Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biomaterials-based approaches to model embryogenesis
Chantelle
Spiteri
a,
Valeria
Caprettini
a and
Ciro
Chiappini
 *ab
*ab
aCentre for Craniofacial and Regenerative Biology, King's College London, London, UK. E-mail: ciro.chiappini@kcl.ac.uk
bLondon Centre for Nanotechnology, London, UK
First published on 26th October 2020
Abstract
Understanding, reproducing, and regulating the cellular and molecular processes underlying human embryogenesis is critical to improve our ability to recapitulate tissues with proper architecture and function, and to address the dysregulation of embryonic programs that underlies birth defects and cancer. The rapid emergence of stem cell technologies is enabling enormous progress in understanding embryogenesis using simple, powerful, and accessible in vitro models. Biomaterials are playing a central role in providing the spatiotemporal organisation of biophysical and biochemical signalling necessary to mimic, regulate and dissect the evolving embryonic niche in vitro. This contribution is rapidly improving our understanding of the mechanisms underlying embryonic patterning, in turn enabling the development of more effective clinical interventions for regenerative medicine and oncology. Here we highlight how key biomaterial approaches contribute to organise signalling in human embryogenesis models, and we summarise the biological insights gained from these contributions. Importantly, we highlight how nanotechnology approaches have remained largely untapped in this space, and we identify their key potential contributions.
Introduction
The largely unexplored molecular and cellular mechanisms accompanying implantation are often referred to as the ‘black box’ of human development.1 Insights into post-implantation events are crucial to comprehend the cellular differentiation of embryonic stem cells (ESCs) accompanied by morphogenesis and growth of the embryo. In fact, a better understanding of embryogenesis (Box 1), more specifically related to the events occurring during gastrulation, can improve our ability to direct cell fate, thereby enabling the engineering of complex cellular architectures. This improved understanding is critical to dissect the complex signalling networks regulating development and to advance biomedical research for tissue regeneration and diseases of developmental derangement such as birth defects and cancer.2 Recent decades have seen rapid and significant improvements in establishing approaches to model and dissect embryonic development. The rise of sophisticated lineage tracing animal models has played a crucial role in shedding light over cell fate, migration and arrangement through morphogenesis and identifying the key driving events that regulate gastrulation. Yet, investigating the early processes of human embryogenesis in vivo is challenging because of the difficulties in accessing and thus visualizing individual cells.3 Furthermore, despite some conserved morphological landmarks between mammalian blastocysts, timing and molecular expression differences exist between species.4 Therefore, the degree of relevance that knowledge obtained from animal models holds for humans remains uncertain.5 For example, contrary to the mechanisms involved in mouse embryogenesis, fibroblast growth factor (FGF) signalling might not be required for hypoblast formation in human, thus suggesting that mechanisms occurring in mice do not directly translate to other mammals.6–8
Box 1 EmbryogenesisEmbryogenesis is the process regulating the formation and development of the embryo. During embryonic development, the first differentiation event leads to the formation of the inner cell mass (ICM) surrounded by a layer of trophectoderm cells. Successively, the ICM shift towards one pole of the blastocysts and just before implantation it segregates in a ‘salt and pepper’ arrangement forming the embryonic ectoderm (epiblast) and the primitive endoderm (PE, hypoblast).9 The resulting epiblast and hypoblast cell types are spatially arranged in two distinct tissues which undergo gastrulation to establish the three embryonic cell layers and sets the basic axes (anterior–posterior, dorsal–ventral and left–right) of the body. In mouse, the epiblast cells express Nodal signalling that acts on the adjacent visceral endoderm which in turn acts as a source of Wnt and Nodal antagonists, migrates towards the anterior domain and form the anterior visceral endoderm (AVE). As the AVE continues to secrete antagonists, an anterior–posterior gradient of Wnt and Nodal signals is established leading to the onset of gastrulation. A groove forms in the cranial end of the epiblast that elongates as epiblast cells proliferate and migrate to form the primitive streak. The epiblast cells experience epithelial to mesenchymal transition to ingress through the primitive streak forming the mesoderm and definitive endoderm.10 The suppression of Nodal and Wnt signalling pathways by their antagonist in the anterior epiblast has proven to be essential for neural induction.11 For a deeper and more exhaustive summary of the current understanding of the molecular and cellular mechanisms occurring during early human development we refer the reader to excellent recent reviews of the subject.12,13 |
GlossaryEmbryo – The early stages of growth, development and differentiation of a multicellular organism.Blastocyst – A hollow spherical structure consisting of: Inner cell mass – a pluripotent cellular mass within one side of the blastocyst's interior forming the embryo. Trophoblast – the outer layer of the blastocyst giving rise to extraembryonic tissues. Epiblast – One of the two layers forming the inner cell mass deriving the three germ layers. Hypoblast – The other layer forming the inner cell mass which gives rise to the yolk sac. Embryoid body – A spherical aggregate of stem cells that can differentiate towards multiple cell lineages. Primitive streak – A temporary elongated depression marking the gastrulation site. Germ layers – The three layers, namely endoderm, mesoderm and ectoderm, that commit to all adult tissues and organs formation as the embryo develops. Gastruloids – In vitro 3D aggregates displaying key embryo-like features similar to those occurring after implantation. Gastrulation – An early embryonic developmental process whereby the three germ layers are established. Extracellular matrix – A 3D structural scaffold consisting of glycoproteins and growth factors among other macromolecules, providing external biochemical and physical support. Autocrine signalling – A form of diffusible signalling secreted from one cell that binds to its own receptors or act on a neighbouring targeted cell of the same type. Paracrine signalling – A form of diffusible signalling secreted from one cell type to a neighbouring targeted cell type. |
Approaches using embryonic tissue explants improve access, visualisation and manipulation of single cells within certain limits, but either cannot address inter-species developmental variations or pose significant ethical issues when using human tissues. Culturing the human embryo in vitro in the absence of maternal tissues established important insights into the early post-implantation phases of human development. These studies displayed a series of key morphogenetic rearrangements which provided a new understanding of the self-organizing capacity of the human blastocysts.14,15 However, the ethical guidelines of the Warnock report limit these experiments to pre-gastrulation stages.16,17
The rapid progress of organotypic embryonic models from human pluripotent stem cells (hPSCs) has contributed to otherwise inaccessible human-relevant insight into implantation and gastrulation events. The establishment of human ESC derived from blastocysts18 has provided an alternative ‘bottom-up’ approach to studying embryogenesis. Additionally, the development of induced pluripotent stem cells (iPSCs) through the reprogramming of adult human fibroblasts, has served as a powerful tool to dissect the fate of pluripotent cells while significantly mitigating the ethical dilemma associated with the use of ESCs.19,20 Both types of pluripotent stem cells can extensively proliferate, and upon manipulation of their environment, they can differentiate into the three embryonic lineages, namely the endoderm, mesoderm and ectoderm. Therefore, stem cells hold a great promise as models to recapitulate processes of early human embryology.21 Indeed, the early use of uniformly distributed morphogens as isolated stimuli to direct embryogenesis in uniform hPSC cultures has provided some valuable insights into key regulatory mechanisms directing embryonic differentiation, but has often proved insufficient to dissect the orchestrated signalling associated with embryonic patterning.22
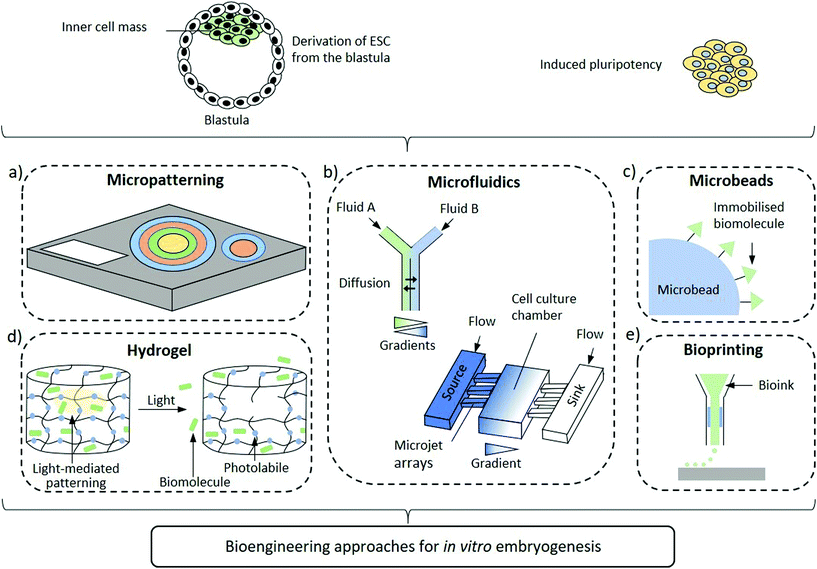
Improved design of the embryonic niche combining spatiotemporally regulated biochemical and biomechanical stimuli are emerging to shed light over the processes regulating axis determination. In fact, the self-organizing capacity of hPSCs needs to be complemented by specific physico-chemical cues to modulate cellular development in an organized, context-dependent manner. As such, self-organization and differentiation are dependent on local cues which comprise, among others, morphogen dynamics23,24 and mechanical stresses.25,26 Even if animal development mechanisms differ from the human ones, a number of vertebrate models suggest that some basic key signalling pathways responsible of determining cell fate and patterning are conserved, including Wnt, transforming growth factor (TGF-β) and FGF.27 Yet, uncovering how the signalling activities are orchestrated in space and time to pattern the epiblast into the three germ layers is still an open challenge. Indeed, controlling differentiation in an organized manner towards the desired outcome is of paramount importance in hPSC research. Initial attempts investigating the morphogen gradients within the embryoid body in vitro consisted in the generation of ESC aggregates followed by exposure to Wnt3a morphogen. These ESC aggregates established the formation of the so-called embryoid bodies (EBs) with a spontaneous self-organizing gradients driving cells to an anteroposterior polarity and primitive streak characteristics.28 These findings suggest that although external signals are required for activation, the ensuing processes are self-reinforcing after initiation. Small aggregates of mouse ESC and more recently human ESC stimulated by CHIR99021 (CHIR), a Wnt agonist, resulted in aggregate elongation and expression of markers resembling the embryonic tail bud. Furthermore, the transcriptional programmes of these gastruloids recapitulated the embryo-like spatiotemporal patterns of gene expression, hence mimicking the processes occurring during symmetry breaking and axial organization.29–31 Undoubtedly, these 3D models are promising substitutes of embryos that will significantly contribute to expand our knowledge of early developmental processes. Nevertheless, fundamental differences persist between these advanced gastruloid models and early embryos. The lack of effective spatiotemporal regulation of stimuli that drive developmental signalling pathways plays a crucial role in the observed differences. Indeed, biomaterial sciences are providing key contributions to recapitulate the developmental niche by supplying these signals. Apart from providing the appropriate biochemical and biomechanical cues, biomaterials can mimic their spatial and temporal organization to recapitulate the stem cell microenvironment and induce the desired differentiation fate. Nanomaterials, thanks to their emergent properties yielding intrinsic multifunctionality, have the yet unfulfilled potential to provide unique contributions to control embryonic development through high resolution, spatiotemporally controlled and environmentally aware delivery of stimuli that regulate biogenesis. In this perspective, we summarise the advances that biomaterial-based approaches offer to the study of human embryonic patterning and highlight how inputs from nanotechnology can further evolve the field (Fig. 1).
2D Micropatterning
Two-dimensional micropatterning can provide effective spatiotemporal control over topographic, biochemical and biophysical cues enabling orchestrating organisation in stem cell colonies. Confining hESCs in vitro within micropatterns of a defined shape and size (Fig. 1a) induces spontaneous, spatially ordered organisation of the colony and patterned differentiation which mimics aspects of embryonic development. Micropatterning can be achieved by topographic modification of the substrate using micro/nanostructures that confine and direct the arrangement and adhesion of cells, or by surface chemical modifications to locally enhance or prevent cell adhesion (Fig. 2a).32,33 These approaches typically comprise photolithography, direct writing, microcontact printing, possibly alongside more exotic microtechnology approaches.34,35 Mouse epiblast-like cells cultured on micropatterned substrates supplemented with appropriate morphogen signalling (Wnt, BMP, ACTIVIN, FGF), induce differentiation events and a spatial arrangement of cell fate comparable to those occurring when establishing the mouse gastrula in vivo.36 Geometrical confinement of pre-streak-like stem cells can control patterning of Brachyury (Bra)/T expression to guide the position of the primitive streak and attain asymmetric patterning of pluripotent stem cells. Indeed, growing cells on an ellipse micropattern instead of circular ones preferentially induces localised T-expressing cells at the tips of the ellipse rather than on the entire circumference (Fig. 2b). The simplicity, robustness, stringent control over key geometrical features and accessibility to molecular readouts of this approach highlights its transformative potential for investigating how geometric asymmetries – such as edges or elongations – in homogenous stem cells colonies influence the colony's response to morphogens and contributes to introduce heterogeneity in cell fate specification.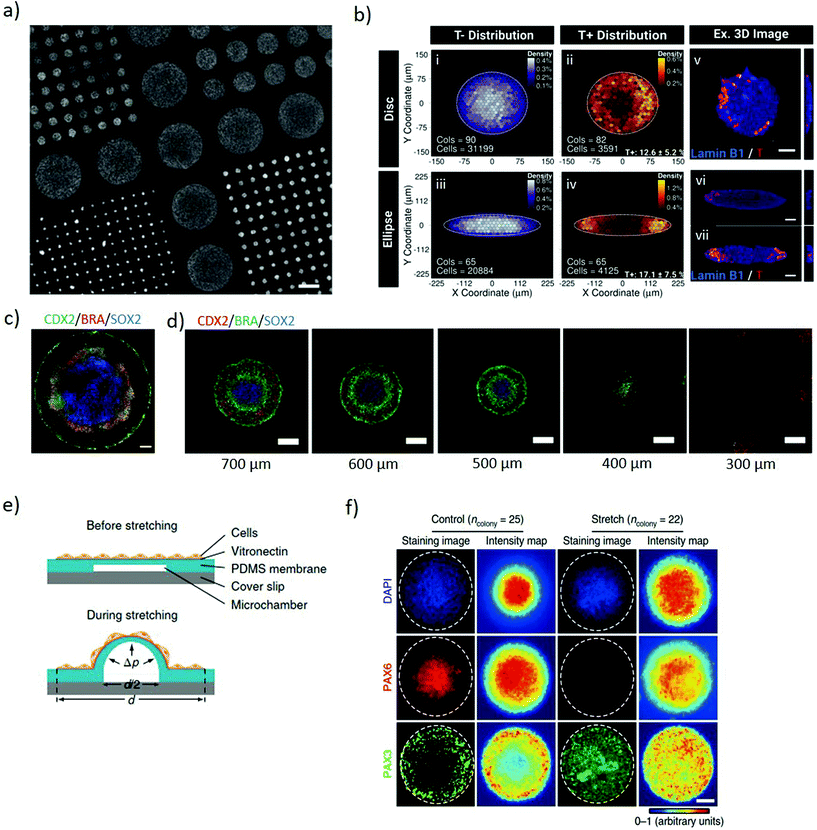 | ||
| Fig. 2 Micropatterning stem cell colonies induces differential germ layer specification. (a) Glass coverslip coated with poly-D-lysine micropatterns of different diameters for selective hESCs adhesion to the functionalised regions. Adapted with permission.39 Copyright 2014, Nature Publishing Group. (b) The geometry of the micropatterned colonies influences the pattern of Brachyury (T) expression. T expression identifies the position a pre-streak like population. Binned density maps (BDMs) images (i–iv) and confocal images (v–vii) show that brachyury positive cells (T+, red) are positioned at the tips of the ellipse micropattern whilst in the disc micropatterns, T+ cells are located on the entire circumference. Cols = number of colonies analysed; cells = number of cells analysed. Scale bars = 50 μm. Adapted with permission.41 Copyright 2018, The Company of Biologists Ltd. (c) Immunofluorescence of micropatterned hESC treated with BMP4, displaying radial organization of the trophectoderm (CDX2) and germ layers markers: mesoderm (BRA) and ectoderm (SOX2). Scale bars = 100 μm. Adapted with permission.39 Copyright 2014, Nature Publishing Group. (d) Immunofluorescence images showing pattern-size dependence of germ-layer domains. As size is reduced SOX2 expression declines and eventually disappears. Scale bars = 200 μm. Adapted with permission.32 Copyright 2017, The Company of Biologists Ltd. (e) Schematic of the micropatterned cell colonies on locally-stretchable PDMS membrane. The central region of the patterned colony stretches upon expansion of the underlying microfluidic channel. (f) Immunofluorescence images and average intensity maps of unstretched (control) and mechanically stretched (stretched) colonies at the central portion at day 8. Staining the colonies with Pax6 (neuroepithelial cell marker) and Pax3 (neural plate border marker), suggest that mechanical stimulation plays a role in regulating neural plate border differentiation. White dashed lines mark the colony periphery. Scale bar = 100 μm. (e and f) Adapted with permission.42 Copyright 2018, Nature Publishing Group. | ||
The size of 2D micropatterned hPSCs is a major determinant in the maintenance of pluripotency within a colony. When exposed to identical stimuli, circular larger colonies maintain pluripotency whereas smaller ones spontaneously differentiate over time. Possibly, the size of hESC colony influences the ratio of pSMAD1 agonist and antagonist expression, important for mediating self-renewal and differentiation. The higher local cell density present in large colonies correlates with higher concentrations of autocrine/paracrine morphogen signals, such as the BMP antagonist GDF3, and induces repression of SMAD1 leading to retaining expression of the pluripotency marker, Oct4. In contrast, small colonies express excess pSMAD1 agonists favouring differentiation.37 When stimulated with BMP2 and activin A, large colonies tend to differentiate towards the mesoderm whilst small colonies differentiate towards the endoderm lineage.38 Uniform soluble BMP4 stimulus applied to patterned circular hPSCs colonies induces concentric differentiation towards the germ layers. The inner portion of the pattern acquires endodermal specification, surrounded by a mesodermal ring, itself encircled by ectoderm whilst the outer layer of the colony acquires a trophectoderm-like phenotype colony (Fig. 2c).39 Size plays an important role for these micropatterns, with the outer ectodermal and mesodermal regions largely independent of the overall colony size, while the inner endodermal specification occupying the remaining of the pattern. Indeed, small colonies of less than 250 μm diameter do not exhibit significant mesodermal or endodermal specification, which progressively arises as the size of the pattern expands (Fig. 2d).32,39 This approach enables investigating the interplay between BMP4 and its inhibitor Noggin and the localization of the BMP4 receptors along a radial symmetry. The sensitivity of BMP4 is reduced from the edge to the centre of the colony, both due to the radial changes in localization of the BMP receptors from the apical to the basolateral surface of the cells and because of differential Noggin activation and diffusion.40 These systems indicate that the size and shape of micropatterns influence the response of cells to uniform diffusible molecules thus establishing local gradients of biochemical cues which induce controlled pattering within stem cell colonies. Identifying that asymmetric micropatterns lead to breaking the radial symmetry suggests a means to further study and secure the positional robustness of the streak.41
Two dimensional micropatterning also enables investigating neuroectoderm patterning. Neural induction of circular micropatterned hPSC colonies induces concentric arrangement of the neural plate border and the neural crest, in accordance with their arrangement during mouse embryo development. Isolated stretching of the central portion of the pattern highlights a local activation of BMP-SMAD signalling in response to mechanical stimulation (Fig. 2e), suggesting its role in the regulation of neural plate border differentiation (Fig. 2f). Overall, this system enables studying the mechanisms inducing neuroectodermal specification with a potential to advance our understanding of neural development and its associated disorders.42 Furthermore, the spatiotemporal regulation of 2D patterns shows promise as a screening platform for teratogenicity. Mesoendoderm generated from micropatterned hPSC treated with known teratogens displays disrupted patterning in a dose-dependent manner, highlighting the role of the drugs in inducing developmental disorders.43
Micropatterning is a reductionist approach that enables isolating and controlling the role of geometrical parameters to study their impact on signalling networks responsible for differentiation and morphogenesis during development. This highly controllable and simple two-dimensional approach shows promising potential for recapitulating key aspects of the early stages of embryogenesis and gastrulation but does not provide a simple avenue to model the temporal regulation of morphogenic cues that happen in vivo. In fact, it is typically challenging and unreliable to modify micropatterns over time after the colony has been established, or to trigger a precise gene expression or inhibition in a time-resolved fashion. This has currently limited micropatterning approaches to probe individual signalling pathways during one key developmental event, hampering more comprehensive investigations of large signalling networks across the developmental timeline.
Microfluidic devices
Soft lithography of silicones by replica moulding – among which polydimethylsiloxane (PDMS) is the most explored – is a largely established approach for rapid prototyping of microfluidic devices, thanks to its low-cost, reliability and ease of processing.44 Microfluidics can combine gradients of soluble factors, providing fine control over the spatial and temporal arrangement of morphogens,45 with technical limits on the number of simultaneous gradients available and their individual spatiotemporal complexity (Fig. 1b). Furthermore, regulating fluid flows allows improved control of nutrients exchange, culture conditions and shear stimuli. This increased complexity in the regulations of cues requires elaborate experimental setups, but can provide the improved versatility necessary to dissect interconnected signalling and events.46Continuous flows can wash autocrine and paracrine factors away from cells in a regulated fashion, hence enabling quantitative investigation of the secreted factors involved in differentiation.47,48 This approach allows investigating the role of FGF4 for neuroectodermal specification in mouse ESCs. Microfluidic delivery of FGF4-supplemented differentiation medium to mESCs is insufficient to induce neuroectoderm when autocrine and paracrine cell-secreted factors are washed away from the system. Instead, the presence of cell-secreted factors in addition to FGF4 recovers neuroectodermal differentiation, highlighting that FGF4 is necessary but not sufficient to establish neuroectodermal fate, thereby providing insight in manipulating diffusible signalling.48 Approaches based on T or Y-shaped laminar flow systems can introduce solutions with different concentrations of desired biochemicals in each side branch, establishing a gradient in the main branch by lateral diffusion. However, as perfusion depletes the autocrine and paracrine factors essential for cell growth, survival or differentiation, continuous flow restricts the study of self-organizing systems. Besides, the continuous laminar flow generates shear stresses that can affect cell behaviour in unpredictable ways.49 Thus, diffusion-based devices are preferable as they avoid active perfusion of the culture. These systems rely on Fickian diffusion to establish gradients over a central culturing chamber interconnected with reservoirs containing desired concentrations of stimulants.50 Such systems can establish stable, opposing linear concentration gradients of multiple signalling molecules, such as sonic hedgehog (SHH) and FGF/BMP, generated using side channels under laminar flows as reservoirs in order to regulate neuronal differentiation.51,52 Exposing neural progenitors to these gradients stimulates a patterned neuronal differentiation with analogies to the dorsoventral specification of the neural tube. A four-channel setup of this system (Fig. 3a) can further introduce opposing retinoic acid and possibly FGF gradients to mimic the distribution of key anteroposterior specification morphogens alongside the dorsoventral SHH/BMP gradients (Fig. 3b).52 Indeed, these systems can induce dorsoventral and anteroposterior patterning as a response to the opposing morphogenic gradients.53
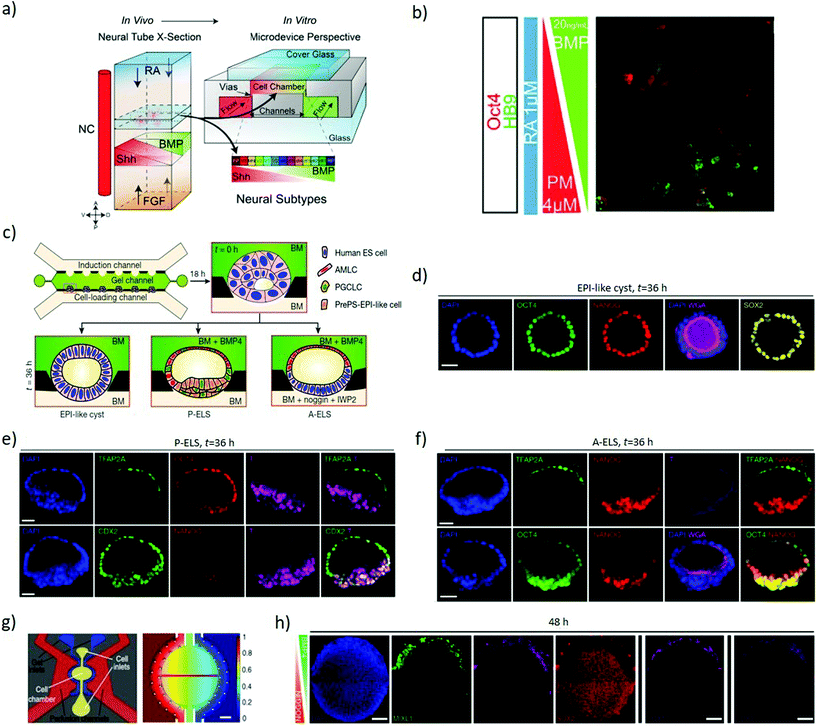 | ||
| Fig. 3 Microfluidics devices establish gradients of morphogens to induce axial specification. (a) Schematic representation of the four primary signals (RA, FGF, SHH and BMP) directing neural tube patterning in vivo and the four-channel setup developed to recapitulate the gradients of these signals in vitro through a source and sink approach. (b) ESCs with HB9-GFP (a gene marker for post-mitotic motor neurons) exposed to opposing BMP4 and PM (SHH analogue) gradients, in the presence of RA. The combined gradients induced dorsoventral patterning as evidenced by localized OCT4 expression (dorsal marker) at the BMP side and HB9 expression (neuronal specification marker) at the PM side. (a and b) Adapted with permission.52 Copyright 2016, The Company of Biologists. (c) A schematic representation of multi-channel microfluidic device with the middle channel preloaded with Geltrex forming concave pockets to host cells and provide an asymmetric environment. The lower channel is used to load cells and flow base medium while the upper channel is used to flow desired concentrations and temporal patterns of chemical inducers. (d) Confocal images showing the lumen developed within the Oct4, Nanog and Sox2 stained epiblast-like cysts. (e) Confocal images showing TFAP2A (a putative amniotic ectoderm marker) detected exclusively at the pole exposed to BMP4. The micrographs represent P-ELS formation as the epiblast-like cells at the opposite pole show the presence of CDX2 and T but a reduction in Nanog. (f) The micrographs represent A-ELS formation with more organized Nanog and Oct4 staining and diminished T staining. (c–f) Adapted with permission.54 Copyright 2019, Nature Publishing Group. (g) Left to right: Schematic of the individual unit within the microfluidic device; and computational simulation of the gradient generated within the active chamber after 48 hours of perfusion. (h) Immunofluorescence analysis of high density hESC colonies exposed for 48 hours to opposing BMP4 (50 ng mL−1) and Noggin (200 ng mL−1) gradients. The colony shows markers of the trophectoderm (CDX2), mesoderm (MIXL1, T) and endoderm (SOX17) at the BMP4 source side, and ectoderm (SOX2) throughout the colony except near the BMP source side. Two rightmost panels display different replicates. Scale bars = 200 μm. (g and h) Adapted with permission.55 Copyright 2019, Nature Publishing Group. | ||
Microfluidics is a key element of a platform that recapitulates the dorsal–ventral patterning of hPSCs epiblast-like cysts (Fig. 3c), showing landmarks of the human epiblast development and recapitulating amniotic ectoderm differentiation with the specification of primordial germ cells. The loaded ESCs reside into gel pockets within a microfluidic device, where they grow to establish a lumen, developing into epiblast-like cysts (Fig. 3d). In the presence of BMP4, the pole of the cysts exposed to BMP4 differentiates into amniotic ectoderm-like cells (AMLC) with an epiblast-like epithelium at the opposite pole not exposed to BMP4 (Fig. 3e), hence resembling a human embryonic sac before its gastrulation onset. Provided that Nanog concentration is low, the cysts acquire a prospective posterior end of the epiblast (PrePS-EPI) phenotype and thus the asymmetric sacs are referred to as posteriorized embryonic-like sacs (P-ELS). Instead, in the presence of Noggin and IWP2 (Wnt-inhibitor) stimuli the cysts retain AMLC patterning on one pole but develop a more organised epiblast-like pole, and become anteriorized embryonic-like sacs (A-ELS) (Fig. 3f).54 To investigate the anterior–posterior symmetry events, microfluidic systems can expose 2D colonies of human pluripotent stem cells to gradients of BMP4 (Fig. 3g) and investigate the influence of cell density and morphogen concentration on the patterning outcome. A low initial cell density combined with the BMP4 gradient can induce an axial arrangement of the germ layers, while a high cell concentration combined with BMP4 and opposing Noggin gradient result in characteristic markers of the trophectoderm (CDX2), mesoderm (MIXL1, T) and endoderm (SOX17) towards the BMP4 source, and ectoderm (SOX2) present throughout the colony except near the source (Fig. 3h).55
Microfluidics also enables shuttling and trapping multiple EBs at precise locations and controlling their interaction, while exposing them differentially to desired combinations of morphogens. With such an approach it is possible to first trap BMP4-free EBs and then direct BMP4-treated EBs within gelatin microparticles towards the trapped EBs in order to induce their fusion. The fused EBs express spatially controlled mesoderm differentiation markers, providing a mean to manipulate and enhance precision in patterning EBs.56
Overall, microfluidics is a powerful tool to control the exposure to morphogens in space and time, which can effectively regulate spatial patterning of cells in 2D and 3D systems alike. These devices can develop organ-on-a-chip and organoid models that emulate key multicellular structures and physiology across key early developmental events. Despite these crucial features, handling 3D systems within microfluidic devices remains challenging, due to diffusion barriers in flow leading to uneven distribution across the tissues. Furthermore, the efficiency of long-term stem cells culturing remains highly variable and there is limited space for tissue development, whilst increasing the channel dimensions and flow rates obliviate the key microfluidic advantage of laminar flow.46,55 Moving forward, platforms integrating microfluidics and hydrogels promise comprehensive control over the biophysical and biochemical niche enabling more accurate modelling of early developmental events in 3D cultures to model embryogenesis. Hydrogels demonstrate a strong potential in tailoring the physicochemical properties of the extracellular matrix in space and time in order to enable in vitro modelling, while microfluidics provides sophisticated control over soluble cues. A hydrogel precursor injected into a PDMS mould followed by gelation generates a hydrogel chip with microchannels. The surface topography of the hydrogel where cells are deposited is fabricated for flat cell cultures or spheroid-based cultures. Exposing mouse EBs to retinoic acid (RA) gradients under spatial and temporal control within this systems results in their neural differentiation, with those EB close to the RA source growing significantly larger.57 Embedding mouse embryoid bodies in a collagen matrix and subjecting the EBs to combined gradients of RA and smoothened agonist (SAG), an SHH activator, creates a range of different microenvironment conditions within the scaffold. The combinatorial effect of the morphogens induces a graded response of motor neurons differentiation in regions of high retinoic acid concentrations similar to in vivo situations.58
Microbeads or microspheres
Immobilizing factors to solid substrates (Fig. 1c) enables investigating the role of signalling that originates from the extracellular matrix as well as induce spatial organization to both matrix and soluble-like cues. The organisation of signalling enables investigating heterogeneity in differentiation and unravelling asymmetric division within the embryoid body. Local presentation of leukaemia inhibitory factor (LIF) to ESCs as an immobilized ligand maintains ESC pluripotency for at least 2 weeks, which is not achievable with soluble LIF. This approach can apply to a range of factors maintaining pluripotency or directing differentiation.59 Wnt coated microbeads can present asymmetric signalling to individual mouse ESC. Cell division in the presence of such asymmetric signalling yields pluripotent daughter cells at the Wnt microbead (Fig. 4a) and differentiated cells away from the microbead (Fig. 4b), enabling investigating the mechanism of asymmetric cell division in ESCs.60,61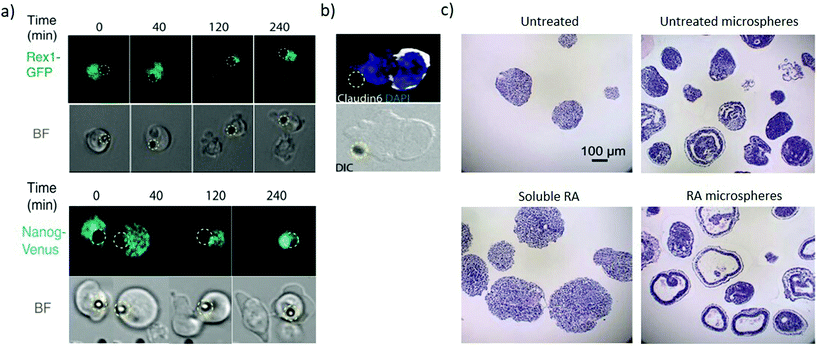 | ||
| Fig. 4 Microbeads provide spatiotemporally regulated display of morphogens. (a) Pluripotency genes expression of Rex1-GFP and Nanog-Venus of selected frame from time-lapse imaging during ES cell division in the presence of Wnt3a beads (represented by the dashed yellow circle). The pluripotency markers are asymmetrically expressed with high expression in the proximal daughter cell than in the distal one. (b) The distal daughter cells expressed Claudin6 as an epiblast stem cell marker suggesting that the cell is differentiating. (a and b) Adapted with permission.60 Copyright 2013, American Association for the Advancement of Science (AAAS). (c) Comparison analysis of the cystic organization of embryoid bodies after 10 days of differentiation under the influence of soluble or microsphere-released RA. Untreated and RA-free microspheres as controls. RA-loaded microspheres induce large cysts within EBs not present when EBs are exposed to soluble RA. Adapted with permission.63 Copyright 2009, Elsevier Ltd. | ||
Biodegradable microspheres enable spatiotemporal control of biomolecule release contributing to recapitulate their regulation during early development.62,63 Poly(lactic-co-glycolic acid) (PLGA) microspheres loaded with RA can be incorporated within embryoid bodies. As the microspheres degrade, the RA induces highly organized cystic regions in a dose-dependent manner, since microspheres with low RA levels (0.3 μg mg−1 PLGA) display fewer cystic EBs than microspheres loaded with high RA levels (30 μg mg−1 PLGA). Interestingly, no cysts are observed across several concentrations of soluble RA (Fig. 4c), hence demonstrating the need for the spatially controlled presentation of the morphogen. The cystic spheroids phenotypically resemble the early streak mouse embryos (E6.75) and could be a useful model to investigate the role of secreted morphogenic factors during early differentiation events.63 Microfluidics approaches can also provide sustained release and gradients of soluble factors, however, currently they require significantly higher complexity and can only generate concentration gradients from outside the cell colony. Contrastingly microbeads enable inside-out gradients with spherical symmetry in a simple package. The material composition of the bead itself also influences cell fate, with equally sized microbeads of agarose, PLGA and gelatin influencing the differentiation proportions in PSCs 3D aggregates.64 These findings suggest that tailoring the physical properties of a bead can influence signalling to the stem cell aggregate and hence direct differentiation.65
Hydrogel and patterning
Bi-dimensional cultures can only mimic a part of the complex and highly three-dimensional scenario that gives rise to the development of a human embryo. Preliminary reports indicate that human ESCs grown on patterned substrates can originate asymmetrical ‘gastrulation-like’ nodes, a model that could dissect the influence of tension and confinement on regulating early development in vitro.66 Importantly, human ESC cultured in disc-shaped colonies within modified polyacrylamide hydrogels exhibit stiffness-dependent response to Wnt stimulation while retaining stiffness-independent self-renewal potential. Taken together these results demonstrate that the mechanical properties of the microenvironment alter the cellular response towards biomolecules influencing the differentiation potential, hence, such an approach can be useful in identifying mechanosensitive molecules and mechanisms regulating embryonic development.67 Extracellular interactions have a substantial influence on morphogenesis, differentiation and proliferation of cells as is well observed in 3D somatic cell reprogramming towards iPSCs.68 Therefore, recapitulating the spatiotemporal milieu of cell–cell interactions and extracellular matrix in vitro are key components needed to properly regulate stem cell fate.69 Three-dimensional microarrays enable high-throughput screening of matrix-initiated cues by combinatorially testing hydrogels parameters (composition, stiffness, bioactivity) to identify their role in stem cell regulation.70 In particular, mechanical forces within the microenvironment are known to regulate embryonic organization, and controlling hydrogel stiffness is a simple approach to regulate the mechano-environment of in vitro models of embryonic development. Indeed, controlling stiffness in fibrin gels regulates the establishment of concentric, organized formation of endoderm, mesoderm and ectoderm in 3D colonies, highlighting that colony-matrix interactions, as demonstrated by the variations in the stiffness, have a significant impact on the spatial patterning of the germ layers.71 These findings indicate that mechanical and biochemical cues act synergistically to recapitulate embryogenic-like processes.Dissecting the chemical and the physical cues guiding mammalian embryogenesis provides a lead to model early developmental events. Indeed, emerging hydrogel approaches enable independent regulation of stiffness, degradation and biochemical cues density allowing improved mimicking of in vivo niches.72 Besides controlling the biophysical environment, hydrogels play a crucial role in presenting matrix-bound biochemical cues, which complement the role of soluble factors. Matrigel can provide support for the co-culture of 3D aggregates of mouse embryonic and extraembryonic trophoblast stem cells enabling their mutual interaction (Fig. 5a). This leads to their fusion and co-development of a common lumen directed through Nodal signalling secreted by the embryonic stem cell compartment (Fig. 5b). Analysis of the symmetry breaking events in the embryo suggests that, unlike embryonic stem cells alone, the presence of the trophoblast stem cell compartment induces regionalized cells of mesoderm lineage which also express mesenchymal markers. Furthermore, Wnt signalling is essential for the expression of mesoderm markers and BMP signalling induces primordial germ-like cells at the boundary between the two compartments.73 The Matrigel provides cues that partially compensate for the visceral endoderm, which is necessary for the mouse ESC to progress towards lumenogenesis and epiblast polarization.74 Further development leads to the asymmetric induction of mesoderm markers similarly positioned to that of the natural embryo.73 Neural tube induction in 3D systems is possible by culturing single mouse ESCs suspensions in Matrigel under neural induction conditions where the cells differentiate along the neural lineage forming apical-basal neuroepithelial cysts with a single lumen. Remarkably, the addition of high concentrations of RA achieves neural tube dorsoventral patterning. PEG hydrogels, which ablate matrix biochemical cues yield similar results although to a lesser extent, indicating that matrix composition is not an essential requirement for such patterning.75 Further investigations on the conditions driving neural tube patterning focused on the combinatorial dissection of cues in the cell microenvironment, allowing elucidating the role of stiffness and extracellular matrix (ECM) composition on neuroepithelial cyst patterning.76 Therefore, this tractable approach enables deciphering some of the complexity that exists in the developmental processes.
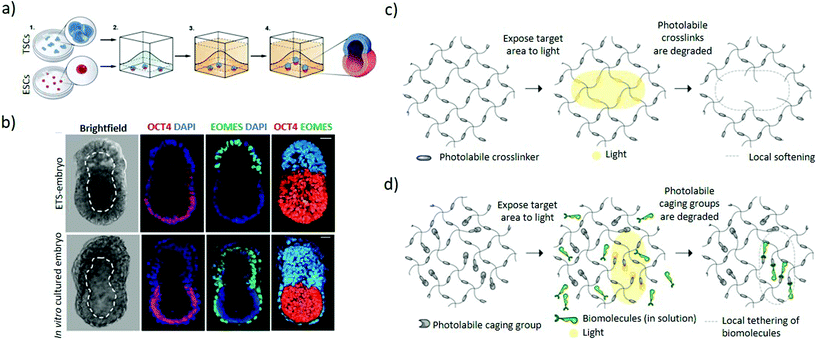 | ||
| Fig. 5 Hydrogel-based approaches to induce asymmetric embryonic microenvironments. (a) Schematic representation of the set-up used to mimic the epiblast architecture. Small clumps of trophoblast stem cells and single embryonic stem cells are suspended in Matrigel in order to establish a dialogue between them through their mutual interaction in a 3D extracellular matrix scaffold. The Matrigel is allowed to solidify followed by culturing in a suitable ETS-embryo medium for the stem cells types to fuse and codevelop into a common lumen. (b) Mouse embryonic stem cells and extraembryonic trophoblast stem cells co-embedded in Matrigel according to (a) interact and codevelop into a common lumen (top panel) resembling the blastocyst stage of mouse (bottom panel), as represented by the ETS-embryo images (upper panel). Staining includes Oct4 as embryonic marker, EOMES as extraembryonic marker and DAPI nuclear counterstaining. Dashed line outlines the cavity. Scale bars = 30 μm. (a and b) Adapted with permission.73 Copyright 2017, American Association for the Advancement of Science (AAAS). (c) Schematic representation of light-mediated patterning for spatially-resolved regulation of mechanical and biochemical signals. Photolabile crosslinkers cleave upon light interaction to locally soften the gel matrix. (d) Bioligands bound to the hydrogel with the active group masked by a photo-sensitive caging group are activated upon exposure to light, leading for patterned tethering of biomolecules. (c and d) Adapted with permission.82 Copyright 2014, The Company of Biologists. | ||
A 3D human ESCs model can achieve anteroposterior symmetry breaking in the absence of extra-embryonic tissues. A single-cell suspension-cultured into polymeric hydrogels supplemented with Matrigel and a precisely controlled uniform dose of soluble BMP4 induces regionalized expression of molecular signatures of the germ layers. In this system, low BMP4 concentrations (0.1 ng mL−1) yield SOX2+ expressing cells whilst high concentrations (5 or 10 ng mL−1) result in morphological and molecular properties changes of the cells, fitting the description of trophectoderm-like cells. Intermediate levels (1 ng mL−1) cause half of the colonies to generate a spatially separated expression of SOX2+ and BRA+ cells, serving as a human model for studying the mechanism leading towards anteroposterior symmetry breaking.77 Apart from modelling post-gastrulation processes, seeding the human pluripotent stem cells at the interface between a Geltrex ECM solution and soft gel bed enables modelling aspects of peri-implantation amniogenesis, a pre-gastrulation event. The physical cues from the soft gel are sufficient to induce squamous cysts, since reducing the thickness of the soft gel bed increases its rigidity and inhibits cysts development. However, in the absence of maternal and extraembryonic biochemical cues, only the combination of soft gel bed and Geltrex ECM induces a combination of transcription factors that resemble differentiation towards amnion-like tissue.78 Concurrently, a small fraction of cysts show asymmetric differentiation with amniotic ectoderm on one side and an opposing epiblast-like pole, resembling the post-implantation amniotic sac. In addition, post-implantation amniotic sac embryoids proceed to develop molecular hallmarks of the posterior primitive streak.79
Light-mediated patterning of hydrogels improves control over the spatiotemporal regulation of cues. Light patterning interacts with photosensitive chemical groups within hydrogels and can induce reversible or irreversible mechanical or chemical changes based on the light pattern (Fig. 1d).80–82 Patterning in the presence of photoinitiators can mediate increase in hydrogel stiffness by triggering additional local crosslinking within the gel.83 In contrast, hydrogels with embedded photolabile chemical groups are cleaved upon light irradiation, resulting in local softening of the gel (Fig. 5c).84,85 Additionally, biochemically patterned hydrogels provide fine 3D control over the distribution and availability of the signals to the cells through light-mediated approaches86 whereby the molecules are presented or removed through photocaging and photocleaving respectively. In photocaging, the bioligand is bound to the hydrogel with its active site masked by a photodegradable moiety and becomes activated upon light irradiation (Fig. 5d).87 Orthogonal photopatterning enables generating photoreversible hydrogels where bioactive proteins are first conjugated to the hydrogel by photopatterning and subsequently removed through the same mechanism, enabling sophisticated spatiotemporal control of extracellular matrix cues.88 These approaches could improve the localization of extracellular cues for differentiation which can promote symmetry-breaking events with spatial control. Nonetheless, many challenges remain to be addressed including identifying hydrogels that reversibly respond to light and improving the efficiency of visible-light photo-reversible and photolabile chemical groups to ensure integrity of the biomolecules and cytocompatibility.87
Although bioconjugation allows effective functionalization of biomolecules, it often reduces their biological activity. Non-covalent binding can provide effective biofunctionalisation with improved biomolecular activity. Several growth factors have a high affinity for heparin and micropatterned dual-crosslinked alginate/heparin hydrogels provide a means to sequester those growth factors in designated spatial locations. The immobilized growth factors can direct migration and differentiation of human mesenchymal stem cells (MSC) encapsulated within the micropatterns.89 However heparin-based systems are subjected to batch-to-batch variability which can hamper their application.90
Hydrogel-based approaches are a versatile tool to study complex and dynamic biological processes and can be applied to investigate phenomena occurring during early embryogenic development. While offering support to the culture, hydrogels can accurately orchestrate the extracellular chemical and physical cues in space and time, mimicking those in vivo. Thanks to the flexibility and ease of use of these approaches it is possible to combinatorially explore the matrix microenvironment during development to identify its effect on embryogenic events and processes.
Bioprinting
Bioprinting is a bottom-up approach to assembling tissues by precisely depositing bioinks, composed of living cells and biomaterials, onto a substrate thus crafting an artificial 3D architecture to mimic physiological organization (Fig. 1e).91 Crucially, bioprinting is a complex process with low throughput and variable cytotoxicity that allows for extremely precise spatial manipulation of cell arrangement.92 Broadly available bioprinting strategies include inkjet and microextrusion bioprinting, typically applicable for materials with low and high viscosities respectively. A less common technique is laser-assisted bioprinting which overcomes the issues of nozzle clogging and the limitations due to material viscosity.93,94 Bioinks can either print biomolecules to mimic the extracellular matrix, cells to provide their desired spatial arrangement or a combination thereof, integrating organisation of cells and matrix. Alternatively, modified microcarriers with bioactive molecules can be included in the bioink to promote lineage commitment.95Importantly, bioprinting has demonstrated its ability in handling stem cells and promoting EBs formation. For instance, a composite bioink composed of alginate, agarose and carboxymethyl-chitosan (CMC) forms a porous 3D scaffold supporting human neural stem cells differentiation to functional neurons and neuroglia.96 Similarly, this bioink can support hiPSCs differentiation. The stem cells maintain their proliferation within the printed construct after gelation and self-assemble to form large pluripotent spheroids. Culturing iPSCs in medium free of basic fibroblast growth factor and without any specific supplements causes the cells to differentiate towards the three germ lineages (Fig. 6a). Whilst culturing in neural induction medium causes EBs to differentiate into functionalised homogenous neural tissues, indicating that cells retain differentiation ability post-printing.97 Provided that there are few instances of iPSCs bioprinting, such a study lays out the initial steps in modelling cells of different lineages. Moreover, the variety of possible combination of cells, soluble factors, inks and spatial organisation, pave the way to high-throughput combinatorial investigations of their role in embryonic development.98 More specifically, valve-based bioprinting of hESCs and hiPSCs maintains their viability and pluripotency comparable to non-printed cells.99,100 Exposing the printed stem cells to hepatic differentiation conditions yields markers characteristic of hepatocyte-like cells.100 Valve-based bioprinting approaches offer very fine control over the number of cell and volumes of biomolecules printed with high spatial precision. Such level of manipulation can be leveraged to craft a variety of systems to investigate embryogenesis and apply these findings for meaningful advancement in tissue engineering.
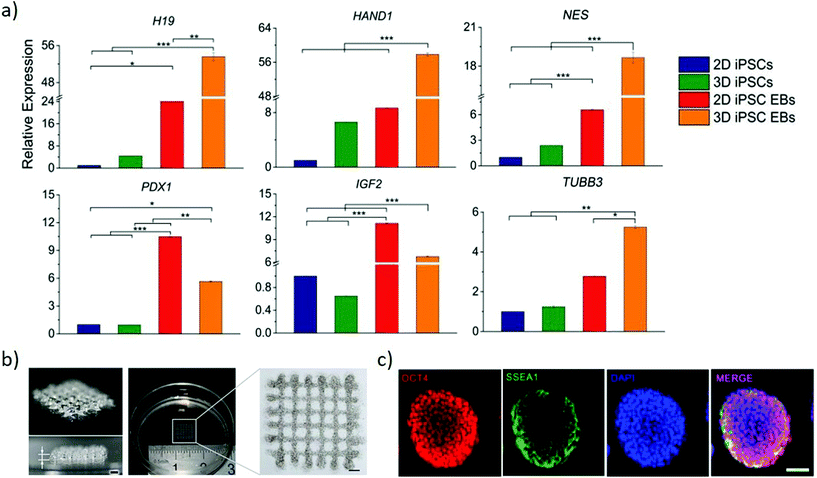 | ||
| Fig. 6 Bioprinting can generate arbitrary 3D stem cell arrangements. (a) Relative genes expression of endodermal markers (H19 and PDX1), mesodermal markers (HAND1 and IGF2) and ectodermal markers (NES and TUBB3) to compare conventional 2D cultured iPSCs and EBs with 3D printed gel-encapsulated iPSCs and EBs after 10 days iPSCs differentiation (15 days post-printing). Increased expression of all the markers emphasizes the potential for the bioprinted cells to differentiate. *P < 0.05; **P < 0.01; ***P < 0.001. Adapted with permission.97 Copyright 2017, Wiley-VCH. (b) Images of 3-D bioprinted grid-like cell-laden hydrogel construct to generate high throughput EBs. (c) Immunofluorescence staining of EBs grown in the 3-D bioprinted cell-laden hydrogel by proliferation (right panel). Oct4 and SSEA1 suggest their pluripotency is maintained at day 7 post-printing. (b and c) Adapted with permission.103 Copyright 2015, IOP publishing group. | ||
The size of EBs/colonies is a critical parameter that determines gene expression, thus influencing the differentiation of the cells. Regulating the shape and size of EB by culturing mouse ESC in concave microwell arrays of different widths yields large EBs preferentially differentiating towards neuronal and cardiomyocyte differentiation than small EBs.101 However, such an approach provides limited control in manipulating the microenvironment of the EBs within the microwells. Direct writing by laser bioprinting enables controlling both the printing density and colony size, allowing exploring these parameters independently. The final diameter of mouse EBs is dependent on the local cell density of the printed colony diameter, highlighting the influence of cell density in predicting and controlling EB size to obtain greater reproducibility during in vitro differentiation.102 A versatile approach generates high throughput EBs of uniform and timely controlled sizes by bioprinting a 3D cell-laden hydrogel system (Fig. 6b). Mouse ESCs are embedded uniformly within hydrogels and cultured for multiple days to obtain pluripotent EBs (Fig. 6c), that are formed through proliferation rather than aggregation. These approaches present a strong potential in generating a high precision tool to engineer EB of the desired size for its later application in differentiation studies and drug screening studies.102,103
Nanomaterials
Nanotechnology is prominently establishing approaches to transform drug delivery, biosensing and stem cell technologies, but its contribution to developing advanced models for dissecting embryonic development remains very limited. Yet, much of the functionality developed for disruptive nanomedicine approaches can translate into key advances for developmental models. Due to their size, nanomaterials display emergent properties with respect to their bulk counterpart, owing to their behaviour as quantum objects. Their dimensions are comparable to biomolecules and subcellular structures enabling direct interaction with the basic constituents of life. Their large specific surface area provides for extreme sensitivity to changes in their surroundings, enabling the design of devices that respond rapidly and efficiently to environmental changes. Overall the combination of these properties allows designing multifunctional nanomaterials with low impact on cell phenotype and capable of effective cellular delivery of multiple biological payloads simultaneously or in precise sequences, while controlling their spatiotemporal localisation either in response to external stimuli or in response to changes in the environment. Such multifunctionality enables delivering combinations of nucleic acids and other impermeable payloads to a precisely controlled pattern of cells with a desired temporal arrangement governed by either external factors or changes to the cellular environment.Using active targeting approaches against stem/differentiating cells markers it is possible to selectively deliver drugs/biologicals to specific cell subtypes, thus controlling their behaviour. The physicochemical properties of RNA nanoparticles can be finely tuned and engineered to favour therapeutic delivery of biomolecules, and a 3 way-junction (3WJ) motifs of RNA packaging has been developed to have a predictable and stable folding.104 Stable 3WJ motifs can assemble into 4 stranded RNA nanoparticles that can selectively bind markers of stemness such as CD133, and mediate RNA interference for gene silencing in a cell-specific fashion.105 Using similar approaches, potent regulators of pathways controlling embryogenesis, including notch, sonic hedgehog and TGF-β, can be specifically directed towards cells based on their surface markers.106,107 In addition, nanoparticles enable co-delivery of biological and non-biological agents which synergise to enhance the effectiveness of delivery and overall treatment.108 Effective co-delivery enables designing complex temporal arrangements of combinations of drugs and biologicals capable of improving the effectiveness of treatments and transferrable to replicate the complex temporal variations of stimuli occurring during embryogenesis.109
Nanomaterials can be designed to couple effectively with physical stimuli such as magnetic, electrical and optical fields to locally induce mechanical forces, increase in heat, and generation of free radicals that can induce biological responses in a spatiotemporally regulated fashion.110 Such interaction enables perturbing both biophysical and biochemical signalling pathways with fine spatiotemporal resolution. Magnetoplasmonic nanoparticles (MPNs) can target mechanoreceptors on the cell surface and be specifically pulled using a magnetic field to interrogate mechanoresponse with subcellular resolution. These mechanical stimuli can regulate the activation of developmentally-relevant pathways such as NOTCH signalling, regulate the spatial distribution of key effector proteins and trigger gene expression with a desired kinetic.111 Similar approaches have the potential to orchestrate changes in the mechanical environment around stem cells with very high spatial resolution.
Inorganic Janus nanoparticles can load both hydrophobic and hydrophilic drugs (docetaxel and doxorubicin hydrochloride) and release them in response to local environmental changes. Since the drugs can be release based on the local pH or by imposing an external near-infrared stimulus (Fig. 7a), these nanomaterials can provide both spatial and temporal control based on the localisation of the endogenous or exogenous stimulus.112 Enzyme-controlled drug delivery systems can be properly tuned to spatiotemporally regulate delivery by releasing morphogens to the target cells on demand. This approach enables releasing morphogens such as BMP2 in response to the proteases secreted by migrating hMSCs, resulting in locally-enhanced osteogenic differentiation.113 This approach also lends itself to combinatorial delivery of stimuli, as evidenced with the spatially-controlled delivery of BMP2 and VEGF stimuli to differentially induce osteogenesis and vasculogenesis towards the development of in vitro vascularised bone tissue. These combined abilities enable nanoparticles to control the delivery of biophysical and biochemical stimuli in space and time by dynamically responding to the changes occurring to cells, their environment or by means of externally applied stimuli. Such extensive dynamic control of stimuli indicates the strong versatility of nanoparticles for the engineering of microenvironments within models of embryogenesis.
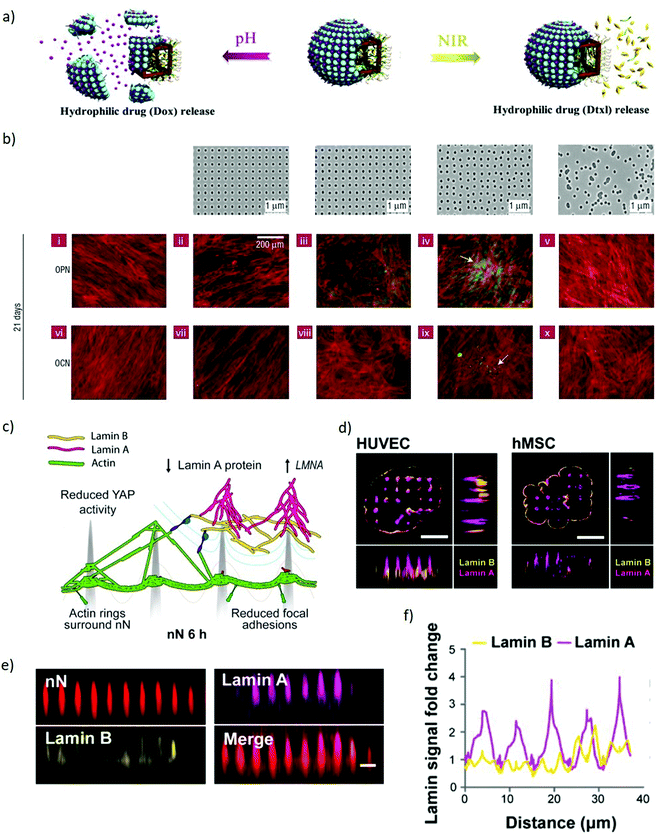 | ||
| Fig. 7 Nanomaterials spatiotemporally regulate biophysical and biochemical signalling demonstrating potential to manipulate models of embryogenesis. (a) A schematic representation of the co-encapsulation of docetaxel and doxorubicin hydrochloride drugs within Janus nanoparticles. The drugs are released independently based either on pH or NIR stimuli. Adapted with permission.112 Copyright 2018, Elsevier. (b) Upper panel, left to right: Flat control, nanotopographies with pits in a square array, pits in an array with random displacement by up to 20 nm from a square pattern (DSQ20, ±20 nm from true center), pits in a displaced square 50 array (DSQ50, ±50 nm from true center), and randomly arranged pits. Lower two panels: After 21 days in culture, only MSCs cultured on DSQ50 (iv and ix) exhibited both osteopontin/osteocalcin (OPN/OCN) positive regions (green) and nodule formation (indicated by the arrows), indicative of osteogenic differentiation. Red = actin, green OPN/OCN as per legend. Adapted with permission.114 Copyright 2007, Nature Publishing Group. (c) Schematic representation of a cell interacting with nanoneedles. The nuclear envelope wraps around the nanoneedles providing mechanical stimulation to multiple intracellular elements. Lamin A reorganises at the nuclear envelope. (d) SIM imaging of human umbilical vein endothelial cells (HUVEC) and human mesenchymal stem cells (hMSC) shows lamin A (magenta) accumulating preferentially at nanoneedles whereas lamin B (yellow) distributes uniformly across the cell membrane. Scale bars = 5 μm. (e) x–z plane from confocal z-stack images showing lamin A and lamin B organization on nanoneedles, scale bar = 2 μm. (f) Quantification of lamin A and B signal intensity distribution along the basal nuclear envelope. Lamin A increases at nanoneedle sites whereas lamin B remaines constant. (c–f) Adapted with permission.139 Copyright 2019, American Chemical Society. | ||
Alongside nanoparticles, extended nanostructures such as thin nanostructured films, nanotopographies and bidimensional arrays serve as effective interfaces to manipulate both the extracellular and intracellular environment. Nanotextured surfaces provide a spatiotemporal control over biophysical stimuli that can regulate stem cell fate, induce signalling clustering and control several phenotypical features including motility, proliferation, morphology and cytoskeletal arrangement. Small changes in the patterns of nanostructured surfaces can provide accurate control over the geometry and reprogramming of stem cells colonies. Slightly disordered nanotopographies induce osteogenic differentiation of hMSc which is not observed in ordered nanotopographies (Fig. 7b).114 Vertical silicon nanostructures counterintuitively promote the formation of mouse iPSCs spheroids, by encouraging cell–cell interaction in contrast with cell–substrate interaction.115 Carbon nanotubes networks promote human dermal fibroblasts reprogramming, demonstrating the ability of nanostructures to induce signs of pluripotency in somatic cells.116 In addition, depositing nanomaterials in a nanopattern on a substrate and attaching specific biomolecules to such nanomaterial results in patterned signalling biomolecules. Using such an approach identified that, by varying the nanospacing between the cell-adhesive peptide arginine-glycine-aspartic acid (RGD) resulted for MSC to exhibit a distinction in the differentiation extents.117
Electrically-active nanostructures, integrated into large scale arrays, can induce highly local electroporation in order to deliver payloads to desired combination of specific cells in a temporally-controlled fashion. Nanowires,118 nanostraws,119 nanoantennas120 and nanoholes121 are particularly well suited to this approach, thanks to the tight interfacing with cells resulting in effective and local poration with relatively small and short electrical pulses, that allow reliable delivery to individual cells while minimising cell toxicity. These systems can integrate reservoirs to enable sustained and repeated delivery of the desired payloads, and/or be integrated within microfluidics systems in order to control nutrient exchange and exposure to payload. This approach delivers nucleic acids, proteins and peptides intracellularly with the potential to generate complex spatiotemporal patterns of autocrine and paracrine signalling, which could orchestrate signals in stem cells.119,120,122–124 Furthermore, nanostraws and other nanoconduits125 can also rapidly sample intracellular fluid with single cell resolution, potentially enabling molecular mapping of complex models of embryogenesis. Alternatively, to electroporation, nanomaterials interacting with light can also induce local permeabilization of the cell membrane, thus enabling spatiotemporally patterned delivery. Nanomaterials interacting with focussed light can locally heat up the surrounding medium inducing local reversible permeabilization of the cell membrane that can be used to gain access to the intracellular environment.126 Plasmonic and meta-plasmonic materials can locally enhance optical fields, inducing ejection of electrons and mechano-acoustic waves in the close surroundings of the so called “hot spot” which also induce local membrane poration enabling intracellular delivery and recording of intracellular electrical activity.127–129 How electroporation and laser-mediated poration physically disrupt the cell membrane and perturb the cytosol around has been investigated,130 confirming the temporary and local modification of the cell membrane making the above approaches suitable to perturb stem cells system to study how different signalling pathways are influencing organogenesis. Alternatively, arrays of complex micromachines can be fabricated to allow controlled-release of multiple payloads on demand, by electrochemical dissolution of thin membranes that otherwise seal microreservoirs containing the desired molecules.131 Such elaborate devices can be applied to stem cells cultures to regulate the triggering of chosen pathways, for example to study how the presence of single signalling molecules affect the development of embryos in time.
When cultured on high aspect ratio nanostructured substrates, the cell membrane wraps around such nanostructures establishing a tight sealing.132–137,149 This physical stimulus results in a local curvature which may trigger the activation of mechanosensing pathways.138,139 Indeed, nanopillars influence the reorganization of intracellular actin leading to a reduction in the cytoskeletal tension140 hence, causing significant morphological alterations. Some nanopillars can also induce deformations in the nuclear envelope which modulates the expression of key mechanosensory regulators (Fig. 7c–f).139,141 In addition to mechanical stimuli, these high aspect ratio nanostructures also enable localised delivery of payloads, potentially highlighting an approach to control the spatial organisation of signalling in embryonic models.142,150 The mechanism of internalization is dominated by endocytic pathways stimulated by the interaction with the nanostructures, whilst physical spontaneous rupture of the cell membrane occurs when tension forces are coupled with high local curvatures of the nanostructures.137,143–147 This permeabilisation of the cell membrane allows the direct intracellular delivery of biomolecules, thus bypassing the cells’ physical barriers.142,151 Interestingly enough, these interaction with nanostructured substrates does not greatly affect the viability of the cells.148,149 This efficient and localised delivery capability can orchestrate signalling by patterned transfection of regulators of gene expression for key morphogens, enabling complex manipulations of stem cell colonies.
Outlook
Controlling symmetry-breaking by gaining precise spatio-temporal control over the physical and biochemical cues provided to the stem cells niche is a yet-unmet key component to accurately model in vitro early stages of embryogenesis and gastrulation. The biomaterials approaches highlighted here have contributed significant progress towards this ambitious goal, yet, there remain significant gaps as currently available technologies only achieve partial control over the organisation of embryoid models. Two-dimensional micropatterning can impact local biochemical signalling gradients, which induce a controlled stem cell differentiation pattern. This pattern can be partly regulated by controlling the size and shape of micropatterns. However, such control is limited, the model is two-dimensional and lacks temporal control over signalling cues, restricting the range of embryogenesis events that can be studied with this approach. Microfluidic devices can establish simple spatiotemporal gradients of soluble cues for 2D and 3D systems and they can modularly integrate with other approaches, such as micropatterning and hydrogels. Despite microfluidics efficacy at establishing patterning, the systems are complex to design and handle, can only achieve a subset of the desired gradients, and encounter challenges to handling tissue growth and long term maintenance. Hydrogels can control both mechanical and biochemical cues in 3D cell culture systems that are essential to collectively coordinate the early developmental organisation. Patterning hydrogels enables controlling the spatial organisation of extracellular mechanical and biochemical cues, providing a high degree of local control to cell patterning. Yet, controlling soluble cues through hydrogels is complex, and temporal regulation of cues is limited to either pre-programmed design through hydrogel degradation or impractical approaches such as reversible photopatterning. Microbeads are a simple and effective approach to present cells with asymmetrical soluble stimuli or to generate symmetrical gradients by allowing biodegradable spheres to release molecules in time. Microbeads elegantly address key challenges of establishing asymmetries by displaying signals to an individual cell or layer and outwards decaying gradients which are complex to achieve otherwise. However, they have very limited scope due to the fixed geometry which only allows displaying cues to cells on a spherical shell or establishing spherical gradients, which alone are insufficient to finely tune the chemical stimuli needed to recapitulate early stages of development. Bioprinting, although at its infant stages, is revealing an extremely powerful tool to shape tissue and embryoid bodies with high spatial and temporal control over the features of the system. Admittedly, bioprinted iPSCs retain their differentiation ability, however, in many instances cell survival, their precise placement and the low throughput of the approaches remain key barriers requiring refinement in order to recreate the microenvironmental conditions directing the stem cells to differentiate to the germ lineages in an organized manner.We highlighted how untapped nanotechnology approaches could address some of the challenges arising from current biomaterials approach to designing niches that break symmetry in models of organogenesis. Nanomaterials can very effectively control the spatial and temporal distribution of soluble cues, using strategies developed with nano delivery vectors for cancer therapy. The topography and composition of nanomaterials can also be tightly spatially controlled and in some instances regulated in time, and their effect as biophysical cues to regulate stem cells differentiation have been established in multiple non-embryonic settings. Indeed, nanomaterials have been extensively effective at delivering biological payloads, including morphogens, with high spatial and temporal control, and they can be programmed to target specific cells, actively respond to environmental cues or be directed by external stimuli. These combined features can enable highly sophisticated design of spatiotemporal arrangement of cues, with unprecedented nanoscale resolution. This improved control and higher resolution over the patterning of cues achievable with nanomaterials, in turn can translate into regulation of stem cell fate approaching single cell resolution. Such sophisticated stem cell control would enable establishing highly localised phenotypes and investigating their role in organising patterning. Arguably, incorporating nanomaterials more effectively within the toolbox available to study embryogenesis can significantly advance our understanding embryonic development.
Conclusion
Understanding and directing the biophysical and biochemical processes regulating human embryogenesis is a very ambitious goal but it is essential to develop tools for designing tissue architecture and functions to be deployed for regenerative medicine. Only with an improved picture of the complex signalling network regulating tissue development and homeostasis, we can develop sufficiently effective bioengineered organs and personalised treatments for chronic and degenerative pathologies. However, ethical restrictions limit experimentation beyond the pre-gastrulation stages and as a result, animal models have been used as a substitute to gain insight on morphogenesis. Yet, differences between animal and human models persist, potentially leading to inaccurate or ineffective hypotheses. The rapidly developing in vitro stem cells technologies have provided a powerful means to decipher key pathways in early human development. Biomaterials have contributed to address challenges arising with instructing stem cell organization to mimic human morphogenesis leading to the development of improved models that can accurately mimic complex in vivo behaviours. Indeed, the broad array of tools available across micropatterning, microfluidics, hydrogels, microbeads and bioprinting approaches provides great flexibility to address the complex and multifactorial issue of designing accurate developmental niches for stem cell colonies. Certainly, integrating multiple approaches is beneficial to enhance the functionality, robustness and versatility of the models. Nevertheless, the available technologies still cannot provide a complete and comprehensive understanding of human early development stages and requiring key advances in our ability to control the spatiotemporal regulation of biophysical and biochemical soluble and immobilised cues which can be provided by combining both emerging technologies to manipulate individual cells and by lateral translation of mature technologies developed in other areas of bioengineering. Indeed, multiple approaches highlight a way towards the application of nanomaterials as rationally designed platforms to locally perturb the microenvironment in order to regulate stem cells organisation, differentiation and migration during the early stages of embryogenesis. The unsurpassed capability of nanomaterials for simultaneous regulation of multiple biochemical and biophysical cues with high spatiotemporal resolution indicates their potential to address outstanding challenges in orchestrating signalling for models of embryogenesis where multifactorial signalling networks can be artificially simulated in order to mimic physiological and pathological development.Conflicts of interest
There are no conflicts to declare.References
- N. S. Macklon, J. P. Geraedts and B. C. Fauser, Conception to ongoing pregnancy: The ‘black box’ of early pregnancy loss, Hum. Reprod. Update, 2002, 8, 333–343 CrossRef CAS.
- C. E. Murry and G. Keller, Differentiation of Embryonic Stem Cells to Clinically Relevant Populations: Lessons from Embryonic Development, Cell, 2008, 132, 661–680 CrossRef CAS.
- S. Nishikawa, L. M. Jakt and T. Era, Embryonic stem-cell culture as a tool for developmental cell biology, Nat. Rev. Mol. Cell Biol., 2007, 8, 502–507 CrossRef CAS.
- A. Piliszek, J. B. Grabarek, S. R. Frankenberg and B. Plusa, Cell fate in animal and human blastocysts and the determination of viability, Mol. Hum. Reprod., 2016, 22, 681–690 CrossRef CAS.
- J. Rossant, Mouse and human blastocyst-derived stem cells: Vive les differences, Development, 2015, 142, 9–12 CrossRef CAS.
- M. Roode, K. Blair, P. Snell, K. Elder, S. Marchant, A. Smith and J. Nichols, Human hypoblast formation is not dependent on FGF signalling, Dev. Biol., 2012, 361, 358–363 CrossRef CAS.
- M. Linneberg-Agerholm, Y. F. Wong, J. A. R. Herrera, R. S. Monteiro, K. G. V. Anderson and J. M. Brickman, Naïve human pluripotent stem cells respond to Wnt, Nodal and LIF signalling to produce expandable naïve extra-embryonic endoderm, Development, 2019, 146 CrossRef CAS , dev.180620.
- E. W. Kuijk, L. T. A. van Tol, H. van de Velde, R. Wubbolts, M. Welling, N. Geijsen and B. A. J. Roelen, The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos, Development, 2012, 139, 871–882 CrossRef CAS.
- T. Brevini and G. Pennarossa, in Gametogenesis, Early Embryo Developmental and Stem Cell derivation, Springer, 2013, pp. 32–35 Search PubMed.
- M. M. Shen, Nodal signaling: Development roles and regulation, Development, 2007, 134, 1023–1034 CrossRef CAS.
- S. M. Chambers, C. A. Fasano, E. P. Papapetrou, M. Tomishima, M. Sadelain and L. Studer, Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling, Nat. Biotechnol., 2009, 27, 275–280 CrossRef CAS.
- K. Taniguchi, I. Heemskerk and D. L. Gumucio, Opening the black box: Stem cell-based modeling of human post-implantation development, J. Cell Biol., 2019, 218, 410–421 CrossRef CAS.
- M. N. Shahbazi and M. Zernicka-goetz, Deconstructing and reconstructing the mouse and human early embryo, Nat. Cell Biol., 2018, 20, 878–887 CrossRef CAS.
- M. N. Shahbazi, A. Jedrusik, S. Vuoristo, G. Recher, A. Hupalowska, V. Bolton, N. M. E. Fogarty, A. Campbell, L. G. Devito, D. Ilic, Y. Khalaf, K. K. Niakan, S. Fishel and M. Zernicka-Goetz, Self-organization of the human embryo in the absence of maternal tissues, Nat. Cell Biol., 2016, 18, 700–708 CrossRef CAS.
- A. Deglincerti, G. F. Croft, L. N. Pietila, M. Zernicka-goetz, E. D. Siggia and A. H. Brivanlou, Self-organization of the in vitro attached human embryo, Nature, 2016, 533, 251–254 CrossRef CAS.
- M. F. Pera, Human embryo research and the 14-day rule, Development, 2017, 144, 1923–1925 CrossRef CAS.
- M. F. Pera, G. De Wert, W. Dondorp, R. Lovell-badge, C. L. Mummery, M. Munsie and P. P. Tam, What if stem cells turn into embryos in a dish?, Nat. Methods, 2015, 12, 917–919 CrossRef CAS.
- J. A. Thomson, Embryonic stem cell lines derived from human blastocysts, Science, 1998, 282, 1145–1147 CrossRef CAS.
- K. Takahashi, K. Tanabe, M. Ohnuki, M. Narita, T. Ichisaka, K. Tomoda and S. Yamanaka, Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors, Cell, 2007, 131, 861–872 CrossRef CAS.
- V. Volarevic, B. S. Markovic, M. Gazdic, A. Volarevic, N. Jovicic, N. Arsenijevic, L. Armstrong, V. Djonov, M. Lako and M. Stojkovic, Ethical and safety issues of stem cell-based therapy, Int. J. Med. Sci., 2018, 15, 36–45 CrossRef CAS.
- M. S. MacAuley, P. R. Crocker and J. C. Paulson, Siglec-mediated regulation of immune cell function in disease, Nat. Rev. Immunol., 2014, 14, 653–666 CrossRef CAS.
- J. Itskovitz-Eldor, M. Schuldiner, D. Karsenti, A. Eden, O. Yanuka, M. Amit, H. Soreq and N. Benvenisty, Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers., Mol. Med., 2000, 6, 88–95 CAS.
- I. Heemskerk, K. Burt, M. Miller, S. Chhabra, M. C. Guerra, L. Liu and A. Warmflash, Rapid changes in morphogen concentration control self-organized patterning in human embryonic stem cells, eLife, 2019, 8, 1–28 CrossRef.
- A. Sagner and J. Briscoe, Morphogen interpretation: concentration, time, competence, and signaling dynamics, Wiley Interdiscip. Rev.: Dev. Biol., 2017, 6, 1–19 Search PubMed.
- A. A. Anlaş and C. M. Nelson, Tissue mechanics regulates form, function, and dysfunction, Curr. Opin. Cell Biol., 2018, 54, 98–105 CrossRef.
- C. J. Chan, C. P. Heisenberg and T. Hiiragi, Coordination of Morphogenesis and Cell-Fate Specification in Development, Curr. Biol., 2017, 27, R1024–R1035 CrossRef CAS.
- S. J. Arnold and E. J. Robertson, Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo, Nat. Rev. Mol. Cell Biol., 2009, 10, 91–103 CrossRef CAS.
- D. Berge, W. Koole, C. Fuerer, M. Fish, E. Eroglu and R. Nusse, Article Wnt Signaling Mediates Self-Organization and Axis Formation in Embryoid Bodies, Cell Stem Cell, 2008, 2, 508–518 CrossRef.
- L. Beccari, N. Moris, M. Girgin, D. A. Turner, P. Baillie-Johnson, A. C. Cossy, M. P. Lutolf, D. Duboule and A. M. Arias, Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids, Nature, 2018, 562, 272–276 CrossRef CAS.
- S. C. Van Den Brink, P. Baillie-johnson, T. Balayo, A. Hadjantonakis, S. Nowotschin, D. A. Turner and A. M. Arias, Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells, Development, 2014, 141, 4231–4242 CrossRef CAS.
- D. A. Turner, M. Girgin, L. Alonso-Crisostomo, V. Trivedi, P. Baillie-Johnson, C. R. Glodowski, P. C. Hayward, J. Collignon, C. Gustavsen, P. Serup, B. Steventon, M. P. Lutolf and A. M. Arias, Anteroposterior polarity and elongation in the absence of extraembryonic tissues and of spatially localised signalling in gastruloids: Mammalian embryonic organoids, Development, 2017, 144, 3894–3906 CrossRef CAS.
- M. Tewary, J. Ostblom, L. Prochazka, T. Zulueta-coarasa and N. Shakiba, A stepwise model of reaction-diffusion and positional information governs self-organized human peri-gastrulation-like patterning, Development, 2017, 144, 4298–4312 CrossRef CAS.
- M. Théry, Micropatterning as a tool to decipher cell morphogenesis and functions, J. Cell Sci., 2010, 123, 4201–4213 CrossRef.
- D. Falconnet, G. Csucs, H. M. Grandin and M. Textor, Surface engineering approaches to micropattern surfaces for cell-based assays, Biomaterials, 2006, 27, 3044–3063 CrossRef CAS.
- S. G. Higgins, M. Becce, A. Belessiotis-Richards, H. Seong, J. E. Sero and M. M. Stevens, High-Aspect-Ratio Nanostructured Surfaces as Biological Metamaterials, Adv. Mater., 2020, 32, 1–44 CrossRef.
- S. M. Morgani, J. J. Metzger, J. Nichols, E. D. Siggia and A. K. Hadjantonakis, Micropattern differentiation of mouse pluripotent stem cells recapitulates embryo regionalized cell fate patterning, eLife, 2018, 7, 1–35 CrossRef.
- R. Peerani, B. M. Rao, C. Bauwens, T. Yin, G. A. Wood, A. Nagy, E. Kumacheva and P. W. Zandstra, Niche-mediated control of human embryonic stem cell self-renewal and differentiation, EMBO J., 2007, 26, 4744–4755 CrossRef CAS.
- L. Haoran, R. Peerani, M. Ungrin, C. Joshi, E. Kumacheva and P. W. Zandstra, Micropatterning of human embryonic stem cells dissects the mesoderm and endoderm lineages, Stem Cell Res., 2009, 2, 155–162 CrossRef.
- A. Warmflash, B. Sorre, F. Etoc, E. D. Siggia and A. H. Brivanlou, A method to recapitualte early embryonic spatial patterning in human embryonic stem cells, Nat. Methods, 2014, 11, 847–854 CrossRef CAS.
- F. Etoc, J. Metzger, A. Ruzo, M. Z. Ozair, A. H. Brivanlou, E. D. Siggia, F. Etoc, J. Metzger, A. Ruzo, C. Kirst, A. Yoney, M. Z. Ozair and A. H. Brivanlou, A Balance between Secreted Inhibitors and Edge Article A Balance between Secreted Inhibitors and Edge Sensing Controls Gastruloid Self-Organization, Dev. Cell, 2016, 39, 1–14 CrossRef.
- G. Blin, D. Wisniewski, C. Picart, M. Thery, M. Puceat and S. Lowell, Geometrical confinement controls the asymmetric patterning of brachyury in cultures of pluripotent cells, Development, 2018, 145, 166025 CrossRef.
- X. Xue, Y. Sun, A. M. Resto-Irizarry, Y. Yuan, K. M. Aw Yong, Y. Zheng, S. Weng, Y. Shao, Y. Chai, L. Studer and J. Fu, Mechanics-guided embryonic patterning of neuroectoderm tissue from human pluripotent stem cells, Nat. Mater., 2018, 17, 633–641 CrossRef CAS.
- J. Xing, Y. C. Toh, S. Xu and H. Yu, A method for human teratogen detection by geometrically confined cell differentiation and migration, Sci. Rep., 2015, 5, 1–13 Search PubMed.
- D. Qin, Y. Xia and G. M. Whitesides, Soft lithography for micro- and nanoscale patterning, Nat. Protoc., 2010, 5, 491–502 CrossRef CAS.
- L. Przybyla and J. Voldman, Probing embryonic stem cell autocrine and paracrine signaling using microfluidics, Annu. Rev. Anal. Chem., 2012, 5, 293–315 CrossRef CAS.
- K. F. Sonnen and C. A. Merten, Microfluidics as an Emerging Precision Tool in Developmental Biology, Dev. Cell, 2019, 48, 293–311 CrossRef CAS.
- D. M. Titmarsh, J. E. Hudson, A. Hidalgo, A. G. Elefanty, E. G. Stanley, E. J. Wolvetang and J. J. Cooper-White, Microbioreactor Arrays for Full Factorial Screening of Exogenous and Paracrine Factors in Human Embryonic Stem Cell Differentiation, PLoS One, 2012, 7, e52405 CrossRef CAS.
- K. Blagovic, L. Y. Kim and J. Voldman, Microfluidic perfusion for regulating diffusible signaling in stem cells, PLoS One, 2011, 6, e22892 CrossRef CAS.
- Kshitiz, D. H. Kim, D. J. Beebe and A. Levchenko, Micro- and nanoengineering for stem cell biology: The promise with a caution, Trends Biotechnol., 2011, 29, 399–408 CrossRef CAS.
- T. Frank and S. Tay, Flow-switching allows independently programmable, extremely stable, high-throughput diffusion-based gradients, Lab Chip, 2013, 13, 1273–1281 RSC.
- J. Y. Park, S. K. Kim, D. H. Woo, E. J. Lee, J. H. Kim and S. H. Lee, Differentiation of neural progenitor cells in a microfluidic chip-generated cytokine gradient, Stem Cells, 2009, 27, 2646–2654 CrossRef CAS.
- C. J. Demers, P. Soundararajan, P. Chennampally, G. A. Cox, J. Briscoe, S. D. Collins and R. L. Smith, Development-on-chip: In vitro neural tube patterning with a microfluidic device, Development, 2016, 143, 1884–1892 CrossRef CAS.
- M. Hashiguchi and M. C. Mullins, Anteroposterior and dorsoventral patterning are coordinated by an identical patterning clock, Development, 2013, 140, 1970–1980 CrossRef CAS.
- Y. Zheng, X. Xue, Y. Shao, S. Wang, S. N. Esfahani, Z. Li, J. M. Muncie, J. N. Lakins, V. M. Weaver, D. L. Gumucio and J. Fu, Controlled modelling of human epiblast and amnion development using stem cells, Nature, 2019, 573, 421–425 CrossRef CAS.
- A. Manfrin, Y. Tabata, E. R. Paquet, A. R. Vuaridel, F. R. Rivest, F. Naef and M. P. Lutolf, Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells, Nat. Methods, 2019, 16, 640–648 CrossRef CAS.
- S. Suri, A. Singh, A. H. Nguyen, A. M. Bratt-Leal, T. C. McDevitt and H. Lu, Microfluidic-based patterning of embryonic stem cells for in vitro development studies, Lab Chip, 2013, 13, 4617–4624 RSC.
- S. Cosson and M. P. Lutolf, Hydrogel microfluidics for the patterning of pluripotent stem cells, Sci. Rep., 2014, 4, 1–6 Search PubMed.
- S. G. M. Uzel, O. C. Amadi, T. M. Pearl, R. T. Lee, P. T. C. So and R. D. Kamm, Simultaneous or Sequential Orthogonal Gradient Formation in a 3D Cell Culture Microfluidic Platform, Small, 2016, 12, 612–622 CrossRef CAS.
- K. Alberti, R. E. Davey, K. Onishi, S. George, K. Salchert, F. P. Seib, M. Bornhäuser, T. Pompe, A. Nagy, C. Werner and P. W. Zandstra, Functional immobilization of signaling proteins enables control of stem cell fate, Nat. Methods, 2008, 5, 645–650 CrossRef CAS.
- S. J. Habib, B. C. Chen, F. C. Tsai, K. Anastassiadis, T. Meyer, E. Betzig and R. Nusse, A localized Wnt signal orients asymmetric stem cell division in vitro, Science, 2013, 339, 1445–1448 CrossRef CAS.
- M. Lowndes, S. Junyent and S. J. Habib, Constructing cellular niche properties by localized presentation of Wnt proteins on synthetic surfaces, Nat. Protoc., 2017, 12, 1498–1512 CrossRef CAS.
- P. N. Dang, N. Dwivedi, L. M. Phillips, X. Yu, S. Herberg, C. Bowerman, L. D. Solorio, W. L. Murphy and E. Alsberg, Controlled Dual Growth Factor Delivery From Microparticles Incorporated Within Human Bone Marrow-Derived Mesenchymal Stem Cell Aggregates for Enhanced Bone Tissue Engineering via Endochondral Ossification, Stem Cells Transl. Med., 2016, 5, 206–217 CrossRef CAS.
- R. L. Carpenedo, A. M. Bratt-Leal, R. A. Marklein, S. A. Seaman, N. J. Bowen, J. F. McDonald and T. C. McDevitt, Homogeneous and organized differentiation within embryoid bodies induced by microsphere-mediated delivery of small molecules, Biomaterials, 2009, 30, 2507–2515 CrossRef CAS.
- A. M. Bratt-Leal, R. L. Carpenedo, M. D. Ungrin, P. W. Zandstra and T. C. McDevitt, Incorporation of biomaterials in multicellular aggregates modulates pluripotent stem cell differentiation, Biomaterials, 2011, 32, 48–56 CrossRef CAS.
- M. A. Kinney and T. C. McDevitt, Emerging strategies for spatiotemporal control of stem cell fate and morphogenesis, Trends Biotechnol., 2013, 31, 78–84 CrossRef CAS.
- J. M. Muncie, N. M. E. Ayad, J. N. Lakins and V. M. Weaver, Mechanics regulate human embryonic stem cell self-organization to specify mesoderm, bioRxiv, 2020, 53, 1–9 Search PubMed.
- L. Przybyla, J. N. Lakins and V. M. Weaver, Tissue Mechanics Orchestrate Wnt-Dependent Human Embryonic Stem Cell Differentiation, Cell Stem Cell, 2016, 19, 462–475 CrossRef CAS.
- M. Caiazzo, Y. Okawa, A. Ranga, A. Piersigilli, Y. Tabata and M. P. Lutolf, Defined three-dimensional microenvironments boost induction of pluripotency, Nat. Mater., 2016, 15, 344–352 CrossRef CAS.
- S. A. Ferreira, M. S. Motwani, P. A. Faull, A. J. Seymour, T. T. L. Yu, M. Enayati, D. K. Taheem, C. Salzlechner, T. Haghighi, E. M. Kania, O. P. Oommen, T. Ahmed, S. Loaiza, K. Parzych, F. Dazzi, O. P. Varghese, F. Festy, A. E. Grigoriadis, H. W. Auner, A. P. Snijders, L. Bozec and E. Gentleman, Bi-directional cell-pericellular matrix interactions direct stem cell fate, Nat. Commun., 2018, 9, 1–12 CrossRef.
- A. Ranga, S. Gobaa, Y. Okawa, K. Mosiewicz, A. Negro and M. P. Lutolf, 3D niche microarrays for systems-level analyses of cell fate, Nat. Commun., 2014, 5, 1–10 Search PubMed.
- Y. Poh, J. Chen, Y. Hong, H. Yi, S. Zhang, J. Chen, D. C. Wu, L. Wang, Q. Jia, R. Singh, W. Yao, Y. Tan, A. Tajik, T. S. Tanaka and N. Wang, Generation of organized germ layers from a single mouse embryonic stem cell, Nat. Commun, 2014, 5, 4000 CrossRef CAS.
- G. M. Jowett, M. D. A. Norman, T. T. L. Yu, P. R. Arévalo, D. Hoogland, S. Lust, E. Read, E. Hamrud, N. J. Walters, U. Niazi, M. W. H. Chung, D. Marciano, O. S. Omer, T. Zabinski, D. Danovi, G. M. Lord, J. Hilborn, N. D. Evans, C. A. Dreiss, L. Bozec, O. P. Oommen, C. D. Lorenz, R. M. da Silva, J. F. Neves and E. Gentleman, ILC1-derived TGFβ1 drives intestinal remodelling, 2020, DOI:10.1101/2020.04.20.051805.
- S. E. Harrison, B. Sozen, N. Christodoulou, C. Kyprianou and M. Zernicka-goetz, Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro, Science, 2017, 356, eaal1810 CrossRef.
- S. E. Harrison, B. Sozen and M. Zernicka-Goetz, In vitro generation of mouse polarized embryo-like structures from embryonic and trophoblast stem cells, Nat. Protoc., 2018, 13, 1586–1602 CrossRef CAS.
- A. Meinhardt, D. Eberle, A. Tazaki, A. Ranga, M. Niesche, M. Wilsch-Bräuninger, A. Stec, G. Schackert, M. Lutolf and E. M. Tanaka, 3D reconstitution of the patterned neural tube from embryonic stem cells, Stem Cell Rep., 2014, 3, 987–999 CrossRef.
- A. Ranga, M. Girgin, A. Meinhardt, D. Eberle, M. Caiazzo, E. M. Tanaka and M. P. Lutolf, Neural tube morphogenesis in synthetic 3D microenvironments, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E6831–E6839 CrossRef CAS.
- M. Simunovic, J. J. Metzger, F. Etoc, A. Yoney, A. Ruzo, I. Martyn, G. Croft, D. S. You, A. H. Brivanlou and E. D. Siggia, A 3D model of a human epiblast reveals BMP4-driven symmetry breaking, Nat. Cell Biol., 2019, 21, 900–910 CrossRef CAS.
- Y. Shao, K. Taniguchi, K. Gurdziel, R. F. Townshend, X. Xue, K. M. A. Yong, J. Sang, J. R. Spence, D. L. Gumucio and J. Fu, Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche, Nat. Mater., 2017, 16, 419–427 CrossRef CAS.
- Y. Shao, K. Taniguchi, R. F. Townshend, T. Miki, D. L. Gumucio and J. Fu, A pluripotent stem cell-based model for post-implantation human amniotic sac development, Nat. Commun., 2017, 8, 1–15 CrossRef CAS.
- W. He, M. Reaume, M. Hennenfent, B. P. Lee and R. Rajachar, Biomimetic hydrogels with spatial- and temporal-controlled chemical cues for tissue engineering, Biomater. Sci., 2020, 8, 3248–3269 RSC.
- T. E. Brown and K. S. Anseth, Spatiotemporal hydrogel biomaterials for regenerative medicine, Chem. Soc. Rev., 2017, 46, 6532–6552 RSC.
- N. Gjorevski, A. Ranga and M. P. Lutolf, Bioengineering approaches to guide stem cell-based organogenesis, Development, 2014, 141, 1794–1804 CrossRef CAS.
- M. Guvendiren and J. A. Burdick, Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics, Nat. Commun., 2012, 3, 792 CrossRef.
- A. M. Kloxin, A. M. Kasko, C. N. Salinas and K. S. Anseth, Photodegradable hydrogels for dynamic tuning of physical and chemical properties, Science, 2009, 324, 59–63 CrossRef CAS.
- M. Lunzer, L. Shi, O. G. Andriotis, P. Gruber, M. Markovic, P. J. Thurner, D. Ossipov, R. Liska and A. Ovsianikov, A Modular Approach to Sensitized Two-Photon Patterning of Photodegradable Hydrogels, Angew. Chem., Int. Ed., 2018, 57, 15122–15127 CrossRef CAS.
- X. Yin, B. E. Mead, H. Safaee, R. Langer, J. M. Karp and O. Levy, Engineering Stem Cell Organoids, Cell Stem Cell, 2016, 18, 25–38 CrossRef CAS.
- L. Li, J. M. Scheiger and P. A. Levkin, Design and Applications of Photoresponsive Hydrogels, Adv. Mater., 2019, 31, 1807333 CrossRef.
- C. A. Deforest and D. A. Tirrell, A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels, Nat. Mater., 2015, 14, 523–531 CrossRef CAS.
- O. Jeon, K. Lee and E. Alsberg, Spatial Micropatterning of Growth Factors in 3D Hydrogels for Location-Specific Regulation of Cellular Behaviors, Small, 2018, 14, 1–6 Search PubMed.
- L. C. Bahlmann, A. Fokina and M. S. Shoichet, Dynamic bioengineered hydrogels as scaffolds for advanced stem cell and organoid culture, MRS Commun., 2017, 7, 472–486 CrossRef CAS.
- S. A. Irvine and S. S. Venkatraman, Bioprinting and differentiation of stem cells, Molecules, 2016, 21, 1188 CrossRef.
- P. Zhuang, A. X. Sun, J. An, C. K. Chua and S. Y. Chew, 3D neural tissue models: From spheroids to bioprinting, Biomaterials, 2018, 154, 113–133 CrossRef CAS.
- S. V. Murphy and A. Atala, 3D bioprinting of tissues and organs, 2014, vol. 32, pp. 773–785 Search PubMed.
- I. Matai, G. Kaur, A. Seyedsalehi and A. Mcclinton, Biomaterials Progress in 3D bioprinting technology for tissue/organ regenerative engineering, Biomaterials, 2020, 226, 119536 CrossRef CAS.
- S. D. Eswaramoorthy, S. Ramakrishna and S. N. Rath, Recent advances in three-dimensional bioprinting of stem cells, J. Tissue Eng. Regen. Med., 2019, 13, 908–924 CAS.
- Q. Gu, E. Tomaskovic-Crook, R. Lozano, Y. Chen, R. M. Kapsa, Q. Zhou, G. G. Wallace and J. M. Crook, Functional 3D Neural Mini-Tissues from Printed Gel-Based Bioink and Human Neural Stem Cells, Adv. Healthcare Mater., 2016, 5, 1429–1438 CrossRef CAS.
- Q. Gu, E. Tomaskovic-Crook, G. G. Wallace and J. M. Crook, 3D Bioprinting Human Induced Pluripotent Stem Cell Constructs for In Situ Cell Proliferation and Successive Multilineage Differentiation, Adv. Healthcare Mater., 2017, 6, 1–11 Search PubMed.
- F. Xu, B. Sridharan, S. Wang, U. A. Gurkan, B. Syverud and U. Demirci, Embryonic stem cell bioprinting for uniform and controlled size embryoid body formation, Biomicrofluidics, 2011, 5, 1–8 Search PubMed.
- A. Faulkner-Jones, C. Fyfe, D. J. Cornelissen, J. Gardner, J. King, A. Courtney and W. Shu, Bioprinting of human pluripotent stem cells and their directed differentiation into hepatocyte-like cells for the generation of mini-livers in 3D, Biofabrication, 2015, 7, 044102 CrossRef.
- A. Faulkner-jones, S. Greenhough, J. A. King, J. Gardner, A. Courtney and W. Shu, Development of a valve-based cell printer for the formation of human embryonic stem cell spheroid aggregates, Biofabrication, 2013, 5, 015013 CrossRef.
- Y. Y. Choi, B. G. Chung, D. H. Lee, A. Khademhosseini, J. H. Kim and S. H. Lee, Controlled-size embryoid body formation in concave microwell arrays, Biomaterials, 2010, 31, 4296–4303 CrossRef CAS.
- A. D. Dias, A. M. Unser, Y. Xie, D. B. Chrisey and D. T. Corr, Generating size-controlled embryoid bodies using laser direct-write, Biofabrication, 2014, 6, 025007 CrossRef CAS.
- L. Ouyang, R. Yao, S. Mao, X. Chen, J. Na and W. Sun, Three-dimensional bioprinting of embryonic stem cells directs highly uniform embryoid body formation, Biofabrication, 2015, 7, 44101 CrossRef.
- D. Shu, E. F. Khisamutdinov, L. Zhang and P. Guo, Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor, Nucleic Acids Res., 2014, 42, 1–9 CrossRef.
- H. Yin, G. Xiong, S. Guo, C. Xu, R. Xu, P. Guo and D. Shu, Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133, Mol. Ther., 2019, 27, 1252–1261 CrossRef CAS.
- V. Chenna, C. Hu, D. Pramanik, B. T. Aftab, C. Karikari, N. R. Campbell, S. M. Hong, M. Zhao, M. A. Rudek, S. R. Khan, C. M. Rudin and A. Maitra, A polymeric nanoparticle encapsulated small-molecule inhibitor of Hedgehog signaling (NanoHHI) bypasses secondary mutational resistance to smoothened antagonists, Mol. Cancer Ther., 2012, 11, 165–173 CrossRef CAS.
- R. K. Verma, W. Yu, S. P. Singh, S. Shankar and R. K. Srivastava, Anthothecol-encapsulated PLGA nanoparticles inhibit pancreatic cancer stem cell growth by modulating sonic hedgehog pathway, Nanomedicine, 2015, 11, 2061–2070 CrossRef CAS.
- Z. Q. Zuo, K. G. Chen, X. Y. Yu, G. Zhao, S. Shen, Z. T. Cao, Y. L. Luo, Y. C. Wang and J. Wang, Promoting tumor penetration of nanoparticles for cancer stem cell therapy by TGF-β signaling pathway inhibition, Biomaterials, 2016, 82, 48–59 CrossRef CAS.
- H. Y. Yoon, S. Son, S. J. Lee, D. G. You, J. Y. Yhee, J. H. Park, M. Swierczewska, S. Lee, I. C. Kwon, S. H. Kim, K. Kim and M. G. Pomper, Glycol chitosan nanoparticles as specialized cancer therapeutic vehicles: Sequential delivery of doxorubicin and Bcl-2 siRNA, Sci. Rep., 2014, 4, 1–12 Search PubMed.
- T. H. Shin and J. Cheon, Synergism of nanomaterials with physical stimuli for biology and medicine, Acc. Chem. Res., 2017, 50, 567–572 CrossRef CAS.
- D. Seo, K. M. Southard, J. W. Kim, H. J. Lee, J. Farlow, J. U. Lee, D. B. Litt, T. Haas, A. P. Alivisatos, J. Cheon, Z. J. Gartner and Y. W. Jun, A mechanogenetic toolkit for interrogating cell signaling in space and time, Cell, 2016, 165, 1507–1518 CrossRef CAS.
- L. Zhang, M. Zhang, L. Zhou, Q. Han, X. Chen, S. Li, L. Li, Z. Su and C. Wang, Dual drug delivery and sequential release by amphiphilic Janus nanoparticles for liver cancer theranostics, Biomaterials, 2018, 181, 113–125 CrossRef CAS.
- S. Kader, M. Monavarian, D. Barati, S. Moeinzadeh, T. M. Makris and E. Jabbari, Plasmin-Cleavable Nanoparticles for On-Demand Release of Morphogens in Vascularized Osteogenesis, Biomacromolecules, 2019, 20, 2973–2988 CrossRef CAS.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS.
- H. Kim, D. H. Kang, K. H. Koo, S. Lee, S. M. Kim, J. Kim, M. H. Yoon, S. Y. Kim and E. G. Yang, Vertical nanocolumn-assisted pluripotent stem cell colony formation with minimal cell-penetration, Nanoscale, 2016, 8, 18087–18097 RSC.
- J. H. Lee, H. K. Kwon, H. J. Shin, G. H. Nam, J. H. Kim and S. Choi, Quasi-Stem Cells Derived from Human Somatic Cells by Chemically Modified Carbon Nanotubes, ACS Appl. Mater. Interfaces, 2018, 10, 8417–8425 CrossRef CAS.
- X. Wang, S. Li, C. Yan, P. Liu and J. Ding, Fabrication of RGD micro/nanopattern and corresponding study of stem cell differentiation, Nano Lett., 2015, 15, 1457–1467 CrossRef CAS.
- N. Jokilaakso, E. Salm, A. Chen, L. Millet, C. D Guevara, B. Dorvel, B. Reddy Jr., A. E. Karlstrom, Y. Chen, H. Ji, Y. Chen, R. Sooryakumar and R. Bashir, Ultra-localized single cell electroporation using silicon nanowires, Lab Chip, 2013, 13, 336–339 RSC.
- X. Xie, A. M. Xu, S. Leal-Ortiz, Y. Cao, C. C. Garner and N. a. Melosh, Nanostraw - Electroporation System for Highly Efficient Intracellular Delivery and Transfection., ACS Nano, 2013, 7, 4351–4358 CrossRef CAS.
- V. Caprettini, A. Cerea, G. Melle, L. Lovato, R. Capozza, J.-A. Huang, F. Tantussi, M. Dipalo and F. De Angelis, Soft electroporation for delivering molecules into tightly adherent mammalian cells through 3D hollow nanoelectrodes, Sci. Rep., 2017, 7, 8524 CrossRef.
- P. E. Boukany, A. Morss, W. Liao, B. Henslee, H. Jung, X. Zhang, B. Yu, X. Wang, Y. Wu, L. Li, K. Gao, X. Hu, X. Zhao, O. Hemminger, W. Lu, G. P. Lafyatis and L. J. Lee, Nanochannel electroporation delivers precise amounts of biomolecules into living cells, Nat. Nanotechnol., 2011, 6, 747–754 CrossRef CAS.
- A. Cerea, V. Caprettini, G. Bruno, L. Lovato, G. Melle, F. Tantussi, R. Capozza, F. Moia, M. Dipalo and F. De Angelis, Selective intracellular delivery and intracellular recordings combined in MEA biosensors, Lab Chip, 2018, 18, 3492–3500 RSC.
- Y. Cao, M. Hjort, H. Chen, F. Birey, S. A. Leal-Ortiz, C. M. Han, J. G. Santiago, S. P. Pasca, J. C. Wu and N. A. Melosh, Nondestructive nanostraw intracellular sampling for longitudinal cell monitoring, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E1866–E1874 CrossRef CAS.
- J. A. Huang, V. Caprettini, Y. Zhao, G. Melle, N. MacCaferri, L. Deleye, X. Zambrana-Puyalto, M. Ardini, F. Tantussi, M. Dipalo and F. De Angelis, On-Demand Intracellular Delivery of Single Particles in Single Cells by 3D Hollow Nanoelectrodes, Nano Lett., 2019, 19, 722–731 CrossRef.
- A. Meister, M. Gabi, P. Behr, P. Studer, J. Voros, P. Niedermann, J. Bitterli, J. Polesel-Maris, M. Liley, H. Heinzelmann and T. Zambelli, FluidFM: Combining Atomic Force Microscopy and Nanofluidics in a Universal Liquid Delivery System for Single Cell Applications and Beyond, Nano Lett., 2009, 9, 2501–2507 CrossRef CAS.
- R. Xiong, K. Raemdonck, K. Peynshaert, I. Lentacker, I. De Cock, J. Demeester, S. C. De Smedt, A. G. Skirtach and K. Braeckmans, Comparison of gold nanoparticle mediated photoporation: Vapor nanobubbles outperform direct heating for delivering macromolecules in live cells, ACS Nano, 2014, 8, 6288–6296 CrossRef CAS.
- M. Dipalo, H. Amin, L. Lovato, F. Moia, V. Caprettini, G. C. Messina, F. Tantussi, L. Berdondini and F. De Angelis, Intracellular and Extracellular Recording of Spontaneous Action Potentials in Mammalian Neurons and Cardiac Cells with 3D Plasmonic Nanoelectrodes, Nano Lett., 2017, 17, 3932–3939 CrossRef CAS.
- G. C. Messina, M. Dipalo, R. La Rocca, P. Zilio, V. Caprettini, R. Proietti Zaccaria, A. Toma, F. Tantussi, L. Berdondini and F. De Angelis, Spatially, Temporally, and Quantitatively Controlled Delivery of Broad Range of Molecules into Selected Cells through Plasmonic Nanotubes, Adv. Mater., 2015, 27, 7145–7149 CrossRef CAS.
- M. Dipalo, G. Melle, L. Lovato, A. Jacassi, F. Santoro, V. Caprettini, A. Schirato, A. Alabastri, D. Garoli, G. Bruno, F. Tantussi and F. De Angelis, Plasmonic meta-electrodes allow intracellular recordings at network level on high-density CMOS-multi-electrode arrays, Nat. Nanotechnol., 2018, 13, 965–971 CrossRef CAS.
- M. Dipalo, V. Caprettini, G. Bruno, F. Caliendo, L. D. Garma, G. Melle, M. Dukhinova, V. Siciliano, F. Santoro and F. De Angelis, Membrane Poration Mechanisms at the Cell–Nanostructure Interface, Adv. Biosyst., 2019, 3, 1–8 Search PubMed.
- J. T. J. Santini, M. J. Cima and R. Langer, A controlled-release microchip, Nature, 1999, 397, 335–338 CrossRef CAS.
- X. Li, L. Matino, W. Zhang, L. Klausen, A. F. McGuire, C. Lubrano, W. Zhao, F. Santoro and B. Cui, A nanostructure platform for live-cell manipulation of membrane curvature, Nat. Protoc., 2019, 14, 1772–1802 CrossRef CAS.
- A. Belu, J. Schnitker, S. Bertazzo, E. Neumann, D. Mayer, A. OffenhÄusser and F. Santoro, Ultra-thin resin embedding method for scanning electron microscopy of individual cells on high and low aspect ratio 3D nanostructures, J. Microsc., 2016, 263, 78–86 CrossRef CAS.
- F. Santoro, W. Zhao, L.-M. Joubert, L. Duan, J. Schnitker, Y. van de Burgt, H.-Y. Lou, B. Liu, A. Salleo, L. Cui, Y. Cui and B. Cui, Revealing the Cell–Material Interface with Nanometer Resolution by Focused Ion Beam/Scanning Electron Microscopy, ACS Nano, 2017, 11, 8320–8328 CrossRef CAS.
- V. Caprettini, J.-A. Huang, F. Moia, A. Jacassi, C. A. Gonano, N. Maccaferri, R. Capozza, M. Dipalo and F. De Angelis, Enhanced Raman Investigation of Cell Membrane and Intracellular Compounds by 3D Plasmonic Nanoelectrode Arrays, Adv. Sci., 2018, 5, 1800560 CrossRef.
- F. A. Pennacchio, F. Caliendo, G. Iaccarino, A. Langella, V. Siciliano and F. Santoro, Three-dimensionally Patterned Scaffolds Modulate the Biointerface at the Nanoscale, Nano Lett., 2019, 19, 5118–5123 CrossRef CAS.
- S. Gopal, C. Chiappini, J. Penders, V. Leonardo, H. Seong, S. Rothery, Y. Korchev, A. Shevchuk and M. M. Stevens, Porous Silicon Nanoneedles Modulate Endocytosis to Deliver Biological Payloads, Adv. Mater., 2019, 31, 1806788 CrossRef.
- H. Y. Lou, W. Zhao, Y. Zeng and B. Cui, The Role of Membrane Curvature in Nanoscale Topography-Induced Intracellular Signaling, Acc. Chem. Res., 2018, 51, 1046–1053 CrossRef CAS.
- C. S. Hansel, S. W. Crowder, S. Cooper, S. Gopal, M. Joaio Pardelha Da Cruz, L. De Oliveira Martins, D. Keller, S. Rothery, M. Becce, A. E. G. Cass, C. Bakal, C. Chiappini and M. M. Stevens, Nanoneedle-Mediated Stimulation of Cell Mechanotransduction Machinery, ACS Nano, 2019, 13, 2913–2926 CrossRef CAS.
- H. Y. Lou, W. Zhao, X. Li, L. Duan, A. Powers, M. Akamatsu, F. Santoro, A. F. McGuire, Y. Cui, D. G. Drubin and B. Cui, Membrane curvature underlies actin reorganization in response to nanoscale surface topography, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 23143–23151 CrossRef CAS.
- L. Hanson, W. Zhao, H. Y. Lou, Z. C. Lin, S. W. Lee, P. Chowdary, Y. Cui and B. Cui, Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells, Nat. Nanotechnol., 2015, 10, 554–562 CrossRef CAS.
- A. K. Shalek, J. T. Robinson, E. S. Karp, J. S. Lee, D. R. Ahn, M. H. Yoon, A. Sutton, M. Jorgolli, R. S. Gertner, T. S. Gujral, G. MacBeath, E. G. Yang and H. Park, Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1870–1875 CrossRef CAS.
- W. Zhao, L. Hanson, H.-Y. Lou, M. Akamatsu, P. D. Chowdary, F. Santoro, J. R. Marks, A. Grassart, D. G. Drubin, Y. Cui and B. Cui, Nanoscale manipulation of membrane curvature for probing endocytosis in live cells, Nat. Nanotechnol., 2017, 1–9 Search PubMed.
- R. Capozza, V. Caprettini, C. A. Gonano, A. Bosca, F. Moia, F. Santoro and F. De Angelis, Cell Membrane Disruption by Vertical Micro-/Nanopillars: Role of Membrane Bending and Traction Forces, ACS Appl. Mater. Interfaces, 2018, 10, 29107–29114 CrossRef CAS.
- X. Xie, A. M. Xu, M. R. Angle, N. Tayebi, P. Verma and N. A. Melosh, Mechanical model of vertical nanowire cell penetration., Nano Lett., 2013, 13, 6002–6008 CrossRef CAS.
- M. Dipalo, A. F. McGuire, H.-Y. Lou, V. Caprettini, G. Melle, G. Bruno, C. Lubrano, L. Matino, X. Li, F. De Angelis, B. Cui and F. Santoro, Cells adhering to 3D vertical nanostructures: cell membrane reshaping without stable internalization, Nano Lett., 2018, 18, 6100–6105 CrossRef CAS.
- Y. Chen, S. Aslanoglou, G. Gervinskas, H. Abdelmaksoud, N. H. Voelcker and R. Elnathan, Cellular Deformations Induced by Conical Silicon Nanowire Arrays Facilitate Gene Delivery, Small, 2019, 15, 1–14 Search PubMed.
- C. N. Prinz, Interactions between semiconductor nanowires and living cells, J. Phys.: Condens. Matter, 2015, 27, 233103 CrossRef.
- C. Chiappini, J. O. Martinez, E. DeRosa, C. Almeida, E. Tasciotti and M. M. Stevens, Biodegradable Nanoneedles for Localized Delivery of Nanoparticles in Vivo: Exploring the Biointerface, ACS Nano, 2015, 9, 5500 CrossRef CAS.
- C. Chiappini, E. DeRosa, J. O. Martinez, X. Liu, J. Steele, M. M. Stevens and E. Tasciotti, Biodegradable silicon nanoneedles delivering nucleic acids intracellularly induce localized in vivo neovascularization, Nat. Mater., 2015, 14, 532 CrossRef CAS.
- C. Chiappini, P. Campagnolo, C. S. Almeida, N. Abbassi-Ghadi, L. W. Chow, G. Hanna and M. M. Stevens, Mapping Local Cytosolic Enzymatic Activity in Human Esophageal Mucosa with Porous Silicon Nanoneedles, Adv. Mater., 2015, 27, 5147 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |