol-based indicators (ARS, bromopyrogallol red (BPR), pyrogallol red (PR), and PV) (Scheme 13).20 The reaction between 3-NPBA with each catechol dye resulted in the formation of a boronate ester complex with an attendant colour change unique to each species. As expected, the addition of a monosaccharide led to dye displacement and optical colour changes that could be readily followed by UV-Vis spectroscopy. Each catechol dye–boronate complex resulted in a colorimetric fingerprint-like response when evaluated against a given monosaccharide. By using linear discrimination analysis (LDA), each monosaccharide could be successfully identified with a classification success rate of 100%. This example highlights the simplicity and cost-effective nature of IDAs that are constructed without the need for synthetic investment.
Pd(II) complexes have been explored as a possible alternative to boronic acid–monosaccharide sensors. Initial studies utilised (en)Pd(OH)2 (en = 1,2-ethylenediamine) and related complexes to bind monosaccharides, including reducing sugars, in aqueous media.21 Unfortunately, the concentrations of monosaccharide required for binding were >100 mM. Severin and co-workers expanded upon this discovery and developed a Pd(II)-based IDA for the selective recognition of monosaccharides with higher sensitivity in aqueous solution.21 Here, a series of Pd-based receptors were synthesised or purchased from commercial sources and screened with an N-methyl anthranilic acid (MAA) indicator (Scheme 14). The formation of each Pd(II)–MAA complex resulted in fluorescence quenching of MAA. Ligand exchange between MAA and glucose, fructose, galactose and α-methylglucopyranoside resulted in a fluorescence turn-on response in an aqueous medium consisting of MOPS buffer (100 mM) at pH 7.4. Screening studies involving various complexes revealed that the di(pyrazol-1-yl)methane complex provided the greatest response. The high affinity for glucose and other sugars was rationalised in terms of the so-called trans effect of the co-ligand. Unfortunately, this IDA strategy displayed a poor selectivity between monosaccharides. While further studies will be required, it is likely that the sensitivity, selectivity, and the fluorescence properties of this platform can be tuned via modulation of the dative ligand steric and electronic properties.
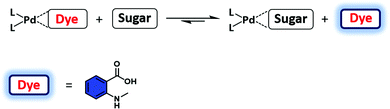 | ||
| Scheme 14 Pd(II)-Based FIDA for the fluorescence detection of monosaccharides using a Pd complex as the host and N-methyl anthranilic acid (MAA) as the indicator. Reproduced from ref. 21 with permission from [The Royal Society of Chemistry], copyright [2011]. | ||
Diol-based species
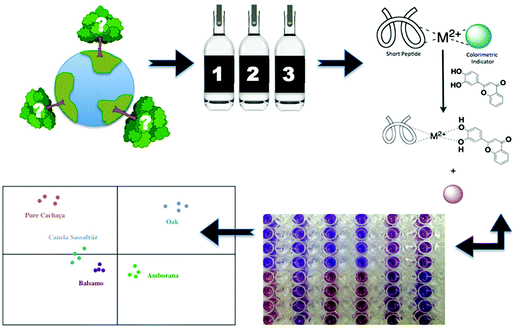
In conjunction with their work on eIDAs, Anslyn and co-workers have pioneered the development of various functional platforms that rely on the use catechol-based species. This includes the development of an IDA sensing array for the high-throughput detection of the naturally occurring polyphenols known as tannins. A first example involved a multi-component, peptide-based sensing ensemble composed of short histidine-containing peptides, a divalent metal ion, and a pH colorimetric indicator. In the presence of tannins, the indicator is displaced resulting in a colour change that could be measured spectrophotometrically (Scheme 15). This peptide–metal–indicator ensemble was used to fingerprint the types of wood used to age the Brazilian beverage Cachaça.22 The composition of tannins in each drink is dictated by the type of wood used to age the beverage. By monitoring wood signatures via tannin composition, it became possible to identify the type of wood used. Remarkably, this peptide-based IDA strategy in combination with LDA, produced a discrimination pattern that was reproducible for oaks obtained from various locations. These researchers were able to classify unknown wood extracts based on the clustering pattern with 60–100% success rate. This strategy might provide a means of identifying and preventing adulteration, as well as the illegal logging and use of endangered tree species. This example further highlights the advantages of IDAs over the use of single molecular sensors.IDA-based sensors for phosphate-based species
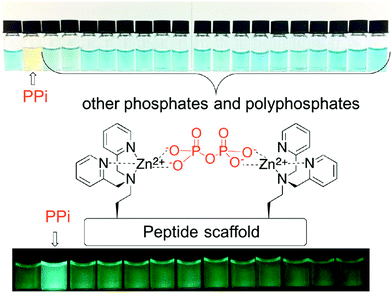
Phosphate and phosphate-functionalised species are essential components of all living cells. Phosphate oxoanions and phosphorylated biomolecules play critical roles in a wide range of biological processes, including genetic information storage, energy transduction, signal processing and membrane transport. There is thus considerable effort being devoted to the development of systems that might allow various phosphate species to be detected and visualised selectively. Unfortunately, the ability to discriminate between phosphate oxo-anion species, i.e., PPi vs. ATP, remains challenging. However, achieving such discrimination is deemed essential if their individual roles and their relationship between one another in various (patho)physiological processes is to be fully elucidated.In this realm, Jolliffe and her team have made substantial contributions with the development of IDAs for the selective detection of PPi using scaffolds containing bis-zinc(II) dipicolylamine (DPA) cores (Fig. 3).23,24
In particular, Jolliffe and co-workers reported the synthesis of a library of cyclic peptide hosts (15–22·Zn2) that contain the bis-Zn(II) dipicolylamine (DPA) motif (Fig. 4).24 Here, the formation of the cyclic peptide–PV ensembles afforded IDA systems that enabled the selective detection of PPi. This remarkable selectivity was attributed to the steric bulk provided by the side chains on the cyclic peptide scaffolds. This observation was confirmed using a simple bis(ZnII-DPA) complex 23·Zn2. Similar to other IDA-based strategies, the selectivity of this IDA strategy could be tuned by changing the indicator with each cyclic peptide scaffold. Overall, these cyclic peptide chemosensor ensembles demonstrated remarkable discrimination between PPi and other polyphosphate ions, with the ability to detect PPi in aqueous solutions containing a greater than 100-fold excess of ATP.
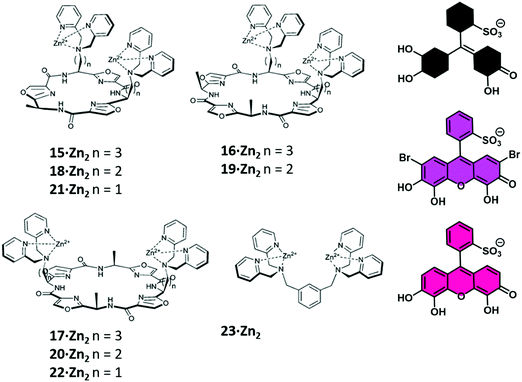 | ||
| Fig. 4 A library of cyclic peptide hosts (15–22·Zn2) functionalised with bis-Zn(II) dipicolylamine (DPA) units that allowed for the naked-eye sensing of pyrophosphate (PPi) in aqueous media. Reproduced from ref. 24 with permission from [The Royal Society of Chemistry], copyright [2013]. | ||
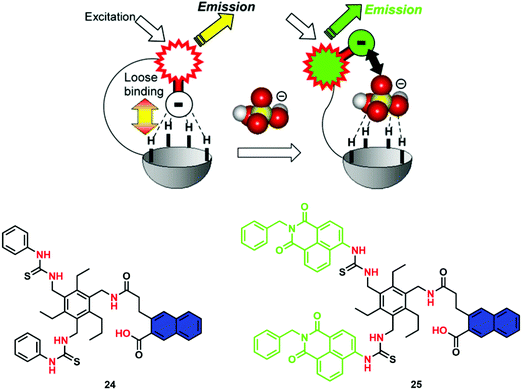
A problematic issue often encountered when developing an IDA is the low signal-to-noise ratio and sensitivity, due to the use of excessive amounts of dye used to form the host–guest ensemble in situ. Anzenbacher and co-workers have recently developed an “intramolecular indicator displacement assay (IIDA)” strategy to overcome such limitations. What is required for the design of an IIDA? IIDAs require the covalent attachment of an indicator to the framework of the synthetic host via a flexible linker. By exploiting the developed knowledge of IDAs and host/guest chemistry, the chosen indicator and receptor must interact to form an intramolecular indicator/host complex. Finally, the target analyte must exhibit a greater affinity for the receptor to allow the intramolecular displacement of the indicator. This particular system by Anzenbacher and co-workers proved efficient for detecting biologically important phosphate-based anions, such as PPi and the phosphate-containing herbicide glyphosate, in aqueous media.25 To demonstrate this concept, two pinwheel-based sensors comprised of amide and thiourea recognition motifs and carboxylate functionalized fluorophores (24 and 25) were developed. Per the design expectations, the carboxylate moieties of the fluorophores were initially bound to the receptor via hydrogen bonding. However, upon the addition of a competing anion with a higher binding affinity, the fluorophore was displaced (Scheme 16). The fact that a receptor–indicator 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio was necessarily enforced endowed the assay with high sensitivity and instant reversibility. Even in the presence of the interferent chloride anion, these IIDA sensors were able to detect phosphate-based anions including glyphosate in aqueous media. Semi-quantitative and quantitative assays for glyphosate showed a limit of detection of 0.2 ppm for glyphosate, which is significantly lower than the maximum contaminant level permitted for drinking water (0.7 ppm). The generality and tunability of this IIDA approach makes it attractive for the further development of new and effective IIDA sensors, including ones that might be amenable to analytes not currently detected by traditional molecular sensors.
1 ratio was necessarily enforced endowed the assay with high sensitivity and instant reversibility. Even in the presence of the interferent chloride anion, these IIDA sensors were able to detect phosphate-based anions including glyphosate in aqueous media. Semi-quantitative and quantitative assays for glyphosate showed a limit of detection of 0.2 ppm for glyphosate, which is significantly lower than the maximum contaminant level permitted for drinking water (0.7 ppm). The generality and tunability of this IIDA approach makes it attractive for the further development of new and effective IIDA sensors, including ones that might be amenable to analytes not currently detected by traditional molecular sensors.
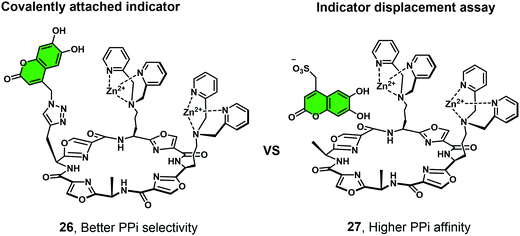
Joliffe and co-workers have explored the IIDA strategy as a means of detecting PPi using the cyclic-based peptide-Zn(II)-DPA receptors 26 and 27.26 In this study, a coumarin-based IIDA platform (26) was evaluated against an analogous IDA system using a comparable fluorescent indicator (Fig. 5). The IIDA analogue displayed a significantly enhanced selectivity for PPi over competing phosphate species; however, a decrease in sensitivity was also seen. The potential application of these fluorescent IIDAs systems were demonstrated through the real time monitoring of pyrophosphatase enzyme activity. This study lends credence to the notion that further IIDAs could be developed that allow for the continuous monitoring of enzymatic activity.
Thus far we have discussed IDA-based strategies that have either used colorimetric and/or fluorometric indicators and which rely on the use of UV-vis and fluorescence spectroscopy. 19F NMR spectroscopy is emerging as an attractive imaging technique due to its high signal sensitivity, wide chemical shift range, total depth penetration and minimal biological/environmental background signal. Smith and co-workers have recently developed an Zn(II)-bis(dipicolylamine) IDA receptor using 19F NMR for PPi detection.27 In this example, the bis-Zn(II)-bis(dipicolylamine) receptor was covalently attached to paramagnetic relaxation agents, a nitroxide motif (28) and Gd(III)–DOTA (29). The addition of the 19F-labeled phosphate indicator resulted in the expected broadening and reduction in height of the corresponding 19F NMR signal. This finding was rationalised in terms of a paramagnetic relaxation effect (PRE). Subsequent addition of PPi to the supramolecular ensemble displaced the 19F-labeled indicator resulting in a concomitant increase in 19F NMR signal, a result fully consistent with the high affinity of PPi for the bis-Zn(II)-bis(dipicolylamine) receptor and its ability to displace the initial 19F-labeled phosphate indicator (Scheme 17).
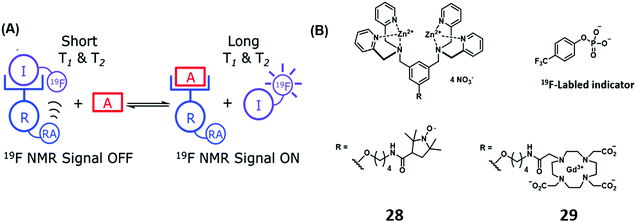 | ||
| Scheme 17 (a) Basic schematic of a 19F NMR-based IDA assay for the detection of PPi. (b) Zn(II)-bis(dipicolylamine) receptor appended with a relaxation agent (RA), a nitroxide motif (28) and Gd(III)–DOTA (29). Reproduced from ref. 27 with permission from [The Royal Society of Chemistry], copyright [2014]. | ||
IDA-based sensors for nucleic acids
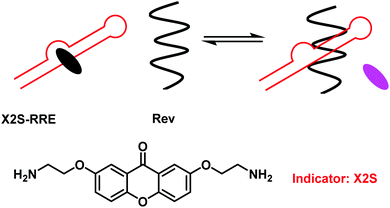
Nucleotides are fundamental building blocks of DNA, RNA, and other bio-relevant macromolecules critical for life.28 For drug discovery purposes it is important to understand the intermolecular interactions between nucleotides and nucleotide-containing species (DNA, RNA, non-canonical triplex, quadruplex and I-motifs) and artificial ligands. The use of IDAs for the evaluation of small molecule binding in combination with principle component analysis (PCA) provides a platform for high-throughput analyses of small molecule therapeutic libraries with target oligonucleotides.29 Traditional IDA strategies for nucleotides/nucleotide containing species have exploited environmentally sensitive intercalating dyes, such as thiazole orange and ethidium bromide.29 These molecules are highly fluorescent when intercalated, but non-fluorescent in free solution. This fluorescence “on–off” response provided a strategy to evaluate the binding properties of potential small molecule nucleotide-based therapeutics.28,30To enhance the sensitivity of these FIDA strategies, Nakatani and co-workers developed a fluorescence “off–on” strategy and used it to determine ligand–RNA interactions.31 In this work, a 2,7-disubstituted 9H-xanthen-9-one (X2S) indicator was developed, which displayed minimal fluorescence when bound to RNA; however, it became fluorescent when it was displaced (Scheme 18). Using X2S, an FIDA was established with an example RNA, the rev responsible element (RRE) of HIV-1 mRNA. This resulted in the formation of a non-fluorescent X2S–RRE complex. The formation of this FIDA is of significance as the binding of the viral protein Rev to the stem loop of RRE is essential to viral replication. Therefore, the identification of ligands that can compete with Rev in binding to RRE have the potential to suppress HIV-1 replication. This FIDA was shown capable of screening new ligands from a chemical library, thus allowing potential ligand binding to RRE to be evaluated.
Unfortunately, most RNA-binding fluorescent dyes were found to have low affinities for RNA. Therefore, to perform the appropriate high-throughput assays, large concentrations of RNA (>1 μM) are required. The development of alternative sensing systems with higher affinities towards RNA are thus desirable. One such system was reported by Hamasaki and co-workers.32 This improved FIDA system exploited the high affinity of the transactivator protein (Tat) for the Tat responsive region of the mRNA, TAR RNA, (a key factor in the replication of human immunodeficiency virus type-1 (HIV-1)).31 The fluorescent reporter was a 16-residue peptide containing a complementary Tat49–57 sequence with fluorescein and tetramethylrhodamine (TAMRA) tags at the N- and C-termini, respectively (Fig. 6). It was found that double dye labelled (FTatRhd) peptide displayed strong fluorescence via a FRET mechanism when bound. In the presence of a competing ligand (small molecule), FTatRhd was displaced from TAR RNA with attendant fluorescence quenching. The interactions of small molecules and TAR RNA could be determined as a function of fluorescence quenching.
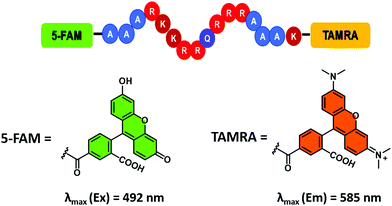 | ||
| Fig. 6 Fluorescein/tetramethylrhodamine (TAMRA) peptide indicator used to develop an FIDA for determining ligand–RNA interactions. Reproduced from ref. 33 with permission from [The Royal Society of Chemistry], copyright [2019]. | ||
More recently, Hargrove and co-workers demonstrated the use of FTatRhd to screen simultaneously small molecular ligands against multiple RNA targets of interest while assessing the associated binding affinities and selectivities using the same assay.33 In this work, similar sized RNA targets (HIV-1-TAR, HIV-2-TAR, A-site, RRE-IIB) possessing different secondary structure were shown to bind to FTatRhd with low nanomolar affinities. This improved system required only low concentrations of RNA (nM) to be effective. Moreover, it proved successful in identifying hits against all four RNA structures while being able to discriminate between different small molecules.
Although IDAs have shown excellent selectivity and sensitivity towards a wide range of analytes, their application in bioimaging remain underexplored. Current difficulties are associated with non-specific binding from biological species present at high concentrations in biological milieus. Zhu and co-workers have recently reported a metal-based supramolecular assembly consisting of the aggregation induced emitter (AIEgen) tetraphenylethylene (TPE) linked to two Zn(II)–cyclen complexes (Scheme 19).34 Formation of the PV-Zn(II)–cyclen (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) complex resulted in fluorescence quenching of the TPE unit. In the presence of thymine-rich single-stranded DNA, the resultant PV-Zn(II)–cyclen complex disassociated and fluorescence was restored. This approach allowed discrimination between T-rich ssDNA and dsDNA and was applied to several human cell lines.
2) complex resulted in fluorescence quenching of the TPE unit. In the presence of thymine-rich single-stranded DNA, the resultant PV-Zn(II)–cyclen complex disassociated and fluorescence was restored. This approach allowed discrimination between T-rich ssDNA and dsDNA and was applied to several human cell lines.
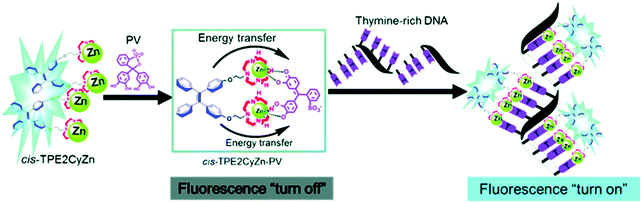 | ||
| Scheme 19 A metal-based supramolecular assembly that relies on the use of tetraphenylethene (TPE)-based Zn2+–cyclen complexes (cis-TPE2CyZn) coordinated to the indicator PV. Thymine-rich single-stranded DNA induces displacement of PV to recover the fluorescence corresponding to TPE. Reproduced from ref. 34 with permission from [The Royal Society of Chemistry], copyright [2018]. | ||
IDA-based sensors for amino acids, peptides and proteins
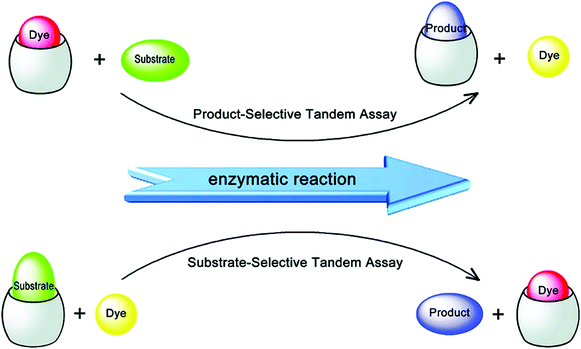
Amino acids and peptides are important in cell growth and development. They also play essential roles in metabolism, signal transmission, immunoregulation, and reproduction. Larger biomolecules, such as proteins, also mediate cell signalling and catalyse transformations required for life, among other processes. Post-translational modification of these species can further determine their precise role in physiological processes. IDA platforms offer imaging modalities for the dynamic monitoring of amino acids, peptides, and proteins. Initial IDA-based strategies targeted the monitoring of enzymatic reactions and were considered as “product-selective supramolecular tandem assays”.34 These systems employed an enzyme-mediated transformation of the substrate to yield a product with higher affinity for the indicator–host complex. Displacement of the indicator provided a direct method to monitor the reaction.Nau and co-workers have recently reported an alternative IDA-based system, described as a “substrate-selective supramolecular tandem assay”.35 This alternative IDA reverses the substrate and product binding affinity, such that the substrate is initially bound to the receptor with a higher affinity than the product. Enzymatic consumption of the substrate enables displacement of the product by a fluorescent dye. This allows for the continuous monitoring of the enzymatic reaction in question. This strategy was designed to provide a general approach to screening various enzyme-based reactions by exploiting commercially available dyes or focusing on the large diversity of host–dye complexes already reported (Scheme 20). In the initial Nau study, two model enzymatic reactions were used. In the first example, p-sulfonatocalix[4]arene (CX4) was used as the macrocyclic host with 1-aminomethyl-2,3-diazabicyclo[2.2.2]oct-2-ene, DBO, being employed as the indicator (guest) (see Fig. 1 for structures). This allowed the arginase-catalysed transformation of arginine to ornithine to be monitored (Scheme 20). In this example, the dye displayed a weaker fluorescence signal in its complexed state. Over the course of the arginase-catalysed reaction, the gradual complexation of DBO within CX4 led to a decrease in the fluorescence signal for DBO. The second example utilised the diamine oxidase-catalyzed oxidation of cadaverine to 5-aminopentanal. Cucurbit[7]uril (CB[7]) was used as the host in conjunction with an acridine orange (AO) indicator. In its complexed state (hydrophobic environment), AO demonstrated a strong fluorescence signal. This allowed for a “turn-on” response when monitoring the enzyme-induced transformation of cadaverine. These systems were demonstrated as effective tools for the evaluation of inhibitors of each enzymatic transformation. Through the coupling of a “product-selective” with a “substrate-selective” assay, it was possible to monitor a multistep biochemical pathway, namely the decarboxylation of lysine to cadaverine by lysine decarboxylase followed by the oxidation of cadaverine by diamine oxidase. This “domino tandem assay” was performed in the same solution with a single reporter pair (CB[7]/AO) (Scheme 20).
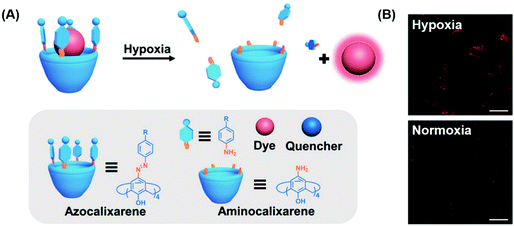
Aberrant cellular function is often associated with changes to the cellular environment. Indeed, most solid tumours are characterised by hypoxic conditions, i.e., a decrease in local O2 levels due to increased O2 consumption. Traditional hypoxia-based sensing strategies utilise covalent approaches and thus typically require extensive synthetic efforts. Guo and co-workers recently developed a simple non-covalent FIDA strategy that exploits host–guest interactions between an easily prepared azocalix[4]arene host and the commercially available dye, rhodamine 123.36 Rhodamine 123 (Rho123) was shown to bind within the azocalix[4]arene (CAC4A) host with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry with attendant fluorescence quenching. In hypoxic environments, bioreductive enzymes capable of reducing a variety of organic functionalities are over-expressed. Upon entering such hypoxic environments, the azo groups in CAC4A–Rho123 are reduced to the corresponding aniline species. This leads to the release of Rho123 and a fluorescence turn-on response (Scheme 21). The construct CAC4A–Rho123 was shown to be capable of differentiating between normoxic and hypoxic conditions in A549 cells.
1 binding stoichiometry with attendant fluorescence quenching. In hypoxic environments, bioreductive enzymes capable of reducing a variety of organic functionalities are over-expressed. Upon entering such hypoxic environments, the azo groups in CAC4A–Rho123 are reduced to the corresponding aniline species. This leads to the release of Rho123 and a fluorescence turn-on response (Scheme 21). The construct CAC4A–Rho123 was shown to be capable of differentiating between normoxic and hypoxic conditions in A549 cells.
The “histone code” is a post-translational enzymatic modification of terminal units on the amino acid side chains within the DNA-packaging proteins called histones. Post-translational modifications occur mainly on the N-terminal tails of the histone proteins. An example of a “histone code” post-translational modification is methylation of the N-terminal lysine and arginine residues. The ability to monitor protein post-translational modification, including histone codes, is essential to understanding the regulation mechanisms within cellular processes. New insights into these processes can also aid the development of effective disease treatments. Current technologies used to detect these “histone code”-based analytes employ antibody-based assays; however, a number of limitations exist within these sensing platforms.37 These include high cost, limited accessibility, high batch to batch variability, and poor selectivity between similar analytes. The problems associated with antibody-based assays are often exacerbated by the presence of modified peptides that make up the histone code.
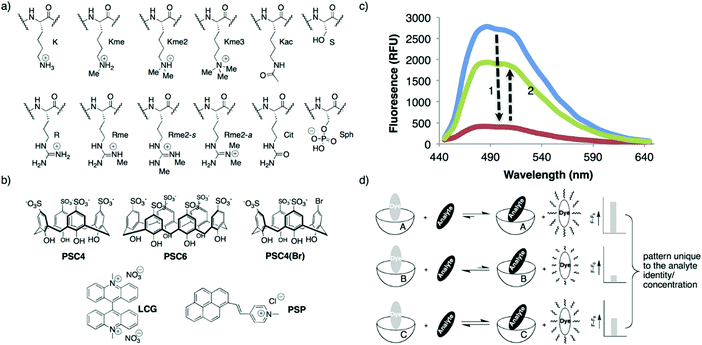
In principle, an IDA-based sensing array would provide a simple, low cost strategy to identify and discriminate individual analytes thus overcoming some of the above limitations. With this vision presumably in mind, Hof and co-workers developed a mix-and-match IDA toolkit designed to respond to a wide variety of histone code analytes, including methylated lysine (Scheme 22).37 The IDA system was comprised of a fluorescent Lucigenin (LCG) and a fluorescent pyrene-modified N-alkylpyridinium (PSP) dye (guests) and calixarene, PSC4, PSC6, and PSC4(Br), (host) molecules. In the presence of a cationic peptide species, the fluorescent dye guests were released leading to a fluorescence turn-on response. These ensembles, when used as a sensing array, produced a fingerprint response that, in combination with LDA, enabled discrimination between unmethylated, mono-, di-, and trimethylated lysine on a single histone tail sequence. Further exploration of this sensing array led to the simultaneous reporting of both the concentration and identity of a range of histone modifications.
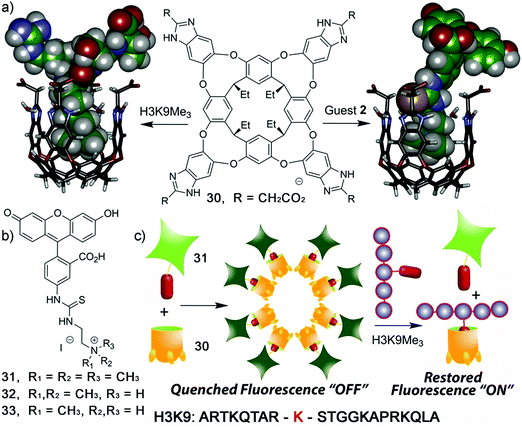
Zhong and Hooley have recently expanded upon this concept with the development of a dual-functional aggregate host–guest displacement assay. This system was able to detect tri-methylated peptides and monitor histone demethylase activity.38 In order to operate in an aqueous environment, a self-folding water-soluble deep cavitand host (30) and the water-soluble fluorescein-based fluorescent indicators 31–33 that differed by degree of methylation were used. The trimethylated fluorescein guest showed the greatest binding affinity for 30 and underwent aggregation induced fluorescence quenching to yield a “turn-on” FIDA. This system was able to selectively detect the trimethylated quaternary histone H3 peptide (H3K9Me3) versus di-/mono-/non-methylated H3K9 (Scheme 23).
Phosphatidylserine (PS) is an important anionic phospholipid found on the inner face of eukaryotic cell membranes. Cell surface exposure of PS is a common marker of cell death and signal of apoptosis. Jolliffe and co-workers have reported an extension of their IIDA platform (Fig. 5) to visualise phosphatidylserine (PS) in live cells.39 This IIDA system, P-IID, consists of a peptide backbone functionalised with two Zn(II) dipicolylamine (ZnDPA) complexes and a 6,7-dihydroxycoumarin fluorophore (Scheme 24). In its resting state, P-IID was found to be non-fluorescent as 6,7-dihydroxycoumarin freely coordinates to one of the ZnDPA moieties resulting in fluorescence quenching. However, in the presence of PS, intramolecular displacement of 6,7-dihydroxycoumarin from a ZnDPA moiety led to a remarkable increase in the fluorescence intensity. This enabled the visualization of cell surface phosphatidylserine on apoptotic cells using both confocal microscopy and flow cytometry. P-IID was also successfully used for time-lapse imaging of cells undergoing apoptosis.
Aqueous solutions, which living organisms are associated with, contain salts, organic molecules, and biological components that can affect binding equilibria and dye emission in ways that diminish the signal intensity or render an IDA inoperative. Hof and co-workers recently reported a new IDA approach designed to overcome such putative limitations. This system was referred to as a DimerDye Disassembly assay (DDA).40 The development of the DDA strategy requires the synthetic integration of an indicator with an identified synthetic receptor that is known to undergo self-assembly in aqueous media. Therefore, the single molecule indicator/receptor hybrid host self-assembled (driven by hydrophobic forces) to form a non-fluorescent dimer through aggregation caused quenching (ACQ). Subsequent analyte binding then triggers disassembly of the dimer to produce host–analyte complexes resulting in changes of the photophysical properties enabling analyte detection.
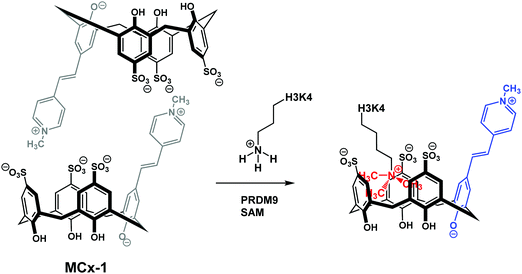
More specifically, two sulfonato-calixarenes containing a solvatochromic Brooker's merocyanine dye were synthesised MCx-1 and MCx-2. Self-assembly in aqueous media to produce the non-emissive dimeric ensembles was then observed. This approach can be seen as similar to IIDAs; however, this extension differs in that the dye is included into the host pocket in an assembled, intermolecular complex. The addition of cationic trimethyllysine (KMe3) to dimeric MCx-1 resulted in the dissociation of the “DimerDye” and gave rise to a highly emissive host–guest complex (Scheme 25). Furthermore, MCx-1 was found to allow for the real time analysis of methyltransferase activity, which is a rarity. Unlike some IDA systems, the DimerDyes remained functional in solutions containing various potential biological interferants, including ones deemed essential for various enzymatic reactions.
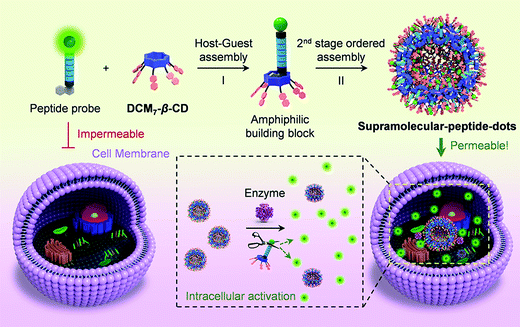
The high specificity of biological peptides towards their corresponding receptors has led to their exploration and use in a range of diagnostic and therapeutic applications. He and co-workers reported a hepta-dicyanomethylene-4H-pyran appended β-cyclodextrin (DCM7-β-CD) as a delivery enhancing “host” for fluorescein and 1-bromonaphthalene-modified functional peptides (P1–P4) (Scheme 26).41 Each fluorescent peptide probe interacted with DCM7-β-CD through the 1-bromonapthalene unit to afford the corresponding host–guest complex. Fluorescence quenching within this host–guest complex was attributed to FRET quenching (fluorescein – donor and DCM7-β-CD – acceptor) and formation of so-called supramolecular-peptide-dots (Spds). Peptides P1 (Spd-1) and P2 (Spd-2) contained the peptide 3Pal-D-βhLeu-F-D and DEVD sequences, respectively. These are substrates for caspase (cysteinyl aspartate specific proteinase), a well-established biomarker for cell apoptosis. In the presence of caspase-3, each peptide in Spd-1 and Spd-2 was cleaved, which led to the release of the fluorescein fluorophore. This inhibited the FRET process and restored the fluorescence emission of fluorescein. Host–guest complexes Spd-1 and Spd-2 were shown to enhance the intracellular delivery of P1 and P2 and visualise cellular apoptosis in HepG2 cells. Peptide P3 (Spd 3) contains the known tubulin binding motif VARVGSPPD. Tubulin is a structural protein essential for cell mitosis. Spd-3 was found to bind tubulin successfully and displaced the fluorescein peptide P3, from the DCM7-β-CD host. Spd-3 allowed cellular mitosis in HeLa cells to be tracked by fluorescence microscopy. As a final demonstration of this host–guest chemistry, a 1-bromonaphthalene-modified antimicrobial peptide (P4, Spd-4) was studied; it displayed enhanced efficacy against both Gram-positive and Gram-negative bacteria as compared to P4 alone. This work served to demonstrate the utility of FIDAs in biological applications, and their potential to serve as new so-called theranostics that permit both diagnostic and therapeutic applications.
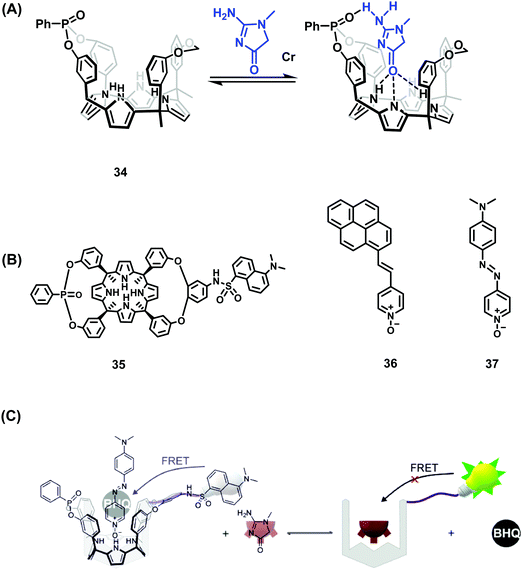
Over the years, Ballester and co-workers have extensively explored the use of calixpyrrole derivatives as synthetic receptors and demonstrated inter alia their affinity for pyridyl N-oxide guests. Recently, this research team reported two calix[4]pyrrole phosphonate-cavitands 34 and 35 that act as receptors for creatinine and its lipophilic derivative hexylcreatinine (Scheme 27).42 Elevated levels of creatinine found in blood serum and urine have been linked to impaired kidney function and kidney disease. As a result, assays for the detection of creatinine are highly desired. In this work, two separate IDA based approaches were employed that exploited the known ability of pyridyl-N-oxides to form host–guest complexes through N–H hydrogen bonding with calix[4]pyrroles. The IDA-based approach employed a fluorescent pyrene pyridyl N-oxide indicator 36 that, upon forming a host–guest complex, was quenched in the bound state. In the presence of creatinine or hexylcreatinine, the indicator was displaced resulting in restoration of the fluorescence emission intensity. A second-generation FRET-based IDA system, involving receptor 35 featuring a covalently attached dansyl fluorophore, was also reported by Ballester and coworkers. The addition of azo-based pyridyl N-oxide 37 resulted in fluorescence quenching of the dansyl unit through FRET. Upon addition of hexylcreatinine, fluorescence emission was restored through the displacement of 37 and inhibition of the FRET process. A putative mechanism was proposed that has as its key principle the favoured binding of creatinine within the phosphonate-functionalised calix[4]pyrroles 34 and 35via complementary hydrogen bonding interactions. Both calix[4]pyrrole systems were found to bind creatine with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry, as inferred from 1H and 31P NMR spectroscopy, isothermal titration calorimetry (ITC), and UV-Vis absorption and fluorescence spectroscopy. The FRET-based IDA approach based on 34 and 35 was characterised by a high sensitivity with a limit of detection (LOD) of ∼110 nM being noted for one of these systems. These levels are in line with what are seen in commercial diagnostic systems used to differentiate healthy and sick patients. Unfortunately, these FRET-based IDAs required halogenated solvents. Nevertheless, this represents an unprecedented example of a fluorescent supramolecular sensor for the detection of creatinine and is expected to inspire work on next-generation systems capable of clinical applications.
1 binding stoichiometry, as inferred from 1H and 31P NMR spectroscopy, isothermal titration calorimetry (ITC), and UV-Vis absorption and fluorescence spectroscopy. The FRET-based IDA approach based on 34 and 35 was characterised by a high sensitivity with a limit of detection (LOD) of ∼110 nM being noted for one of these systems. These levels are in line with what are seen in commercial diagnostic systems used to differentiate healthy and sick patients. Unfortunately, these FRET-based IDAs required halogenated solvents. Nevertheless, this represents an unprecedented example of a fluorescent supramolecular sensor for the detection of creatinine and is expected to inspire work on next-generation systems capable of clinical applications.
Expanding on the use of GO for the development of QDAs, He and co-workers developed supramolecular sensor arrays.43 This sensing platform consisted of GO and the fluorescent peptides T-RS5, T-QY7, and T-ED17 containing the peptide fragments, RS5, QY7 and ED17, respectively, each of which was derived from a patented anti-glycoprotein antibody (Scheme 28). Glycoproteins (GPs) exist on the capsid of a number of viral species, therefore, each peptide had a high affinity for viral species. The simple mixing of GO and with each fluorescent peptide led to the formation composites T-RS5/GO, T-QY7/GO and T-ED17/GO with concomitant fluorescence quenching. In presence of three pseudoviruses, Marburg virus (MARV), and the Ebola virus (EBOV), and the vesicular stomatitis virus (VSV), a concentration-dependent fluorescence increase was observed. This increase was attributed to the removal of each peptide from the GO surface and formation of peptide/virus complexes. Owing to the sensitivity differences between each GO-based peptide probe, a sensing array was constructed and in combination with PCA enabled the differential sensing between virus species. This QDA platform represents an important sensing strategy that should be subject to ready generalisation. That makes it particularly relevant during the current COVID-19 outbreak.
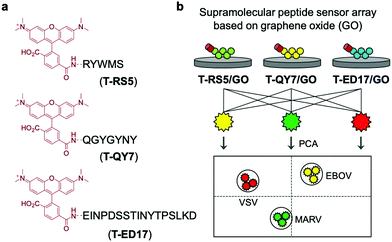 | ||
| Scheme 28 Graphene oxide (GO) and rhodamine-functionalized peptides used for the fluorescence-based detection of pseudoviruses, Marburg virus (MARV), EBOV, and the vesicular stomatitis virus (VSV). Reproduced from ref. 43 with permission from [The Royal Society of Chemistry], copyright [2020]. | ||
IDA-based sensors for reactive oxygen (ROS) and nitrogen species (RNS)
Reactive oxygen (ROS) and reactive nitrogen species (RNS) play important roles as signalling molecules that regulate a wide variety of biological processes. Aberrant production of these species has been associated with inflammation, Alzheimer's diseases, and cancer, among other diseases. Imaging agents capable of sensing of RNS/ROS in vitro and in vivo are thus of great importance and may allow their precise role in these diseases to be better understood. Current ROS and RNS sensing approaches rely on reaction-based fluorescent probes with limited development of IDA technologies being seen to date.6Inspired by the area of reaction-based molecular sensors, James and co-workers developed an IDA-based strategy that utilised the ROS/RNS-mediated transformation of boronic acids to phenols to release a bound indicator. This method was referred to as a RIA (reaction-based indicator displacement assay).44 Like IDAs, RIAs involve the initial formation of the host–indicator system. However, in the case of an RIA, the addition of an analyte requires a chemical transformation to either the host or indicator to trigger the disassembly of the system and provide a measurable change to the optical signal.
The first reported example utilised the well-known self-assembly of 2-(N,N-dimethylaminomethyl)phenylboronic acid (NBA) with ARS to afford a highly fluorescent ARS–NBA complex (Scheme 29). In the presence of peroxynitrite (ONOO−), a highly reactive RNS, a large change in UV absorption and fluorescence spectroscopic features was observed. This system produced a minimal colorimetric or fluorescence response towards hydrogen peroxide (H2O2) and other ROS/RNS. This system required an excess of ARS and, as result, lacked sufficient sensitivity towards ONOO – for use in biological evaluation protocols. Nevertheless, this system represented a new strategy that may in due course allow for the development of IDAs suitable for the detection of biologically important species under in vitro or in vivo conditions.
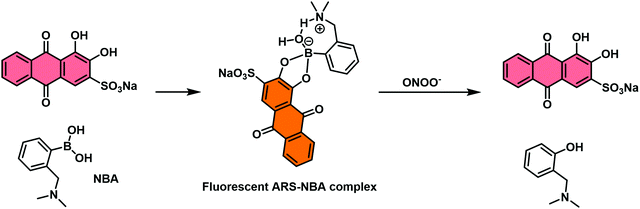 | ||
| Scheme 29 Schematic illustration of the proposed “reaction-based indicator displacement assay” (RIA) and sensing mechanism of the ARS–NBA complex when used as a probe for peroxynitrite (ONOO−). | ||
James and co-workers reported a second RIA system that proved suitable for H2O2 detection. This system relied on the use of a cetyl trimethylammonium bromide (CTAB) surfactant to facilitate boronate ester formation between phenylboronic acid (PBA) and ARS in aqueous solution.45 Similar to previous observations, the resulting construct, PBA–ARS, produced orange coloured solutions with an easily detectable fluorescence signal (Scheme 30). Exposure to H2O2 resulted in an easy-to-visualise orange-to-purple colour change, indicative of the release of the indicator (ARS), with concomitant turn-off in fluorescence. In addition, a “turn-on” electrochemical signal was used to detect the conversion of the boronic acid to its corresponding phenol. These examples highlight potential applications of RIA strategies in ROS/RNS sensing. Nevertheless, extensive efforts will be needed before these IDAs begin to rival the traditional reaction-based sensors.
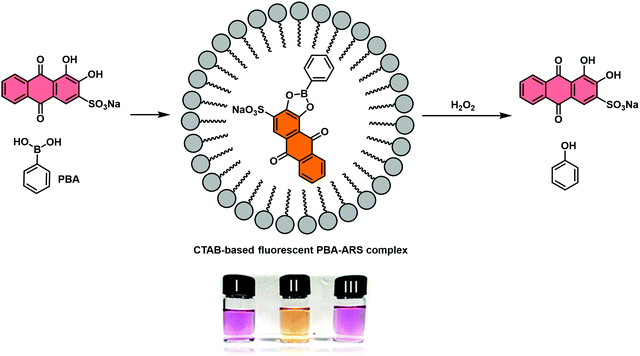 | ||
| Scheme 30 Schematic illustration of the proposed “reaction-based indicator displacement assay” (RIA) and sensing mechanism of a CTAB-based ARS–PBA complex used as a probe for the detection of hydrogen peroxide (H2O2). Reproduced from ref. 44 with permission from [The Royal Society of Chemistry], copyright [2015]. | ||
Jenkins and co-workers recently combined the RIA strategy with borogels to effect the H2O2-mediated release of ARS (Scheme 31).46 In this case, UV-Vis absorption analyses indicated the ARS-borogel underwent a dose-dependent release of ARS in the presence of H2O2. This PBA–ARS hydrogel system was also applied for the inhibition of methicillin-resistant staphylococcus aureus (MRSA) biofilm formation at the lag phase, a result ascribed to the combined toxicity of ARS and H2O2. Such constructs afford an opportunity to combine dual modal RIA with stimuli-responsive smart materials to afford covalently masked boronic acid pro-drugs materials.
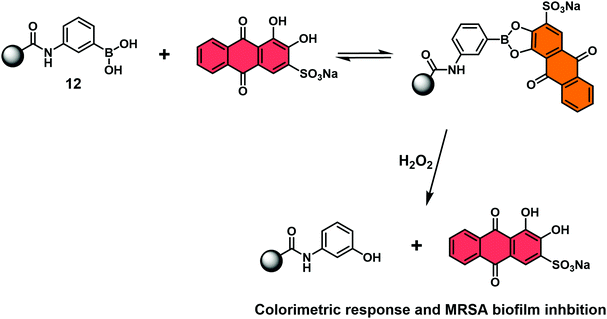 | ||
| Scheme 31 Schematic view of a hydrogel bound reaction-based indicator displacement assay (RIA) that allows for the simultaneous detection of H2O2 and inhibition of MRSA biofilm formation. | ||
IDA-based sensors for small molecules, gases and mechanical forces
The continued evolution of supramolecular architectures available for molecular sensing is allowing the development of new IDA systems. Recent IDA strategies include methods for detecting small molecule therapeutics, gases, mechanical force, and alkylamines.Isaacs and co-workers reported the complexation of amine functionalized trans-azobenzenes with CB[7] in NaOAc buffer (pH 4.76). The formation of these supramolecular ensembles was believed to result from a combination of strong hydrogen bonding and hydrophobic interactions that together served to promote the thermal isomerization of the trans-azobenzene to the corresponding cis-azobenzene form. This resulted in a change to the colour of the solution from yellow to purple. With this observation, the authors rationalised the design of an IDA for the detection of cationic biological active amines (Scheme 32).47 Of particular note was the finding that CB[7]·cis-azobenzene complex 38 was effective as an IDA and could be used to quantify pseudoephedrine content in Sudafed tablets. These researchers also exploited differences in the pH sensitivity of CB[7] and various cationic amines to effect efficient amine differentiation by PCA over a narrow pH range.
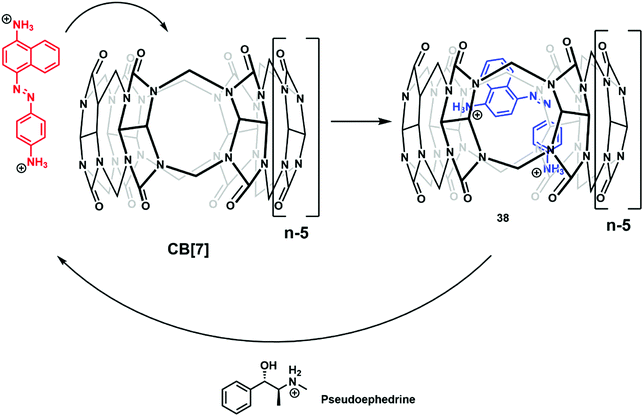 | ||
| Scheme 32 The formation of IDA CB[7]·cis-azobenzene 38, a construct that proved effective for the detection of biologically relevant cationic amines, such as pseudoephedrine. | ||
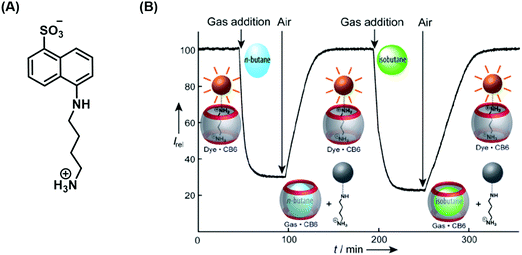
Nau and co-workers developed a remarkable IDA strategy that allows for the real-time monitoring of volatile hydrocarbons in aqueous solution (Scheme 33).48 The active host–guest complex used for these studies consisted of a water-soluble cucurbit[6]urils (CB[6]) (receptor) and 1-naphthylamine-5-sulfonic acid (indicator) functionalised with a putrescine anchor. Exposure of the FIDA in water to various hydrocarbon gases led to the displacement of the fluorescent indicator with attendant fluorescence “turn-off”. Subsequent purging with air demonstrated the reversibility of this active IDA system. Fluorescence and binding constant analyses of the interaction between the gaseous hydrocarbon analytes and CB[6] revealed that the smaller hydrocarbons were characterised by higher binding affinities for CB[6]. This identified hydrocarbon selectivity allowed CB[6] to be used as a purification strategy for commercially available neopentane with a purity of 99%. 1H NMR spectroscopic analyses revealed a trace unknown impurity in neopentane-saturated D2O. However, neopentane proved to be too large to bind effectively to CB[6]. On the other hand, the impurity was found to bind well, allowing for its removal from the initial hydrocarbon stock and production of purified neopentane.
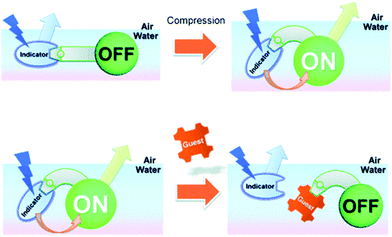
Ariga and Anslyn reported a mechanically controlled indicator displacement assay (MC-IDA), in which the fluorescence signal of the host–indicator complex can be switched on through surface compression (Scheme 34).49 A multimodal peptide host, consisting of a boronic acid receptor, cholesterol (air–water directing group), and a carboxyfluorescein unit, was constructed. Diol-containing 4-ML was used as the fluorescent indicator for the complementary binding to the boronic acid receptor and facilitating FRET between 4-ML (FRET donor) and carboxyfluorescein (FRET acceptor) moieties. To this design, when the FRET pairs became in close proximity to each other, the excitation of 4-ML (373 nm) resulted in fluorescence emission at 530 nm, which corresponded to carboxyfluorescein emission. Surface compression from 10 mN m−1 to 20 mN m−1 at the air–water interface induced FRET presumably as the result of the modifications in the distance and orientation between the FRET pairs. The addition of D-glucose led to the displacement of 4-ML (FRET donor) and inhibition of the FRET process, resulting in a ratiometric change (R = F525/F450) in fluorescence emission intensity. This work stands as an interesting example of an IDA-based strategy that could be applied to interfacial sensing applications. Therefore, to design a MC-IDA, the indicator must be bound to a receptor unit, which has the ability to change the spatial orientation under external forces (i.e. increase in pressure) resulting in a measurable change of the optical signal. Subsequent analyte-mediated displacement of the indicator expedites measurement of the analyte concentration using changes in the optical output.
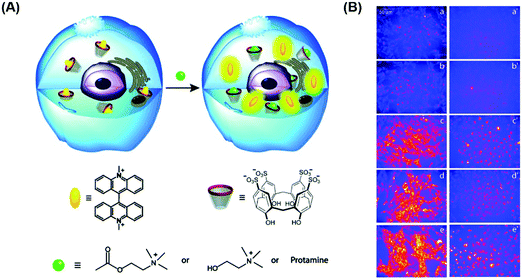
As noted previously, due to the poor selectivity of IDAs with species present at high concentrations in biological systems, their applications in vitro and in vivo have been limited. However, Nau and co-workers recently demonstrated that a simple combination of the macrocycle CX4 and fluorescent dye LCG results in the formation of an FIDA that could be used within live cells. These researchers showed it was possible to image the cellular uptake of cationic amine analytes in live V79 and CHO cells.50 The CX4–LCG host–guest complex displayed an initial low fluorescence emission intensity, but subsequent addition of choline, acetylcholine, or protamine resulted in a fluorescence turn-on response (Scheme 35). This example thus highlights a new direction for IDAs, namely their exploration in cellular applications and may define a general approach for monitoring the cellular uptake and trafficking of medicinally relevant compounds.
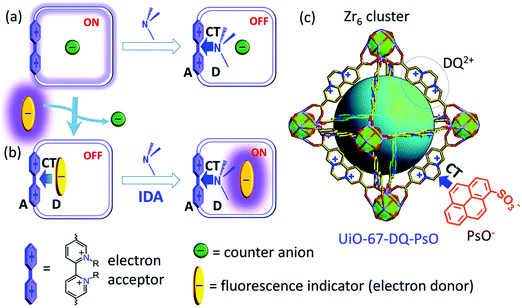
Gao and co-authors recently reported a metal–organic framework (MOF)-based IDA for the fluorescence detection of alkylamines.51 Here a Zr(IV) ion MOF (UiO-67) was constructed that contained the electron-deficient diquat (DQ2+) dicarboxylate as a secondary building unit and provide electrostatic interactions for the electron rich fluorescent anionic 1-pyrenesulfonate (PsO−) indicator allowing for the formation of a UiO-67-DQ–PsO host–guest complex. The PsO− guest readily underwent displacement with concomitant enhancement of fluorescence emission intensity when exposed to alkylamines. In contrast to a traditional IDA, this MOF IDA demonstrated several advantages, including the ability to carry out IDA in aqueous media with enhanced selectivity and sensitivity. This work marked a first example of the use of confined pore IDAs in small molecular sensing applications (Scheme 36) and thus stands as an exciting augury for future developments.
 | ||
| Scheme 36 Development of a metal organic framework (MOF) IDA for the fluorescence detection of alkylamines. (a) The Zr(IV) ion MOF (UiO-67-DQ–PsO) containing electron-deficient diquat receptor units (DQ2+) is used to bind electron-rich 1-pyrenesulfonate anions (PsO−). (b) This results in a fluorescent indicator displacement assay (FIDA) that gives rise to an increase in fluorescence intensity when exposed to alkylamines. Reproduced from ref. 51 with permission from [The Royal Society of Chemistry], copyright [2019]. | ||
Conclusion
Over the past few decades, the supramolecular community has attempted to mimic host–guest interactions found in nature with the development of synthetic molecular-based receptors. This led to the identification of a number of host–guest interactions that are capable of forming unique supramolecular ensembles. This acquired knowledge was exploited to design supramolecular-based molecular sensors, known as indicator displacement assays (IDAs). Early examples of IDAs were introduced by the groups of Inouye, Shinkai, and Anslyn using synthetic hosts that had complementary binding to their corresponding indicator. Due to the previously identified high affinities of each target species for their receptors, in the presence of the target analyte, the indicator was displaced leading to a change to the optical properties and facilitate the analyte's detection. The initial IDA examples inspired researchers worldwide to develop a plethora of novel indicator IDAs. These extensions have led to the development of enantioselective indicator displacement assays (eIDAs), fluorescent indicator displacement assays (FIDAs), reaction-based indicator displacement assays (RIAs), DimerDye disassembly assays (DDAs), intramolecular indicator displacement assays (IIDA), allosteric indicator displacement assays (AIDAs), mechanically controlled indicator displacement assays (MC-IDAs) and quencher displacement assays (QDAs). The continued development of synthetically accessible receptor scaffolds and commercialisation of dyes has led to all these variations of the original IDAs principle.Further, IDAs lend themselves readily to the creation of sensing arrays because several indicators and be paired with several receptors. This allow for pattern-based recognition of individual analytes and complex mixtures. It has also afforded sensing platforms that are able to identify and discriminate unknown sample mixtures. This strategy is highlighted by the work of Anslyn and co-workers that gave rise to peptide–metal–indicator ensembles. These latter IDAs proved capable of fingerprinting the types of wood used to age the Brazilian beverage Cachaça and classify unknown wood extracts. This overcomes the limitations of the “lock and key” approach with reaction-based molecular sensors (irreversible)/chemosensors (reversible). Minami and co-workers further exemplified this sensing array advantage with the sole use of commercially available reagents for the detection and discrimination of monosaccharides. Taken in aggregate, these contributions highlight the simplicity and cost-effective nature of IDAs, in that often no synthetic effort is required. In addition, IDAs provide a sensing strategy that often cannot be achieved by traditional molecular sensors. This benefit is underscored by the work of Nau and co-workers, in which an FIDA was shown to image the cellular uptake of medicinally relevant cationic amines in live V79 and CHO cells.
In summary, the exploration of IDA-based systems has led to the identification and demonstration of a number of significant advantages for IDAs over more traditional reaction-based molecular sensors (irreversible) and chemosensors (reversible). These include:
• Minimal synthetic efforts and the use of commercially available chromophore components
• Easy operation with high throughput analysis “sensing arrays” that can be combined with linear discrimination analysis (LDA) or principle component analysis (PCA)
• An ability to identify and discriminate unknown sample mixtures
• Facile detection of analytes via IDA-based platforms
• Tunable optical properties with the ability to use several indicators on the same system
• In situ formation of the reporter–host system for use in biological milieus.
Despite the promise shown by IDAs, a number of limitations exist, which need to be overcome if the full potential of the method is to be realised. For example, eIDAs are limited to the determination of the ee of a sample only and the determination of the de of a sample remains a significant challenge. This has led to the dynamic covalent self-assembly approach for determining both ee and de. One future direction for IDAs may involve the development of reversible indicator-based dynamic covalent approaches that facilitates the high throughput determination of ee and de to complement high throughput syntheses of relevance to the pharmaceutical industry. More recently, Anzenbacher and co-workers have addressed the known limitations of the low signal-to-noise ratio and sensitivity of IDAs, with the development and implementation of intramolecular indicator displacement assays (IIDA). Despite their promise, the number of reported IIDAs remains limited. In addition, a number of researchers have noted the difficulties of designing IDA-based platforms for use in biological applications. However, recent examples have demonstrated the ability to overcome this limitation, which includes the development of DimerDye disassembly assay (DDA).
We believe the recent examples highlighted illustrate how IDAs are still “new” to the field of molecular sensing with limitations only now being addressed. However, we hope that as the result of this Tutorial Review, the significance and excitement of IDAs is more apparent to the reader and that this understanding will provide an inspiration to pursue the development of new and improved IDAs whose limitations appear constrained only by our imaginations.
Abbreviations
| AC | Alizarin complexone |
| ACQ | Aggregation caused quenching |
| ARS | Alizarin Red S |
| AIDA | Allosteric indicator displacement assay |
| AIE | Aggregation induced emission |
| AO | Acridine orange |
| BA | Boronic acid |
| BPR | Bromopyrogallol red |
| CB6 | Cucurbit[6]uril |
| CB7 | Cucurbit[7]uril |
| Cu | Copper |
| CTAB | Cetyl trimethylammonium bromide |
| CX4 | p-Sulfonatocalix[4]arene |
| DBO | 1-Aminomethyl-2,3-diazabicyclo[2.2.2]oct-2-ene |
| DPA | Dipicolylamine |
| ee | Enantiomeric excess |
| eIDA | Enantiomeric indicator displacement assay |
| en | 1,2-Ethylenediamine |
| FIDA | Fluorescent indicator displacement assay |
| FRET | Fluorescence resonance energy transfer/Förster resonance energy transfer |
| GO | Graphene oxide |
| H2O2 | Hydrogen peroxide |
| IIDA | Intramolecular indicator displacement assay |
| IDA | Indicator displacement assay |
| ITC | Isothermal titration calorimetry |
| LDA | Linear discrimination analysis |
| LCG | Lucigenin |
| LOD | Limit of detection |
| M | Molar |
| MAA | N-Methyl anthranilic acid |
| ML | Methylesculetin |
| MOPS | 3-(N-Morpholino)propanesulfonic acid |
| MOF | Metal–organic framework |
| MRSA | Methicillin-resistant staphylococcus aureus |
| NPBA | 3-Nitrophenylboronic acid |
| NIR | Near-infrared |
| NMR | Nuclear magnetic resonance |
| ONOO− | Peroxynitrite |
| PCA | Principle component analysis |
| Pd | Palladium |
| PeT | Photoinduced electron transfer |
| PLP | Pyridoxal phosphate |
| PPi | Pyrophosphate |
| ppm | Parts per million |
| PR | Pyrogallol red |
| PS | Phosphatidylserine |
| PSO | 1-Pyrene sulfonate |
| PSP | Pyrene-modified N-alkylpyridinium |
| PV | Pyrocatechol violet |
| Rho123 | Rhodamine 123 |
| RIA | Reaction-based indicator displacement assay |
| RNA | Ribonucleic acid |
| RRE | Rev responsible element |
| Spds | Supramolecular-peptide-dots |
| TAMRA | Tetramethylrhodamine |
| TPE | Tetraphenylethylene |
| UD | Uranine dye |
| Zn | Zinc |
| Zr | Zirconium |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
XLS thanks the National Natural Science Foundation of China No. 21907080 and Natural Science Foundation of Shaanxi No. 2020JM-069. TDJ wishes to thank the Royal Society for a Wolfson Research Merit Award. EVA and JLS are grateful for continued support throughout their careers from the Robert A. Welch Foundation, and most recently the Welch Regents and Doherty-Welch Chairs (F-0046 to EVA and F-0018 to JLS).Notes and references
- D. Wu, A. C. Sedgwick, T. Gunnlaugsson, E. U. Akkaya, J. Yoon and T. D. James, Chem. Soc. Rev., 2017, 46, 7105–7123 RSC.
- J. M. Lehn, Chem. Soc. Rev., 2017, 46, 2378–2379 RSC.
- J. S. Wu, B. Kwon, W. M. Liu, E. V. Anslyn, P. F. Wang and J. S. Kim, Chem. Rev., 2015, 115, 7893–7943 CrossRef CAS.
- B. T. Nguyen and E. V. Anslyn, Coord. Chem. Rev., 2006, 250, 3118–3127 CrossRef CAS.
- A. T. Wright and E. V. Anslyn, Chem. Soc. Rev., 2006, 35, 14–28 RSC.
- J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973–984 CrossRef CAS.
- B. T. Herrera, S. L. Pilicer, E. V. Anslyn, L. A. Joyce and C. Wolf, J. Am. Chem. Soc., 2018, 140, 10385–10401 CrossRef CAS.
- L. Zhu, Z. L. Zhong and E. V. Anslyn, J. Am. Chem. Soc., 2005, 127, 4260–4269 CrossRef CAS.
- X. L. Sun and T. D. James, Chem. Rev., 2015, 115, 8001–8037 CrossRef CAS.
- K. L. Wen, S. S. Yu, Z. Huang, L. M. Chen, M. Xiao, X. Q. Yu and L. Pu, J. Am. Chem. Soc., 2015, 137, 4517–4524 CrossRef CAS.
- S. L. Pilicer, P. R. Bakhshi, K. W. Bentley and C. Wolf, J. Am. Chem. Soc., 2017, 139, 1758–1761 CrossRef CAS.
- M. Pushina, S. Farshbaf, E. G. Shcherbakova and P. Anzenbacher, Chem. Commun., 2019, 55, 4495–4498 RSC.
- S. Sheykhi, L. Mosca, J. M. Durgala and P. Anzenbacher, Chem. Commun., 2019, 55, 7183–7186 RSC.
- B. T. Herrera, S. R. Moor, M. McVeigh, E. K. Roesner, F. Marini and E. V. Anslyn, J. Am. Chem. Soc., 2019, 141, 11151–11160 CrossRef CAS.
- S. Arimori, C. J. Ward and T. D. James, Tetrahedron Lett., 2002, 43, 303–305 CrossRef CAS.
- W. M. J. Ma, M. P. P. Morais, F. D'Hooge, J. M. H. van den Elsen, J. P. L. Cox, T. D. James and J. S. Fossey, Chem. Commun., 2009, 532–534, 10.1039/b814379j.
- X. L. Sun, B. Zhu, D. K. Ji, Q. B. Chen, X. P. He, G. R. Chen and T. D. James, ACS Appl. Mater. Interfaces, 2014, 6, 10078–10082 CrossRef CAS.
- S. Gamsey, N. A. Baxter, Z. Sharrett, D. B. Cordes, M. M. Olmstead, R. A. Wessling and B. Singaram., Tetrahedron, 2006, 62, 6321–6331 CrossRef CAS.
- A. Jose, M. Elstner and A. Schiller, Chem. – Eur. J., 2013, 19, 14451–14457 CrossRef.
- Y. Sasaki, Z. Zhang and T. Minami, Front. Chem., 2019, 7, 49 CrossRef CAS.
- V. Janowski and K. Severin, Chem. Commun., 2011, 47, 8521–8523 RSC.
- E. Ghanem, S. Afsah, P. N. Fallah, A. Lawrence, E. LeBovidge, S. Raghunathan, D. Rago, M. A. Ramirez, M. Telles, M. Winkler, B. Schumm, K. Makhnejia, D. Portillo, R. C. Vidal, A. Hall, D. Yeh, H. Judkins, A. A. da Silva, D. W. Franco and E. V. Anslyn, ACS Sens., 2017, 2, 641–647 CrossRef CAS.
- K. A. Jolliffe, Acc. Chem. Res., 2017, 50, 2254–2263 CrossRef CAS.
- X. J. Liu, H. T. Ngo, Z. J. Ge, S. J. Butler and K. A. Jolliffe, Chem. Sci., 2013, 4, 1680–1686 RSC.
- T. Minami, Y. L. Liu, A. Akdeniz, P. Koutnik, N. A. Esipenko, R. Nishiyabu, Y. Kubo and P. Anzenbacher, J. Am. Chem. Soc., 2014, 136, 11396–11401 CrossRef CAS.
- X. J. Liu, D. G. Smith and K. A. Jolliffe, Chem. Commun., 2016, 52, 8463–8466 RSC.
- A. J. Plaunt, K. J. Clear and B. D. Smith, Chem. Commun., 2014, 50, 10499–10501 RSC.
- W. C. Tse and D. L. Boger, Acc. Chem. Res., 2004, 37, 61–69 CrossRef CAS.
- R. del Villar-Guerra, R. D. Gray, J. O. Trent and J. B. Chaires, Nucleic Acids Res., 2018, 46, e41 CrossRef CAS.
- D. L. Boger and W. C. Tse, Bioorg. Med. Chem., 2001, 9, 2511–2518 CrossRef CAS.
- J. H. Zhang, S. Umemoto and K. Nakatani, J. Am. Chem. Soc., 2010, 132, 3660–3661 CrossRef CAS.
- C. Matsumoto, K. Hamasaki, H. Mihara and A. Ueno, Bioorg. Med. Chem. Lett., 2000, 10, 1857–1861 CrossRef CAS.
- N. N. Patwardhan, Z. G. Cai, C. N. Newson and A. E. Hargrove, Org. Biomol. Chem., 2019, 17, 1778–1786 RSC.
- D. Tian, F. Li, Z. Zhu, L. Zhang and J. Zhu, Chem. Commun., 2018, 54, 8921–8924 RSC.
- W. M. Nau, G. Ghale, A. Hennig, H. Bakirci and D. M. Bailey, J. Am. Chem. Soc., 2009, 131, 11558–11570 CrossRef CAS.
- W. C. Geng, S. R. Jia, Z. Zheng, Z. H. Li, D. Ding and D. S. Guo, Angew. Chem., Int. Ed., 2019, 58, 2377–2381 CrossRef CAS.
- S. A. Minaker, K. D. Daze, M. C. F. Ma and F. Hof, J. Am. Chem. Soc., 2012, 134, 11674–11680 CrossRef CAS.
- Y. Liu, L. Perez, M. Mettry, C. J. Easley, R. J. Hooley and W. W. Zhong, J. Am. Chem. Soc., 2016, 138, 10746–10749 CrossRef CAS.
- V. E. Zwicker, B. L. Oliveira, J. H. Yeo, S. T. Fraser, G. J. L. Bernardes, E. J. New and K. A. Jolliffe, Angew. Chem., Int. Ed., 2019, 58, 3087–3091 CrossRef CAS.
- M. A. Beatty, J. Borges-Gonzalez, N. J. Sinclair, A. T. Pye and F. Hof, J. Am. Chem. Soc., 2018, 140, 3500–3504 CrossRef CAS.
- J.-B. Jiao, G.-Z. Wang, X.-L. Hu, Y. Zang, S. Maisonneuve, A. C. Sedgwick, J. L. Sessler, J. Xie, J. Li, X.-P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 1925–1932 CrossRef CAS.
- A. F. Sierra, D. Hernandez-Alonso, M. A. Romero, J. A. Gonzalez-Delgado, U. Pischel and P. Ballester, J. Am. Chem. Soc., 2020, 142, 4276–4284 CrossRef CAS.
- M.-Q. Fu, X.-C. Wang, W.-T. Dou, G.-R. Chen, T. D. James, D.-M. Zhou and X.-P. He, Chem. Commun., 2020, 56, 5735–5738 RSC.
- X. Sun, K. Lacina, E. C. Ramsamy, S. E. Flower, J. S. Fossey, X. Qian, E. V. Anslyn, S. D. Bull and T. D. James, Chem. Sci., 2015, 6, 2963–2967 RSC.
- X. L. Sun, M. L. Odyniec, A. C. Sedgwick, K. Lacina, S. Y. Xu, T. T. Qiang, S. D. Bull, F. Marken and T. D. James, Org. Chem. Front., 2017, 4, 1058–1062 RSC.
- B. L. Patenall, G. T. Williams, L. Gwynne, L. J. Stephens, E. V. Lampard, H. J. Hathaway, N. T. Thet, A. E. Young, M. J. Sutton, R. D. Short, S. D. Bull, T. D. James, A. C. Sedgwick and A. T. A. Jenkins, Chem. Commun., 2019, 55, 15129–15132 RSC.
- J. Wu and L. Isaacs, Chem. – Eur. J., 2009, 15, 11675–11680 CrossRef CAS.
- M. Florea and W. M. Nau, Angew. Chem., Int. Ed., 2011, 50, 9338–9342 CrossRef CAS.
- K. Sakakibara, L. A. Joyce, T. Mori, T. Fujisawa, S. H. Shabbir, J. P. Hill, E. V. Anslyn and K. Ariga, Angew. Chem., Int. Ed., 2012, 51, 9643–9646 CrossRef CAS.
- A. Norouzy, Z. Azizi and W. M. Nau, Angew. Chem., Int. Ed., 2015, 54, 792–795 CrossRef CAS.
- N. N. Yang, L. J. Zhou, P. Li, Q. Sui and E. Q. Gao, Chem. Sci., 2019, 10, 3307–3314 RSC.
| This journal is © The Royal Society of Chemistry 2021 |