Open Access Article
Open Access ArticleInvestigation of metallodrug/protein interaction by X-ray crystallography and complementary biophysical techniques
Giarita
Ferraro
and
Antonello
Merlino
 *
*
Department of Chemical Sciences, University of Naples Federico II, Complesso Universitario di Monte Sant'Angelo, via Cinthia, 21, 80126, Naples, Italy. E-mail: antonello.merlino@unina.it
First published on 10th March 2025
Abstract
Protein metalation, the process by which a metal compound (or a metal ion) reacts with a protein to produce a metal/protein adduct, is at the basis of many biological events; the knowledge of this process at the atomic level is important in the design and development of new metallodrug-based therapeutic approaches. Recently, single crystal X-ray diffraction experiments have been frequently used to characterize the structures of the adducts formed upon the reaction of Pt, Au, Ru, Rh, Ir, Cu, Mn and V-based drugs with proteins. Although X-ray crystallography is certainly useful to determine the structure of these adducts, the combination with other biophysical techniques provides insights into the system behavior in solution, the reactivity of metal compounds with proteins, fate and stability of the metal/protein adduct and is often helpful for the rationalization of ambiguous or unexpected crystallographic data. Here we describe the results of selected studies carried out in the field of protein metalation, where the structural information achieved by X-ray crystallography has been complemented by data collected using mass spectrometry, vibrational spectroscopy, electron paramagnetic resonance, and computational methods, including density functional theory, docking and molecular dynamics simulations. These works allow us to define the protein metalation process at the molecular level, providing information on the factors responsible for the formation and stability of metal/protein adducts.
Introduction
The interaction between metal ion/metal complexes and proteins often results in the formation of metal/protein adducts, where metal centers are generally coordinated with specific residue side chains. This process has a major role in biology1 since it is involved in the correct folding and function of metalloproteins, which are a large fraction of proteins within the cells.2 Protein metalation significantly influences the absorption, transportation and storage of metallodrugs within the body and has a major role in the design of synthetic artificial metalloenzymes3 and in the development of protein-based metallodrug delivery systems. The interplay between metal compounds and proteins also contributes to the assessment of adverse reactions associated with the use of metal-based anticancer treatments. Indeed, the exploration of protein metalation induced by anticancer metal-based agents enables the elucidation of fundamental aspects underlying the occurrence of side effects and activation/inactivation mechanisms of these compounds, which are frequently pro-drugs. The formation of metal/protein adducts also alters the catalytic properties of various enzymes.4 Therefore, a detailed understanding of the mechanisms that form the basis of the recognition of metal compounds by proteins has become one of the most stimulating goals in recent years.5–7 X-ray diffraction (XRD) experiments are routinely used for the determination of structures of adducts formed upon the reaction of metal compounds with proteins.8–12 In an XRD experiment, X-rays interact with the electrons of atoms in a crystal, providing an experimental dataset that is related to the electron density (e.d.) map, which allows localizing of electrons of the crystal asymmetric unit and thus determination of the molecular structure. In protein X-ray crystallography, the e.d. map shows a time- and space-average of the e.d. of the atoms in the crystal. When a protein region is highly flexible, it is associated with a poor e.d. map and is difficult to model. Thus, it is sometimes omitted by the final protein structure model. Similarly, molecules that are disordered or present in crystals with low occupancy, i.e. only in a fraction of the unit cell, are associated with weak e.d. and are often difficult to identify. This also happens in the structures of metal/protein adducts. The position of the metal is generally well defined, but metal ligands cannot be unambiguously modelled. Furthermore, in the formation of metal/protein adducts, the molecular species that bind the protein are not those reacting with the macromolecules in solution or those used to treat protein crystals. Indeed, an important role in the definition of the final metal/protein adduct is played by the reaction that occurs in solution (ligand exchange, hydrolysis, speciation, reduction/oxidation, etc.) or by the reaction occurring upon the metal-containing fragment binding to the protein. The combination of X-ray crystallography with other biophysical techniques helps to define details of the molecular processes that occur before or upon the formation of the metal/protein adduct. In this context, spectroscopic analysis, UV-vis absorption spectroscopy and nuclear magnetic resonance (NMR), for example, are fundamental techniques to study the metal compound behavior in solution alone and in the presence of a protein. On the other hand, circular dichroism or fluorescence can be helpful in unveiling protein secondary and tertiary structural alterations induced by the metal complex binding. Interesting information has also been obtained by using small angle X-ray scattering (SAXS)13,14 or perturbed angular correlation spectroscopy.15–17Here, selected examples of the combined use of XRD and specific biophysical techniques (mass spectrometry, IR and Raman spectroscopy, electron paramagnetic resonance, dynamic light scattering (DLS) and computational methods, including density functional theory, docking and molecular dynamics simulations) in the definition of the protein metalation process with several different metallodrugs are discussed. The selected works reflect the authors’ personal perspective; thus, the paper does not pretend to be a comprehensive study.
Protein metalation studied by a combination of XRD and mass spectrometry
XRD data provide experimental evidence on the metal coordinates and on the protein residues involved in metal recognition. However, sometimes the definition of the e.d. map does not allow us to precisely identify the exact metal-containing fragments that bind a protein.18 In these cases, mass spectrometry techniques can be used to characterize the metal/protein adducts formed upon the reaction of metal compounds with proteins.19,20 Electrospray ionization mass spectrometry (ESI-MS) is one of the most widely used mass spectrometry techniques. It is based on the ionization into the gas phase of polar molecules dissolved in a liquid.21 The samples are transferred to the ion source either directly (through a pump22 or a flow-injection unit23) or indirectly (via a separation system, such as liquid chromatography, LC, high-performance liquid chromatography, HPLC24 or capillary electrophoresis).25 Mass spectra report the intensity values as a function of corresponding mass-to-charge ratios (m/z). ESI-MS is a so-called ‘soft ionization’ technique, since it produces very little fragmentation, thus allowing the determination of molecular masses and chemical structures of metal/protein adducts, the metal-containing fragment/protein stoichiometry, and the protein binding sites and their number, even preserving non-covalent interactions of metal complexes with proteins.26 ESI-MS is fast, sensitive and specific. It can investigate the metal compound/protein interaction over time and at different metal to protein molar ratios and under different experimental conditions,27 also defining the effect of protein post-translation modifications on the metal compound binding.28 The main disadvantages of this technique are the difficulty in ionizing metal adducts of large proteins or of proteins that tend to aggregate forming higher oligomers, causing unclear or multiple mass peaks, and their sensitivity to pH.29 Examples of studies combining ESI-MS and X-ray crystallography to characterize the formation of metal/protein adducts include different metals (Pt, Ru, Au, Ir, and V, among others, see for example complexes shown in Fig. 1) and diverse proteins (examples are shown in Fig. 2). This approach has been already summarized in previous works.27,30,31 Cisplatin (Fig. 1) binding to proteins has been studied both from the crystallographic and mass spectrometry points of view in a number of cases.9 It has been shown that Pt2+, [Pt(NH3)2]2+, and [Pt(NH3)2Cl]+ or the product of its hydrolysis [Pt(NH3)2(H2O)]2+ bind Nδ1 or Nε2 atoms of His15 of hen egg white lysozyme (HEWL, Fig. 2).32 Pt binding sites have been found close to Met29 or both Met29 and Gln28,33 and His105 and His119 side chains of bovine pancreatic ribonuclease (RNase A,33,34Fig. 2). Cys12 and Cys15 have been identified as Pt binding sites in Atox-135 (Fig. 2), while Met65 and Glu61 are the residues involved in the metal recognition by cytochrome c36 (Fig. 2). Cisplatin fragments bind His68 of ubiquitin29 (Fig. 2), His105, His128, His146, His247, His288, Met298, Met329, His305, His338, His440 + Lys436, and Met548 of human serum albumin37–39 (HSA, Fig. 2).It has been shown that the same metal containing fragments can bind β-lactoglobulin,40 transferrin41 and angiogenin42 (Fig. 2).
Using the same approach, the interaction of some of these proteins with carboplatin28 and oxaliplatin (Fig. 1)38–41 has also been characterized. In the latter case, [Pt(dach)(H2O)]2+, [Pt(dach)]2+ (dach = 1,2-diaminocyclohexane) or the intact oxaliplatin binds proteins43–46 close to the side chains of Asp, Met, His or Cys residues, in metal binding sites that are in some cases distinct from those of cisplatin. Using the combined crystallographic/ESI-MS approach it has also been possible to analyze in detail the protein binding of other potential anticancer Pt-based drugs, like picoplatin (Fig. 1),47 the diiodido analogue of cisplatin cis-PtI2(NH3)233 (Fig. 1), trans-Pt derivatives (like trans-(dimethylamino) (methylamino) dichloroplatinum(II), [t-PtCl2(dma)(ma)], Fig. 1),48 a tetranuclear Pt-thiosemicarbazone complex49 and Pt-compounds containing O,S-bidentate ligands (like compound 9b in Fig. 1).50
Binding of Ru-,51,52 Au-,53,54 V-, and homo (Ru–Ru55 and Rh–Rh,56–58Fig. 1) and hetero-bimetallic-(like Pt–Au59 and [Pt(μ-NHC(CH3)O)2ClAs(OH)2], AP-160,61 in Fig. 1) based drugs to HEWL and RNase A62,63 has also been investigated. In the case of the antimetastatic metallodrug imidazolium (Im) trans-[tetrachlorido(S-dimethyl sulfoxide)(1H-imidazole)ruthenate(III)] (NAMI-A, Fig. 1) and its derivative AziRu (Fig. 1),64 a complete hydrolysis associated with the release of all the metal ligands has been observed in the reaction with HEWL,51 carbonic anhydrase (hCAII),65 and human H-chain ferritin (h-H-Ft)66 (Fig. 2). The HEWL ruthenation mechanism by NAMI-A has been studied in detail by Papakyriakou and coworkers, who solved the X-ray structures of the protein with NAMI-A at different soaking times.67 The structures highlight a series of events that ultimately lead to the final “ruthenated” protein: NAMI-A non-covalently binds the protein (after 1.5 h of soaking, Fig. 3A), then it exchanges all Ru ligands except for Im (8 h and 26 h of soaking, Fig. 3B and C) and finally the aquated Ru ion coordinates to His15 and Arg14 (98 h) (Fig. 3D). More recently, a comparative structural analysis of the HEWL adducts with two RuIII complexes [HIsq][trans-RuCl4(dmso)(Isq)] (1, Fig. 1) and [H2Ind][trans-RuCl4(dmso)(HInd)] (2, Fig. 1) (where HInd – indazole and Isq – isoquinoline are analogues of NAMI-A), together with in-solution studies, has demonstrated that the hydrolytic release of the N-heterocyclic ligand is one of the main factors in determining the interaction of Ru-containing fragments to proteins.68 From these works it emerges that anticancer Ru agents are often prodrugs, which have to be activated before they can react with the final protein target.69 In the case of Ru compounds the activation could be a simple aquation reaction.
The selectivity for different binding sites on HEWL of a series of isostructural N-heterocyclic carbene (NHC) compounds with the formula [M(L)(dmb/dbb)Cl2] (M = Ru/Os/Rh/Ir; L = η6-p-cymene [cym], η5-pentamethylcyclopentadienyl [Cp*]; dmb = 1,3-dimethylbenzimidazol-2-ylidene; dbb = 1,3-dibenzylbenzimidazol-2-ylidene, Fig. 1) was investigated by Sullivan and co-workers70 choosing this combined approach. The X-ray structures of the adducts highlighted a diverse behavior of the analyzed metal complexes. Indeed, the Ru and Rh compounds showed a tendency to bind the surface exposed His15 upon loss of the p-cymene or the NHC ligands, while the Ir and Os derivatives were identified close to the electronegative peptidoglycan-binding pocket HEWL, showing a preferential binding for Asn103 and Asp101. In these cases, the binding occurred upon replacement of the chloride ligand with the side chain of the protein residues.
Gold-based drugs also must be activated before reaching their final targets. Crystallographic and ESI-MS studies carried out to unveil the reactivity of anticancer AuIII-based drugs with proteins have suggested that for these compounds the protein binding occurs after or concomitantly to metal compound degradation and metal center reduction, with the coordination of AuI to His or Met side chains.54,71 Indeed, in the reaction of Auoxo6 ([Au2(bipy2Me)2(μ-O)2][PF6]2, where bipy2Me = 6,6′-dimethyl-2,2′-bipyridine, Fig. 1), Au2phen ([Au2(phen2Me)2(μ-O)2][PF6]2, where phen2Me = 2,9-dimethyl-1,10- phenanthroline, Fig. 1) and AuSac2 ([NH4][Au(Sac)2], where Sac = deprotonated saccharinate ligand, Fig. 1) with HEWL, only one AuI atom bound to His15 has been observed.54 An additional structure of the HEWL adduct with AuSac2, obtained under a different experimental condition, has revealed two AuI ions close to the side chain of His15, and an additional gold center close to the side chain of Met105, in a buried hydrophobic pocket. These crystallographic data are in good agreement with ESI-MS results indicating that the protein can bind up to three AuI ions.71 Degradation of the AuIII compound and binding to AuI to a Gln side chain has been observed in the reaction of HEWL with Aubipyc ([(bipydmb-H)Au(OH)][PF6],53Fig. 1), in the reactivity of Auoxo6 with RNase A62 and of the medicinal AuIII dithiocarbamato complex AuL12 (Fig. 1) with bovine serum albumin.72 ESI-MS/XRD data have been collected also for adducts formed by proteins with vanadium compounds. ESI-MS recorded on the system containing bis(maltolato)oxidovanadium(IV) ([VIVO(malt)2], Fig. 1) and HEWL with a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 metal to protein molar ratio at pH 5.0 and pH 6.5 showed the formation of HEWL-VO-malt adducts, with [VIVO]2+, [VIVO(malt)]+ and [VIVO(malt)2] that bind the protein. These results agree well with crystallographic data showing that in the HEWL-VO-malt adduct, cis-[VIVO(malt)2(H2O)] (Fig. 4) and [VIVO(malt)(H2O)3]+ bind the protein non-covalently, while [VIVO(H2O)3–4]2+, cis-[VIVO(malt)2] and other V-containing fragments are coordinated to the side chains of Glu35, Asp48, Asn65, Asp87, and Asp119 and to the C-terminal carboxylate.73 Both covalent and non-covalent binding modes have also been predicted by ESI-MS and then verified by X-ray crystallography in the adduct formed when [VIVO(8-HQ)2]74 (8-HQ = 8-hydroxyquinoline) and [VIVO(empp)2] (where Hempp is 1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone) react with the protein75 (Fig. 4).
1 metal to protein molar ratio at pH 5.0 and pH 6.5 showed the formation of HEWL-VO-malt adducts, with [VIVO]2+, [VIVO(malt)]+ and [VIVO(malt)2] that bind the protein. These results agree well with crystallographic data showing that in the HEWL-VO-malt adduct, cis-[VIVO(malt)2(H2O)] (Fig. 4) and [VIVO(malt)(H2O)3]+ bind the protein non-covalently, while [VIVO(H2O)3–4]2+, cis-[VIVO(malt)2] and other V-containing fragments are coordinated to the side chains of Glu35, Asp48, Asn65, Asp87, and Asp119 and to the C-terminal carboxylate.73 Both covalent and non-covalent binding modes have also been predicted by ESI-MS and then verified by X-ray crystallography in the adduct formed when [VIVO(8-HQ)2]74 (8-HQ = 8-hydroxyquinoline) and [VIVO(empp)2] (where Hempp is 1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone) react with the protein75 (Fig. 4).
 | ||
| Fig. 4 Examples of covalent and non-covalent binding of V compounds to RNase A (panels A and B) and HEWL (panels C–F). | ||
A similar approach has been used to characterize the interaction of the VIVO-picolinato complex ([VIVO(pic)2(H2O)], pic = picolinate) with RNase A (Fig. 4): the structure of the adduct formed when the V complex reacts with the bovine enzyme at acidic pH revealed that [VIVO(pic)2] binds the Glu111 side chain by replacing the water equatorial ligand. ESI-MS confirms this result.76
Metal/protein adducts ionized by electrospray can also be monitored by ion mobility mass spectrometry (IM-MS). In IM-MS, the ions are separated thanks to their different mobility through an inert gas: compact ions move faster than ions with the same m/z values but less compact.77 IM-MS can give insights into the effect of metalation on protein folding, stability and dynamics and can distinguish between different protein conformations, which might be indistinguishable by traditional mass spectrometry techniques. The main disadvantage of this technique is its limited availability, since it is not diffuse as the other mass spectrometry techniques. Other disadvantages are related to limits in treating data from large proteins, that often produce peak overlap or broadening.
IM-MS has been used to study the binding of [Ru(cym)Cl2]2 (Fig. 1) to HEWL.78 X-ray crystallography revealed that [Ru(cym)Cl2] binds the side chains of His15 and Asp101, after cleavage of the dimeric structure. IM-MS data indicated that the number of higher charge state HEWL ions is increased by “ruthenation”, suggesting that metal coordination to HEWL destabilizes the protein structure. Additionally, ion mobilograms indicated possible protein unfolding and a compaction of the unfolded state due to the metal-containing fragment binding. The reduced stability of the Ru/HEWL adduct when compared to metal-free HEWL is confirmed by differential scanning calorimetry (DSC) experiments.71 Compaction upon metalation was observed in the metalation of other proteins.79–81
Recently, the combined XRD/mass spectrometry approach has also been used for systems of large dimensions. ESI-MS has been used, in combination with proteolytic assays and site directed mutagenesis studies, to identify h-H-Ft residues that bind gold atoms from sodium aurothiomalate, an FDA-approved anti-arthritic drug, in the Au-h-H-Ft adduct formed upon the reaction of the protein with the drug.82 In this study, the incubation of sodium aurothiomalate with h-H-Ft under near-physiological solution conditions led to the formation of the Au/h-H-Ft adduct with enhanced biological activity when compared to the gold-compound alone. In Au/h-H-Ft, a cluster of gold atoms are bound to Cys90 and Cys102. These residues have been identified as gold binding sites in the adduct formed by h-H-Ft with auranofin.83,84
There are other mass spectrometry techniques that have been frequently used to characterize the formation of metal/protein adducts. As an example, the protein interaction of Pt complexes has been characterized by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry (ESI-FT-ICR-MS).85 Collision-induced dissociation (CID), higher energy collision-induced dissociation (HCD), electron transfer dissociation (ETD) or electron capture dissociation (ECD) have also been used to elucidate specific residues involved in metal compound binding to proteins.86,87
Inductively coupled plasma (ICP)-MS is an element-specific analytical method, which allows quantitative determination of metallodrugs in biological samples through the evaluation of their metal content.88 The technique features high specificity, in the ng L−1 concentration range, and can be used to determine the metal to protein molar ratio in a metal/protein adduct and its stability over time.20 Interferences from isotopic patterns and problems with the calibration and standardization of the instrument, which could lead to limited sensing ability towards specific metals, are the main disadvantages of this technique. ICP-MS measurements have been employed to define the presence of Ru within crystals of binuclear trithiolato-bridged arene ruthenium complex (diruthenium-1)/horse spleen L-chain ferritin (hs-L-Ft) nanocages obtained using a protein sample that was disassembled and reassembled in the presence of the anticancer compound diruthenium-1 (DiRu-1 in Fig. 1)89 and within crystals of the adduct of indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019, Fig. 1) with HSA.90 In the structure of diruthenium-1/hs-L-Ft, Ru atoms were not observed in the e.d. maps, thus suggesting that the anticancer compound was in the bulk within the cage.89 In the structure of the KP1019/HSA adduct, Ru atoms were coordinated to His146 and His242. Solvent molecules complete the Ru octahedral coordination sphere, suggesting dissociation of both indazole ligands from the metal center.90
ICP-MS measurements have been also used to quantify the amount of Ru in the bionanocomposite formed when recombinant L-chain apo-ferritin from horse liver (hl-L-Ft) was treated with [Ru(CO)3Cl2]2 (CORM-2, Fig. 1)91 and the amount of Au upon encapsulation of Auoxo3 ([Au2(bipyMe)2(μ-O)2][PF6]2, bipyMe = 6-methyl-2,2′-bipyridine, Fig. 1),92 Auoxo4 ([Au2(bipynP)2(μ-O)2][PF6]2, bipynP = 6-neo-pentyl-2,2′-bipyridine, Fig. 1), and Au2phen93 within hs-L-Ft nanocages. A similar analysis has been also carried out to evaluate the amount of Pt within the cage of the same protein94 and the amount of Pt and As within AP-1/h-H-Ft.95 The structure of the RuCO/hl-L-Ft nanocomposite shows Ru atoms coordinated to the side chains of His114, Glu130 and His132;91 the structures of the Au/hs-L-Ft adducts show binding of Au centers to Cys48, His49, His114, His114 and Cys126, Cys126, His132, His147. Pt binds the side chains of His49, His114 and His132.94 AP-1 binds the side chain of His49 of hs-L-Ft.95
Notably, ICP-MS has also been used to characterize the Au/apo-R168H/L169C-hl-L-Ft system where gold nanoclusters are formed.96,97 Ten97 and 8–1296 atoms of Au in clusters were found (see for example Fig. 5). Using the same approach, Fujita et al.98 studied the role of the R52C mutation in hl-L-Ft when it reacted with Mn(CO)5Br to produce a ferritin-based nanocomposite containing a manganese–carbonyl complex. When the nanocomposite is designed starting from the wild-type protein, only a few Mn atoms could be detected within the cage (about 3 atoms), while the mutated Ft could accumulate about 48 Mn atoms.
 | ||
| Fig. 5 A gold nanocluster found within a hl-L-Ft nanocage (PDB code 7VIT). Gold atoms are in yellow, and protein chains involved in the recognition of gold atoms in pink. | ||
A novel approach, developed by Timerbaev et al.,99 matching capillary electrophoresis with ICP-MS enabled a deep characterization of the affinity of cisplatin and two of its novel analogues, 2-ABDP, (SP-4-2)-bis((R)-(–)-2-aminobutanol)dichloroplatinum(II), and 4-ABDP, (SP-4-2)-bis(4-aminobutanol)dichloroplatinum(II) (Fig. 1) for HSA. The coupling of these two techniques turned out to be a useful tool to measure the rate constant of the binding reactions and to determine the number of drug molecules attached to the protein. The binding of cisplatin to HSA was a pseudo-first order reaction. When the protein is incubated in the presence of a huge excess of metallodrug, it can bind up to 10 Pt atoms per protein chain.
ICP-MS is suitable to be combined with different types of techniques. Indeed, the combination of the separation capacity of SDS PAGE with the specificity of ICP-MS has been employed to characterize oligomers of different orders formed upon interaction of HSA with cisplatin.100 It was found that increasing amounts of cisplatin push the serum albumin to form oligomers bigger than its dimeric form, which is generally the most abundant. In fact, as the reaction proceeds, dimeric HSA is drained from the reaction environment leaving the monomer as the most representative species.
Alternatively, the amount of metals (Pd,101,102 Rh,103 Ru,66 Au,83 Pt104) within Ft nanocages with solved X-ray structures has been evaluated using inductively coupled plasma-optical emission spectrometry (ICP-OES). ICP-OES can detect heavy metal concentrations, even in the range of parts per billion (ppb) or parts per trillion (ppt), with high accuracy. It can analyze a wide spectrum of metals, even simultaneously, but it is a rather expensive technique. Although the ICP-OES is generally resistant to interference, the chemical composition of the sample matrix can affect the results, requiring the use of internal standards or corrections.
Matrix-assisted laser desorption/ionization-mass spectrometry (MALDI-MS) is an ionization technique that is based on the use of a laser energy-absorbing matrix to create ions from large molecules, with minimal fragmentation.105 MALDI-MS is similar to ESI-MS since both are ‘soft ionization’ techniques, but it typically produces far fewer multi-charged ions. MALDI-MS can be used for samples of metal/protein adducts treated with denaturants, like urea, or with reducing (like dithiothreitol) and alkylating (like iodoacetamide) agents or using digested samples.
Recently, MALDI-MS combined with chromatographic separation techniques has served for studying an unusual reaction product obtained when HEWL crystals have been treated with [VIVO(malt)2] in 1.1 M NaCl and 0.1 M CH3COONa at pH 4.0.106 In this work, crystals of the adduct formed by the V compound with the protein have been dissolved and analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel showed that HEWL formed an SDS-resistant dimer in the presence of [VIVO(malt)2] (Fig. 6A), together with a metalated monomeric species. Metalated monomeric and dimeric protein forms were characterized and found to be in agreement with the crystallographic model: [VIVO(malt)2] degrades within HEWL crystals, the malt ring is broken, and two V centers are bound to a modified N-terminal Lys side chain, with one of the two metal centers that also acts as a crosslinker of two protein molecules within the crystal (Fig. 6B). Gel and mass spectrometry have also been used to test the stability of metal–protein bonds in five platinated proteins by Moreno-Gordaliza et al.107
 | ||
| Fig. 6 (A) SDS-PAGE of dissolved crystals of HEWL (lane 2) and its adduct with [VIVO(malt)2] (lane 3). Markers are in lane 1. The gel indicates that upon incubation of HEWL crystals with the V compound, a metalated cross-linked HEWL dimer is formed. (B) Modification of Lys1 in crystals of HEWL grown in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 and treated with [VIVO(malt)2] (PDB code 9FMY). A 2Fo − Fc e.d. map is reported at the 1.0σ level in grey; anomalous difference e.d. map is colored in yellow and reported at the 3.0σ level. *Indicates atoms from a symmetry related molecule. These atoms are in grey; their e.d. map at the 1.0σ level is in red. | ||
A MALDI-TOF-MS-X-ray crystallography combined approach was used to characterize the adduct formed upon reaction of a series of Cu(II) compounds with 6-methyl-2-formylpyridine-4N-substituted thiosemicarbazone compounds (Fig. 1) with HSA previously complexed with palmitic acid (PA).108 The presence of PA in the IB subdomain of the protein strengthens the affinity of copper for HSA preventing its early release.
Protein metalation studied by dynamic light scattering
The formation of peptide and protein dimers or higher oligomers or a change in the overall conformation of large protein systems upon metalation has been observed in many cases.109–111 To study the formation of metal-based cross-linked oligomeric protein states, SDS-PAGE or size exclusion chromatography can be used.Cross-linking mass spectrometry (XL-MS) has been widely used in the analysis of metal-induced cross-linked products. Nie et al. exploited this methodology to elucidate structural differences between the allosteric enzymes, glycogen phosphorylases (GP) b and a.112 It is a two-stage analysis which couples a cross-linking experiment, in this case triggered by cisplatin, to an MS experiment. This approach has revealed itself to be worthwhile particularly for the analyses of the N-terminus and the helix interfaces, which were the uncrystallized regions.
Another technique that has been used for this type of characterization is dynamic light scattering (DLS). DLS studies carried out on fatty acid-free and bound forms of HSA, in the absence and in the presence of cisplatin have shown that the tendency of the protein to aggregate increases in the presence of the Pt-based drug, providing a precise estimate of the size of the aggregates.37 DLS analysis has also confirmed the formation of large aggregates of caspase cleaved prostate apoptosis response-4 protein (cl-Par-4) in the presence of cisplatin.113
Protein metalation studied by X-ray crystallography complemented with spectroscopic techniques
Infrared (IR) spectroscopy and Raman spectroscopy are two variants of vibrational spectroscopy, which provide information on the vibrational transitions which strictly depend on chemical composition, structure, symmetry, electronic environment and bonding of molecules. Infrared spectroscopy has been combined with crystallographic data to characterize the formation of the [Re(CO)3(H2O)2]+/HEWL adduct114 upon the reaction of [Re(CO)3(H2O)3]+ with the protein and to monitor a time-resolved series of HEWL-fac-[Re(CO)3]+-imidazole (HEWL-Re-Im) crystals.115 For these systems, carbonyl stretching frequencies can be used as spectroscopic probes to determine metal coordination.The presence of [Re(CO)3]+ within HEWL crystals in the [Re(CO)3(H2O)2]+/HEWL adduct was confirmed by ν(CO) stretching bands at 1890, 2001 and 2013 cm−1. The appearance of the 2001 and 2013 cm−1 bands suggests the presence of multiple rotamers of the [Re(CO)3]+ adduct.114
In the IR spectra of HEWL-Re-Im crystals ν(CO) stretching bands (2017, 1896 cm−1) started to appear from week 1, while a slight shoulder appeared at 1896 cm−1 from week 15. Longer soaking (up to 67 weeks) was associated with an increase in the wavenumber (ν(CO) = 2019, 1899, 1883 (shoulder) cm−1), suggesting a continuation of movement of the Re complexes within the protein structure (Fig. 7).115 Along the same line, carbonyl stretching frequencies in ATR-IR spectra have been used to monitor the presence of [Mn(CO)3] within HEWL microcrystals soaked with [Mn(CO)3(acetone)3](CF3SO3) for 4–5 days in the dark116 and the presence of CORM-2 derivatives in the RuCO/hl-L-Ft adduct formed when hl-L-Ft was treated with the Ru carbonyl complex.91 Fujita et al.98 employed ATR-FTIR spectroscopy to confirm the presence of CO ligands in the ferritin composite containing a manganese–carbonyl complex designed reacting the R52C-hl-L-Ft mutant with Mn(CO)5Br previously described. In the crystal structure there was no electron density map assignable to the CO ligands around the metal center. However, the ATR-FTIR spectrum of MnCO·R52C-hl-L-Ft showed the peaks at 2028, 2011, and 1917 cm−1 in the CO-stretching vibrational region which were assigned to a fac-Mn(CO)3 structure. Similarly, for the [Mn(CO)3]/HEWL system, the ν(CO) stretching bands at 2030 cm−1 and 1938 cm−1 have been noted. In the RuCO/hl-L-Ft adduct, the observed bands were at 2038 and 1956 cm−1.
 | ||
| Fig. 7 IR spectrum of HEWL-Re-Im crystals. ν(CO) stretching bands have been highlighted. Insets of this region for crystals at weeks 1, 12, 15 and 67 after crystallization have been added to the spectrum. Reproduced from ref. 115 with permission from IUCr Journals, copyright 2024. | ||
Raman spectroscopy uses the inelastic diffusion of monochromatic incident radiation to gain knowledge about molecular vibrations of the molecules of the sample. In confocal Raman microscopy, an optical microscope focuses the laser beam onto the sample.117 The technique is very efficient in the analysis of the distribution of chemical moieties within several matrices, including protein crystals.118 For example, Raman microspectroscopy has been used to evaluate the kinetics of the binding of platinochloride ions within HEWL single crystals treated with a tetrachloroplatinate salt (K2PtCl4).
Raman spectra were collected at different times from the HEWL crystal during 14 h of soaking. The spectra revealed growing characteristic Pt–Cl stretching bands in the 305–330 cm−1 region (Fig. 8).119 The crystal structure then confirmed the presence of four anomalous map signals at the surface of the protein, attributed to Pt atoms.
 | ||
| Fig. 8 Raman spectra of a HEWL crystal soaked in 10 mM solution of K2PtCl4 for 12 min (dark), 2 h (blue), 4 h (violet), 6 h (pink) and 8 h (red). The Pt–Cl stretching band at 330 cm−1 ((1) in the spectrum) and Trp ring breathing band at 760 cm−1 ((2) in the spectrum) are shown. Reproduced from ref. 118 with permission from IUCr Journals, copyright 2007. | ||
A similar approach has been used to study the release or the persistence of CO bound to [IrCl5CO]2− (Fig. 1) within HEWL or RNase A crystals.
Raman spectra have been collected during [IrCl5CO]2− soaking into HEWL crystals (647 nm line), using spectra of Cs2[IrCl5CO] powder, of the protein and of the crystallization conditions as references.120 The diffusion of the Ir complex into HEWL crystals has been followed by observing the appearance of low frequency bands (around 300 cm−1) in the spectra of the crystal, during the first hours of soaking. As in the case of IR spectra, Raman carbonyl frequencies can be conveniently used as spectroscopic probes for metal compound binding to the protein. In the spectrum of Cs2[IrCl5CO], one signal at about 2060 cm−1 is found. When [IrCl5CO]2− soaks into HEWL crystals, two new bands (2173 and 2076 cm−1) and a new shoulder (at 2122 cm−1) appear. The two bands have been attributed to free CO trapped into the protein crystal and to a mixture of unbound [IrCl5CO]2− and [IrCl4CO(H2O)]− ions. The shoulder at 2122 cm−1 has been attributed to the metal containing fragment coordinated to the His15 imidazole.
A similar analysis has been carried out monitoring the diffusion of [IrCl5CO]2− into RNase A crystals. In this case, the entrance into the crystal of the Ir compound was verified by the appearance of a band at 2073 cm−1, attributed to CO bound to the metal center.121
Hunter et al.122 coupled NMR spectroscopy to X-ray crystallography to obtain structural information on the interaction of the anti-HIV drugs Cu-cyclam and Cu2-xylyl-bicyclam (Fig. 1) with HEWL, taken as the model protein for CXCR4 coreceptor, which is the entrance way for HIV into cells. The X-ray structures revealed that Cu-cyclam and Cu2-xylyl-bicyclam protein recognition occurs through polar and non-polar interactions, involving H bonding between the cyclam ring and protein carboxylate groups, as already reported,123 and hydrophobic interactions between the cyclam ring and the indole ring of tryptophan. The latter interaction was confirmed by the decrease of line-broadening of tryptophan indole NH resonances in the 1H NMR spectra induced by Cu-cyclam.
Protein metalation studied by a combination of X-ray crystallography and electron paramagnetic resonance
The impact of other biophysical techniques to support X-ray crystallography in the definition of the protein metalation process is illustrated using EPR in achieving the definition of the interaction of VIV, RuIII and CuII compounds with HEWL. EPR can be in principle used to elucidate electronic structures of transition metal complexes with one or more unpaired electrons.124,125The EPR spectra of a monometallic paramagnetic compound with one unpaired electron are characterized by two spin Hamiltonian parameters: the g factor and the hyperfine coupling (HFC) constant between the unpaired electron and the nucleus (giso and Aiso in an isotropic spectrum, and gx, gy, gz and Ax, Ay, Az in an anisotropic spectrum).126 When the crystal field has axial symmetry, the splitting between two spin energy states is described by g⊥ and g‖. g⊥ = gx = gy, g‖ = gz. The g and A tensors provide information on the metal chemical environment.
VIV (d1 configuration), RuIII (d5 configuration) and CuII are paramagnetic and thus can be monitored by EPR, while VV, RuII and CuI are diamagnetic and thus do not exhibit EPR signals. For this reason, the reduction of RuIII and CuII and oxidation of VIV can be detected by the loss of EPR signal intensity.
EPR data have been collected for several VIV compounds in the presence of proteins (see for example, ref. 127 and 128) but only in a few cases these experiments have been combined with crystallographic data.
Crystal structure of the adducts formed by VIVOSO4 and phen/bipy (L) with HEWL when protein crystals have been treated with these molecules showed binding of the VIVOL2+ moiety to the protein. EPR data collected on these systems confirmed the presence of the adducts with spin Hamiltonian parameters in agreement with equatorial (Nbipy/phen, Nbipy/phen; COO-Asp; COAsn) coordination.129
EPR experiments also verified the formation of adducts between [VIVO(pic)2] and [VIVO(8-HQ)2] with RNase A.130 For these systems, crystallographic data have been collected at acidic pH, indicating the binding of V to Glu111. EPR data indicated the binding of V centers to carboxylate groups, stemming from Asp or Glu residues, at acidic pH and of an imidazole nitrogen from His residues at physiological pH,76,130 (Fig. 9). Spectroscopic data also suggested that His-N or Ser/Thr-O− are possible metal binding sites at basic pH (Fig. 9).130
 | ||
Fig. 9 In solution EPR spectra at 120 K of the systems: (A) [VIVO(8-HQ)2]-RNase A (metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein molar ratio = 1 protein molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) x with x = 0–1) and (B) [VIVO(8-HQ)2]-RNase A (metal x with x = 0–1) and (B) [VIVO(8-HQ)2]-RNase A (metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein molar ratio = 1 protein molar ratio = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) y with y = 0–1). The spectra were recorded with VIVO2+ y with y = 0–1). The spectra were recorded with VIVO2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8-HQ 8-HQ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RNase A 1 RNase A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 at pH 3.0 (trace a), VIVO2+ 0 at pH 3.0 (trace a), VIVO2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8-HQ 8-HQ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RNase A 1 RNase A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 at pH 5.1 (trace b), at pH 6.4 (trace c) and pH 7.4 (trace d); VIVO2+ 1 at pH 5.1 (trace b), at pH 6.4 (trace c) and pH 7.4 (trace d); VIVO2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8-HQ 8-HQ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RNase A 1 RNase A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 at pH 4.0 (trace e); VIVO2+ 0 at pH 4.0 (trace e); VIVO2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8-HQ 8-HQ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RNase A 1 RNase A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 at pH 5.1 (trace f), at pH 7.4 (trace g) and at pH 8.4 (trace h). V concentration is 1.0 mM in traces a and e, and 0.8 mM in traces b–d and f–h. I indicates MI = 5/2, 7/2 resonances of [VIVO(8-HQ)(H2O)2]+, II of the adduct [VIVO(8-HQ)(H2O)]+-RNase A with Asp/Glu-COO-coordination, IIIa and IIIb of the two isomers cis-[VIVO(8-HQ)2(H2O)], IV of the adduct [VIVO(8-HQ)(H2O)]+-RNase A with His-N coordination, and V of the adduct [VIVO(8-HQ)(OH)]-RNase A with His-N coordination or [VIVO(8-HQ)(H2O)]+-RNase A with Ser/Thr-O− coordination, while aq denotes MI = 7/2 resonances of the aqua ion [VIVO(H2O)5]2+. The positions of MI = 5/2, 7/2 resonances of I, II, IIIa, IIIb, IV, and V species are also indicated by the dotted lines. Reproduced from ref. 130, which is licensed under a Creative Commons Attribution 3.0 Unported Licence, with permission from RSC, copyright 2023. 1 at pH 5.1 (trace f), at pH 7.4 (trace g) and at pH 8.4 (trace h). V concentration is 1.0 mM in traces a and e, and 0.8 mM in traces b–d and f–h. I indicates MI = 5/2, 7/2 resonances of [VIVO(8-HQ)(H2O)2]+, II of the adduct [VIVO(8-HQ)(H2O)]+-RNase A with Asp/Glu-COO-coordination, IIIa and IIIb of the two isomers cis-[VIVO(8-HQ)2(H2O)], IV of the adduct [VIVO(8-HQ)(H2O)]+-RNase A with His-N coordination, and V of the adduct [VIVO(8-HQ)(OH)]-RNase A with His-N coordination or [VIVO(8-HQ)(H2O)]+-RNase A with Ser/Thr-O− coordination, while aq denotes MI = 7/2 resonances of the aqua ion [VIVO(H2O)5]2+. The positions of MI = 5/2, 7/2 resonances of I, II, IIIa, IIIb, IV, and V species are also indicated by the dotted lines. Reproduced from ref. 130, which is licensed under a Creative Commons Attribution 3.0 Unported Licence, with permission from RSC, copyright 2023. | ||
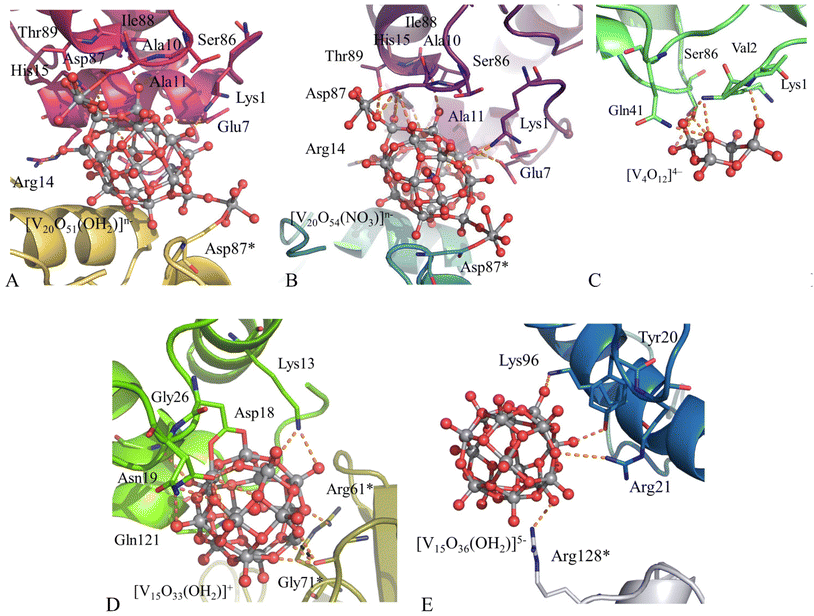
EPR has been used to confirm the formation of mixed valence polyoxidovanadate species in HEWL crystals treated with [VIVO(acac)2] (acac is acetylacetonato)131,132 (Fig. 1). During soaking of this compound in HEWL crystals, under different experimental conditions, [VIVO(acac)2] degrades, acac− ligand is released and at least a part of the VIV centers oxidizes to VV forming [VV4O12]4−, [V20O54(NO3)]n−, based on the {V18O46} cage,131 [VV7VIV8O36(OH2)]5−, [VV7VIV8O33(OH2)]+ and [V20O51(OH2)]n− (the latter is based on an unusual {V18O43} cage).132 Details of the interactions formed by these unusual compounds with HEWL are shown in Fig. 10.
These data enrich the repertoire of structures of polyoxometalate/protein structures reported in the PDB.133–137 Since POVs can act as catalysts in a wide range of reactions and can act as antiviral, antibacterial, and antitumor agents, these studies open the way to future research on the reactivity of POV/protein adducts.
EPR has also been used to investigate the binding of [dichlorido(dmb)(cym)ruthenium(II)] compound (where dmb = 1,3-dimethylbenzimidazol-2-ylidene and cym = p-cymene)(3, Fig. 1) to HEWL.138 The structure reveals the formation of a bidentate [Ru(dmb)(OHx)Cl2]/HEWL adduct (x is not determined) close to the side chain of Arg14 and His15, the binding of [Ru(dmb)(OHx)2Cl2] to the side chain of Lys33 and of a [RuCl(OHx)4] fragment close to the carbonyl oxygen of Ala107, thus suggesting that the arene ligand is lost upon the HEWL binding, while the dmb remains coordinated to the metal. EPR has been used to evaluate the effect of the cym ligand exchange on the Ru oxidation state.138 The adduct formed by 3 with HEWL is paramagnetic, consistent with the presence of a mononuclear RuIII species,139 as observed also in the case of HEWL crystals treated with AziRu.64
EPR sensitivity to structural changes around the CuII centers has been used to obtain structural information on the binding to HEWL of the CuII complex with a threonine derivative (CuST), a superoxide dismutase mimic. EPR spectra collected on this system have been compared to those registered for CuST-Im, an imidazole(Im)-bound Cu(II) complex formed when CuST reacts with Im. The g‖ and g⊥ values for CuST-Im were 2.25 and 2.07. The adduct with HEWL (CuST@HEWL) exhibited g‖ and g⊥ values of 2.24 and 2.06, respectively. These data suggest that the Cu(II) centers of CuST-Im and in the adduct adopt a square planar or square pyramidal geometry. The comparison between g‖and |A‖| values of CuST@HEWL and CuST-Im indicates smaller distortion of the metal coordination sphere in CuST@HEWL than in CuST-Im, in agreement with the crystal structure, where the Cu center binds the His15 side chain.140
Protein metalation studied by a combination of X-ray crystallography and quantum chemistry (density functional theory)
Although the combined use of spectrometric and spectroscopic techniques with XRD data permits us to collect a lot of information about the metal compound/protein interactions, computational methods were often necessary to define the origin of the products obtained by the reaction of metal compounds with proteins, fate and reactivity of the metal/protein adducts when in contact with additional ligands. Density functional theory (DFT) is a quantum-mechanical (QM) method that is used to calculate electronic and structural properties of multi-electron system modeling structures up to several hundreds of atoms.141 Proteins are too complicated systems for this computational technique. For this reason, simplified structural models,142 like those obtained using the cluster approach recently reviewed by Himo and co-workers,143,144 have been used to study the metalation process by computational methods. Using DFT calculations and a larger cluster model able to represent the protein environment of the metal binding site, T. Marino and coworkers studied the metalation process of RNase A by AP-1145 (Fig. 1), comparing the computational results with those obtained solving the structure of the metal/protein adduct.61 According to these authors, the binding to histidine of the AP-1 fragment found in the structure of the AP-1/RNase A adduct takes place by the aquated AP- 1 form and not by AP-1 itself, following the scheme reported in Fig. 11.145 DFT has also been used to gain insights into the reactivity of the metal/protein adduct with additional ligands. It has been shown that metal/protein adducts within protein crystals can still react with ligands. The post-protein binding reactivity of the [Re(CO)3(OH2)2]+ fragment bound to His15 of HEWL has been investigated by Zobi and Spingler.146 These authors have shown that the [Re(CO)3(OH2)2]+/HEWL adduct reacts with Im, pyridine-2-carboxylic acid or L-serine leading to metal ligand substitution.146 Along the same line, XRD data have demonstrated that when dirhodium(II,II) tetraacetate ([Rh2(μ-O2CCH3)4], Fig. 1) reacts with RNase A an adduct with the dirhodium center bound to the side chains of His105 and His119 is formed.12,56 This adduct can react with Im, leading to the formation of an unexpected product with Im that binds the dirhodium center at an equatorial site rather than at the expected axial site.147 The origin of this surprising experimental evidence has been studied using DFT. A small portion of the structure of the dirhodium/protein adduct around the His105 residue has been used as a model (cluster model) to investigate the reactivity of Im to the three available sites: (1) the only free axial site (Route 1 in Fig. 12), (2) the equatorial position to the Rh atom that was not coordinated to His105 (r2) (Route 2 in Fig. 12), and (3) the equatorial position to the Rh atom that was directly bound to the His (r) (Route 3 in Fig. 12). A corresponding minimal model has been used for comparison. The results suggest that Im favourably replaces the water molecule at the (r) equatorial site of the hydrated species [Rh2(Im)(μ-O2CCH3)(H2O)7]3+, where Im mimics the His105 side chain. The electronic properties of [Rh2(Im)(μ-O2CCH3)(H2O)7]3+, where the two metal centers are not equivalent, are responsible for such unexpected Im binding.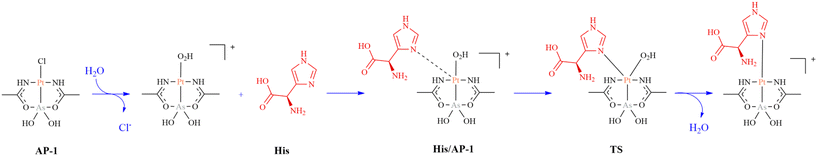 | ||
| Fig. 11 Proposed mechanism of the metalation process of RNase A by AP-1. TS stands for transition state. His in red is used to mimic His of the protein. | ||
 | ||
| Fig. 12 Substitution reactions with inclusion of Im for the dirhodium/protein adduct (cluster model). | ||
The binding of [Ru2Cl(D-p-CNPhF)(O2CCH3)3] (D-p-CNPhF– = N,N′-bis(4-cyanophenyl)formamidinate, Fig. 1) to His105 of RNase A has been recently studied using a similar approach. The structure of the adduct formed upon reaction of the diruthenium compound with the protein showed that the diruthenium center anchors to the His side chain at the axial site.148 In the adduct, two water molecules replace an acetate equatorial ligand. To understand the origin of the formation of the hydrolyzed species, quantum chemical calculations have been carried out.148 QM calculations have also been performed to estimate the O− disproportionation mechanism catalyzed by CuST@HEWL140 and to study phenylacetylene polymerization by Rh complex encapsulated in hl-L-Ft.103,149
Protein metalation studied by a combination of XFEL and QM/MM
In recent years, we have seen the advancement of X-ray-free electron lasers (XFELs) and their use in various scientific domains. XFELs produce powerful X-ray femtosecond (10−15 s) pulses that can be used to solve the structure and dynamics of matter.150 The structures can be merged into “molecular movies”, which can provide information on the biological macromolecules in action. Recently, time-resolved serial femtosecond crystallography (TR-SFX) using XFEL has been used to characterize a light-induced CO-release from Mn to HEWL within porous metal/protein adduct microcrystals.116 In this study, conformational variations of the metal/protein adduct during the CO release from the metal center upon light exposure have been evaluated. In darkness, crystals of HEWL in the presence of [Mn(CO)3(acetone)3](CF3SO3) (Fig. 1) show a [Mn(CO)3(wat)2]2+ fragment bound to the side chain of His15. At this site, the metal adopts an octahedral geometry, with two CO and two water ligands equatorially coordinated (Mn–COeq and Mn–OH2,eq bonds) and Im of His and the third CO that are axially coordinated (Mn–COax bond). The structures refined using data collected after 10 ns, 100 ns, and 1 μs of light excitation using a 20 μJ pump laser indicate a greater susceptibility to radiation of the Mn–COax bond when compared to the Mn–COeq bond. Comparison between the solved structures permits them to capture the real-time intermediate formation. Interestingly, the paper also describes the structure of the protein after the complete release of CO. This structure does not show the presence of Mn coordinated to His15. After complete CO release, the Mn-aqua species that probably forms is unstable and immediately releases from the protein. CO release from metal-based CO releasing molecules bound to proteins had previously been described by collecting snapshots from different crystal structures of Ru-CORMs/HEWL adducts.52,151QM/molecular mechanics (QM/MM) calculations has been used to elucidate the detailed reaction processes in photodissociation.116 The QM/MM calculations combine the accuracy of ab initio QM calculations and the speed of the classic approaches, thus allowing the study of chemical processes in large systems as proteins.152 The QM/MM results predict that the release of the Mn(CO)-unit is faster than the exchange of third CO in the equatorial position with water molecules, in agreement with crystallographic observation.
Protein metalation studied by a combination of X-ray crystallography and other computational approaches
Molecular docking is the computational method that allows the prediction of small molecule binding sites on a biological macromolecule structure. Docking is frequently used in structure-based drug design, but it has been recently combined with DFT calculations and crystallographic experiments for a better definition of the stability of the metal/protein adduct and to highlight the role of specific interactions on the metal compound/protein recognition process. For example, in the structure of the adduct formed upon the reaction of [VIVO(8-HQ)2] with RNase A, a [VIVO(8-HQ)(H2O)]+ ion binds the protein close to the side chain of Glu111.130 Docking experiments have been used to refine the metal binding site revealing the reasons behind the ligand exchange process, not observed for similar complexes like [VIVO(pic)2]76 and not predicted by thermodynamic considerations. The computational data show that the [VIVO(8-HQ)(H2O)]+/RNase A adduct forms from [VIVO(8-HQ)2]/RNase A and that its formation is facilitated by the presence of the Gln69 side chain.Similarly, docking experiments have been performed to study the binding of the same compound to HEWL. X-ray diffraction analyses showed that [VIVO(8-HQ)(H2O)]+ coordinates to the Asp119 side chain. The binding is stabilized by interactions with Asn103 and Trp62 from a symmetry-related molecule.74 First, the structure of [VIVO(8-HQ)(H2O)2]+ has been optimized by DFT. In the second step, the coordinative interaction of [VIVO(8-HQ)(H2O)]+ with the protein has been investigated to unveil the molecular reasons why the binding of [VIVO(8-HQ)(H2O)]+ to HEWL occurs at the Asp119 side chain and not at other potential binding sites such as Glu35, Asn46, Asp48 and Asp52 side chains. To assess the effect of the crystal lattice or, in general, of protein–protein interactions on the binding of [VIVO(8-HQ)(H2O)]+ to the protein, two different docking approaches have been applied: (1) An ensemble docking, denoted as the “classic model”, which uses one metal-containing fragment and the three superimposed structures of HEWL; and (2) a docking denoted as the “symmetry mate model”, which uses one metal-containing fragment with two symmetry-related protein chains. This calculation has been carried out to model the crystal lattice or protein–protein interactions. According to ensemble docking results, [VIVO(8-HQ)(H2O)]+ should interact with Glu35 or Asp52 side chains. Thus, protein–protein stabilization has to be invoked to explain the experimental data. Indeed, the coordination of the V center to the carboxylate-O of Asp119 is observed using the “symmetry mate model”. Overall, these data indicate that protein–protein stabilization may facilitate unexpected binding of metal-based compounds to proteins, offering new insights into the interaction mechanisms of metal-based drugs with proteins.74
A series of docking simulations have also been performed to gain insights into the V compound chiral discrimination by HEWL. The computational data indicate that the formation of the HEWL-VO-bipy adduct is driven by the presence of structured water molecules, suggesting the pivotal role of microsolvation for chiral discrimination of the binding region and in stabilizing the final structure of the metal/protein adduct.129
Molecular dynamics (MD) simulations also gave a contribution in defining the interaction of metal compounds with proteins, in combination with X-ray crystallography. Recently, for example, the structures of HEWL adducts with isostructural NHC complexes with Ru, Os, Rh, and Ir were further studied using constant-pH molecular dynamics (CpHMD) simulations. The X-ray structures of these adducts reveal that [Ru(dmb)(H2O)Cl2] binds His15 and Arg14, [Os(cym)(dmb)Cl] interacts with Asn103; [Rh(Cp*)Cl2] binds His15, while [Ir(Cp*)(dmb)Cl] the side chain of Asp101.
CpHMD simulations allow us to evaluate pKa values of the metal-binding residues. CpHMD indicates that His15 and Lys33 have pKa of 6.67 and 9.91, respectively, while Asp87 and Asp101 have pKas of 2.73 and 0.44. This result suggests that these two Asp have pKa values significantly lower than those observed in the case of solvent-exposed Asp (pKa = 4) and provides an explanation for the hydrogen bonds that Asp87 forms with His15. This interaction probably affects the electropositivity of His15, providing insights into the observed specificity of His15 for Ru and Rh compounds vs. Os and Ir, which prefer electronegative binding pockets.70
Classic MD simulations have also been used in other studies. The X-ray structures of HEWL, proteinase K and thaumatin (Fig. 2) in the presence of Tb-Xo4 (G1, Fig. 1) and three variants (Tb-Xo4-NMet2, Tb-Xo4-OH and Tb-Xo4-SO3, denoted as V1, V2 or V3, Fig. 1) have been solved. To gain insights into the role of pendant arms in the structure of G1, V1, V2 or V3 on the binding to proteins, MD simulations (200 ns) have been carried out on systems containing metal compounds and four HEWL molecules. These starting models mimic the protein crystal packing. Results indicate that V1, V2 or V3 can bind one protein chain through interaction formed by carboxylates with the metal; the pendant arms can interact with surface residues of the other protein molecules with ammonium or sulfonate moieties that form salt bridges with acid residues and the OH group that forms hydrogen bonds, both as H donor or acceptor.153
Computational studies have also been used to help the refinement process of crystal structures of metal/protein adducts. In the adduct formed upon soaking of VIVOSO4 in HEWL crystals, a metal center binds a water molecule, an Ooxido, a Cl− ion, and the side chains of Asp46 and Asp52. The sixth ligand, needed to complete the octahedral coordination sphere, is not unambiguously identified by the authors, since it cannot be seen in the electron density map at the 1.0σ level. Refinement of this site addressed by docking and QM/MM calculations suggests that the sixth coordination position is probably occupied by a water molecule.129
Conclusions
In the last 15 years, X-ray crystallography has provided a detailed description of the protein metalation process in several cases. However, significant progress can be made in this field since many aspects of the protein metalation process remain still unknown.8 The combined use of X-ray crystallography and biophysical techniques, as described in this work, has certainly allowed substantial progress in this specific research area. Of course, there are many other biophysical techniques that can be used to complement X-ray crystallography in the definition of the structure of metal/protein adducts. Examples are synchrotron terahertz and neutron scattering spectroscopy,154 and capillary electrophoresis.155 Putting together the results obtained using various physicochemical methods, the molecular mechanisms underlying the metallodrug/protein recognition process and structural/functional features of metal/protein adducts can be elucidated with high accuracy.Data that have been collected so far demonstrate that the binding of metallodrugs to proteins does not alter the overall in-crystallo conformation of the biological macromolecules156 and only in a few cases it alters their stability.38,78 However, the binding can significantly alter the catalytic activity of enzymes62 and their recognition by other protein partners. The binding of metal containing fragments to proteins can result in a modification of the metal local environment, significantly affecting its reactivity with additional ligands and sometimes creating a novel catalytic activity. Although there are many potential metal binding sites on protein surfaces, the coordination of metals to residue side chains takes place preferentially only in a few positions, suggesting that the protein metalation is a selective process. The preferential choice of some residues with respect to others is determined by different parameters: solvent accessibility and flexibility of the protein residues involved in the first steps of the metal compound/protein recognition process, steric/electronic effects due to metal ligands or protein residues, and potential interactions that metal ligands can form with protein surface patches. Hard–hard and soft–soft interactions of course contribute to driving the metallodrug towards its final target. In this respect, it is worth noting that the reactivity of metal complexes with proteins is certainly regulated by the experimental conditions used for the analysis. Thus, the pictures that emerge using different techniques that require different solutions and conditions can sometimes diverge.
Further studies will greatly accelerate the comprehension of reaction mechanisms of metallodrugs with protein targets, thus aiding the design of novel therapeutics. In this respect, recent reviews have summarized the methods that can be used for target identification by metal-based anticancer agents.157,158 The accumulated knowledge will also facilitate the design of novel protein-based drug delivery systems, allowing the development of systems with precise mechanism of metallodrug release, and the production of new artificial metalloenzymes, allowing the development of systems that catalyze new reactions. Artificial intelligence and machine learning are groundbreaking tools that can be integrated with existing methods to accelerate this process even further. The study of the interaction of metal compounds with proteins in real cellular systems is certainly enormously more difficult. There are thousands of proteins within a cell and a significant number of additional low molecular weight metabolites at variable concentrations.159 Nonetheless, we believe that studying the protein metalation process using different techniques can certainly represent a useful basis for the understanding of this process within cells.
Data availability
Data are all available in the literature or deposited in the PBD.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. Merlino thanks MIUR PRIN 2022-Cod. 2022JMFC3X, “Protein Metalation by Anticancer Metal-based Drugs” for financial support.References
- D. Osman and N. J. Robinson, Protein metalation in a nutshell, FEBS Lett., 2023, 597, 141–150 CAS.
- A. W. Foster, D. Osman and N. J. Robinson, Metal Preferences and Metallation, J. Biol. Chem., 2014, 289, 28095–28103 CAS.
- H. J. Davis and T. R. Ward, Artificial Metalloenzymes: Challenges and Opportunities, ACS Cent. Sci., 2019, 5, 1120–1136 Search PubMed.
- A. Bergamo and G. Sava, Ruthenium complexes can target determinants of tumour malignancy, Dalton Trans., 2007, 1267 CAS.
- E. J. Anthony, E. M. Bolitho, H. E. Bridgewater, O. W. L. Carter, J. M. Donnelly, C. Imberti, E. C. Lant, F. Lermyte, R. J. Needham, M. Palau, P. J. Sadler, H. Shi, F.-X. Wang, W.-Y. Zhang and Z. Zhang, Metallodrugs are unique: opportunities and challenges of discovery and development, Chem. Sci., 2020, 11, 12888–12917 RSC.
- A. Frei, J. Zuegg, A. G. Elliott, M. Baker, S. Braese, C. Brown, F. Chen, C. G. Dowson, G. Dujardin, N. Jung, A. P. King, A. M. Mansour, M. Massi, J. Moat, H. A. Mohamed, A. K. Renfrew, P. J. Rutledge, P. J. Sadler, M. H. Todd, C. E. Willans, J. J. Wilson, M. A. Cooper and M. A. T. Blaskovich, Metal complexes as a promising source for new antibiotics, Chem. Sci., 2020, 11, 2627–2639 Search PubMed.
- X. Xu, F. Dai, Y. Mao, K. Zhang, Y. Qin and J. Zheng, Metallodrugs in the battle against non-small cell lung cancer: unlocking the potential for improved therapeutic outcomes, Front. Pharmacol., 2023, 14, 1242488 CrossRef CAS PubMed.
- A. Merlino, Recent advances in protein metalation: structural studies, Chem. Commun., 2021, 57, 1295–1307 RSC.
- L. Messori and A. Merlino, Cisplatin binding to proteins: A structural perspective, Coord. Chem. Rev., 2016, 315, 67–89 CAS.
- A. Giorgio and A. Merlino, Gold metalation of proteins: Structural studies, Coord. Chem. Rev., 2020, 407, 213175 Search PubMed.
- A. Merlino, Metallodrug binding to serum albumin: Lessons from biophysical and structural studies, Coord. Chem. Rev., 2023, 480, 215026 CAS.
- D. Loreto and A. Merlino, The interaction of rhodium compounds with proteins: A structural overview, Coord. Chem. Rev., 2021, 442, 213999 CAS.
- J. G. Grossmann, M. Neu, E. Pantos, F. J. Schwab, R. W. Evans, E. Townes-Andrews, P. F. Lindley, H. Appel, W.-G. Thies and S. S. Hasnain, X-ray solution scattering reveals conformational changes upon iron uptake in lactoferrin, serum and ovo-transferrins, J. Mol. Biol., 1992, 225, 811–819 CAS.
- J. G. Grossmann, M. Neu, R. W. Evans, P. F. Lindley, H. Appel and S. S. Hasnain, Metal-induced Conformational Changes in Transferrins, J. Mol. Biol., 1993, 229, 585–590 CAS.
- R. Bauer, A. Atke, E. Danielsen, J. Marcussen, C. E. Olsen, J. Rehfeld, T. Saermark, D. Schneider, H. Vilhardt and M. Zeppezauer, The potential of perturbed angular correlation of gamma rays as a tool for dynamic studies of peptides/proteins, Int. J. Radiat. Appl. Instrum., Part A, 1991, 42, 1015–1023 CAS.
- J. M. Legendre, A. Turzo, J. F. Morin and P. P. Morin, Influence du pH et de l'anion de liaison sur l'interaction In3+-transferrine: étude spectroscopique de la corrélation angulaire perturbée des raies γ 172–245 keV de l'indium 111, Biochimie, 1984, 66, 429–436 CAS.
- P. J. Marsden, F. A. Smith and R. W. Evans, Evidence of conformational changes in the non-equivalent binding sites of human serum transferrin, Int. J. Radiat. Appl. Instrum., Part A, 1989, 40, 715–722 CAS.
- I. Russo Krauss, G. Ferraro, A. Pica, J. A. Márquez, J. R. Helliwell and A. Merlino, Principles and methods used to grow and optimize crystals of protein–metallodrug adducts, to determine metal binding sites and to assign metal ligands, Metallomics, 2017, 9, 1534–1547 CAS.
- C. G. Hartinger, M. Groessl, S. M. Meier, A. Casini and P. J. Dyson, Application of mass spectrometric techniques to delineate the modes-of-action of anticancer metallodrugs, Chem. Soc. Rev., 2013, 42, 6186 CAS.
- S. Theiner, A. Schoeberl, A. Schweikert, B. K. Keppler and G. Koellensperger, Mass spectrometry techniques for imaging and detection of metallodrugs, Curr. Opin. Chem. Biol., 2021, 61, 123–134 CAS.
- N. Potier, H. Rogniaux, G. Chevreux and A. Van Dorsselaer, in Methods in Enzymology, Elsevier, 2005, vol. 402, pp. 361–389 Search PubMed.
- K. Wilson and J. M. Walker, in Principles and techniques of biochemistry and molecular biology, Cambridge University Press, Cambridge (Mass.), 7th edn, 2010 Search PubMed.
- S. Vaidyanathan, D. B. Kell and R. Goodacre, Flow-injection electrospray ionization mass spectrometry of crude cell extracts for high-throughput bacterial identification, J. Am. Soc. Mass Spectrom., 2002, 13, 118–128 CAS.
- M. C. McMaster, in HPLC, a practical user's guide, Wiley-Interscience, Hoboken, N.J, 2nd edn, 2007 Search PubMed.
- J. A. Olivares, N. T. Nguyen, C. R. Yonker and R. D. Smith, On-line mass spectrometric detection for capillary zone electrophoresis, Anal. Chem., 1987, 59, 1230–1232 CAS.
- G. R. D. Prabhu, E. R. Williams, M. Wilm and P. L. Urban, Mass spectrometry using electrospray ionization, Nat. Rev. Methods Primers, 2023, 3, 23 CAS.
- L. Messori and A. Merlino, Protein metalation by metal-based drugs: X-ray crystallography and mass spectrometry studies, Chem. Commun., 2017, 53, 11622–11633 CAS.
- N. Zhang, M. Cui, Y. Du, Z. Liu and S. Liu, Exploring the interaction of cisplatin with β2-microglobulin: new insights into a chemotherapeutic drug, RSC Adv., 2014, 4, 2300–2305 CAS.
- H. Li, J. R. Snelling, M. P. Barrow, J. H. Scrivens, P. J. Sadler and P. B. O'Connor, Mass Spectrometric Strategies to Improve the Identification of Pt(II)-Modification Sites on Peptides and Proteins, J. Am. Soc. Mass Spectrom., 2014, 25, 1217–1227 CAS.
- T. Marzo, G. Ferraro, A. Merlino and L. Messori, in Encyclopedia of Inorganic and Bioinorganic Chemistry, ed. R. A. Scott, Wiley, 2nd edn, 2020, pp. 1–17 Search PubMed.
- A. Casini, A. Guerri, C. Gabbiani and L. Messori, Biophysical characterisation of adducts formed between anticancer metallodrugs and selected proteins: New insights from X-ray diffraction and mass spectrometry studies, J. Inorg. Biochem., 2008, 102, 995–1006 CrossRef CAS PubMed.
- A. Casini, G. Mastrobuoni, C. Temperini, C. Gabbiani, S. Francese, G. Moneti, C. T. Supuran, A. Scozzafava and L. Messori, ESI mass spectrometry and X-ray diffraction studies of adducts between anticancer platinum drugs and hen egg white lysozyme, Chem. Commun., 2007, 156–158 RSC.
- L. Messori and A. Merlino, Cisplatin Binding to Proteins: Molecular Structure of the Ribonuclease A Adduct, Inorg. Chem., 2014, 53, 3929–3931 CrossRef CAS PubMed.
- L. Messori, T. Marzo and A. Merlino, Interactions of carboplatin and oxaliplatin with proteins: Insights from X-ray structures and mass spectrometry studies of their ribonuclease A adducts, J. Inorg. Biochem., 2015, 153, 136–142 CrossRef CAS PubMed.
- S. B. Howell, R. Safaei, C. A. Larson and M. J. Sailor, Copper Transporters and the Cellular Pharmacology of the Platinum-Containing Cancer Drugs, Mol. Pharmacol., 2010, 77, 887–894 CrossRef CAS PubMed.
- G. Ferraro, L. Messori and A. Merlino, The X-ray structure of the primary adducts formed in the reaction between cisplatin and cytochrome c, Chem. Commun., 2015, 51, 2559–2561 RSC.
- S. Chen, C. Yuan, L. Jiang, Z. Luo and M. Huang, Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid, Int. J. Biol. Macromol., 2022, 216, 172–178 CrossRef CAS PubMed.
- I. Moraleja, E. Moreno-Gordaliza, D. Esteban-Fernández, M. L. Mena, M. W. Linscheid and M. M. Gómez-Gómez, A shotgun approach for the identification of platinum–protein complexes, Anal. Bioanal. Chem., 2015, 407, 2393–2403 CrossRef CAS PubMed.
- W. Hu, Q. Luo, K. Wu, X. Li, F. Wang, Y. Chen, X. Ma, J. Wang, J. Liu, S. Xiong and P. J. Sadler, The anticancer drug cisplatin can cross-link the interdomain zinc site on human albumin, Chem. Commun., 2011, 47, 6006 Search PubMed.
- N. Balasco, G. Ferraro, D. Loreto, I. Iacobucci, M. Monti and A. Merlino, Cisplatin binding to β-lactoglobulin: a structural study, Dalton Trans., 2020, 49, 12450–12457 Search PubMed.
- R. Troisi, F. Galardo, G. Ferraro, F. Sica and A. Merlino, Cisplatin Binding to Human Serum Transferrin: A Crystallographic Study, Inorg. Chem., 2023, 62, 675–678 Search PubMed.
- G. Ferraro, V. Sanfilippo, L. Chiaverini, C. Satriano, T. Marzo, A. Merlino and D. La Mendola, Cisplatin binding to angiogenin protein: new molecular pathways and targets for the drug's anticancer activity, Dalton Trans., 2023, 52, 9058–9067 CAS.
- H. Li, S. A. Wells, J. E. Jimenez-Roldan, R. A. Römer, Y. Zhao, P. J. Sadler and P. B. O'Connor, Protein flexibility is key to cisplatin crosslinking in calmodulin, Protein Sci., 2012, 21, 1269–1279 CrossRef CAS PubMed.
- D. M. Monti, D. Loreto, I. Iacobucci, G. Ferraro, A. Pratesi, L. D'Elia, M. Monti and A. Merlino, Protein-Based Delivery Systems for Anticancer Metallodrugs: Structure and Biological Activity of the Oxaliplatin/β-Lactoglobulin Adduct, Pharmaceuticals, 2022, 15, 425 CrossRef CAS PubMed.
- T. Marzo, G. Ferraro, L. M. Cucci, A. Pratesi, Ö. Hansson, C. Satriano, A. Merlino and D. La Mendola, Oxaliplatin inhibits angiogenin proliferative and cell migration effects in prostate cancer cells, J. Inorg. Biochem., 2022, 226, 111657 CAS.
- D. Picone, F. Donnarumma, G. Ferraro, I. Russo Krauss, A. Fagagnini, G. Gotte and A. Merlino, Platinated oligomers of bovine pancreatic ribonuclease: Structure and stability, J. Inorg. Biochem., 2015, 146, 37–43 CAS.
- G. Ferraro, T. Lyčková, L. Massai, P. Štarha, L. Messori and A. Merlino, Picoplatin binding to proteins: X-ray structures and mass spectrometry data on the adducts with lysozyme and ribonuclease A, Dalton Trans., 2024, 53, 8535–8540 Search PubMed.
- C. Mügge, T. Marzo, L. Massai, J. Hildebrandt, G. Ferraro, P. Rivera-Fuentes, N. Metzler-Nolte, A. Merlino, L. Messori and W. Weigand, Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein BindingClick to copy article link, Inorg. Chem., 2015, 54, 8560–8570 Search PubMed.
- T. Marzo, F. Navas, D. Cirri, A. Merlino, G. Ferraro, L. Messori and A. G. Quiroga, Reactions of a tetranuclear Pt-thiosemicarbazone complex with model proteins, J. Inorg. Biochem., 2018, 181, 11–17 Search PubMed.
- J. Delasoie, A. Pavic, N. Voutier, S. Vojnovic, A. Crochet, J. Nikodinovic-Runic and F. Zobi, Highly Potent rhenium(I) Tricarbonyl Complexes with Dual Anticancer and Anti-Angiogenic Activity Against Colorectal Carcinoma, 2020, preprint, DOI:10.26434/chemrxiv.12012840.v1.
- L. Messori and A. Merlino, Ruthenium metalation of proteins: the X-ray structure of the complex formed between NAMI-A and hen egg white lysozyme, Dalton Trans., 2014, 43, 6128 CAS.
- G. Tamasi, A. Merlino, F. Scaletti, P. Heffeter, A. A. Legin, M. A. Jakupec, W. Berger, L. Messori, B. K. Keppler and R. Cini, {Ru(CO)x}-Core complexes with benzimidazole ligands: synthesis, X-ray structure and evaluation of anticancer activity in vivo, Dalton Trans., 2017, 46, 3025–3040 CAS.
- L. Messori, M. A. Cinellu and A. Merlino, Protein Recognition of Gold-Based Drugs: 3D Structure of the Complex Formed When Lysozyme Reacts with Aubipy c, ACS Med. Chem. Lett., 2014, 5, 1110–1113 CAS.
- L. Messori, F. Scaletti, L. Massai, M. A. Cinellu, C. Gabbiani, A. Vergara and A. Merlino, The mode of action of anticancer gold-based drugs: a structural perspective, Chem. Commun., 2013, 49, 10100 CAS.
- L. Messori, T. Marzo, R. N. F. Sanches, Hanif-Ur-Rehman, D. de Oliveira Silva and A. Merlino, Unusual Structural Features in the Lysozyme Derivative of the Tetrakis(acetato)chloridodiruthenium(II,III) Complex, Angew. Chem., Int. Ed., 2014, 53, 6172–6175 CAS.
- G. Ferraro, A. Pratesi, L. Messori and A. Merlino, Protein interactions of dirhodium tetraacetate: a structural study, Dalton Trans., 2020, 49, 2412–2416 CAS.
- D. Loreto, B. Maity, T. Morita, H. Nakamura, A. Merlino and T. Ueno, Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts, Inorg. Chem., 2023, 62, 7515–7524 CAS.
- D. Loreto, G. Ferraro and A. Merlino, Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme, Int. J. Mol. Sci., 2021, 22, 1496 CAS.
- M. Serratrice, L. Maiore, A. Zucca, S. Stoccoro, I. Landini, E. Mini, L. Massai, G. Ferraro, A. Merlino, L. Messori and M. A. Cinellu, Cytotoxic properties of a new organometallic platinum(II) complex and its gold(I) heterobimetallic derivatives, Dalton Trans., 2016, 45, 579–590 CAS.
- G. Ferraro, D. Cirri, T. Marzo, A. Pratesi, L. Messori and A. Merlino, The first step of arsenoplatin-1 aggregation in solution unveiled by solving the crystal structure of its protein adduct, Dalton Trans., 2021, 50, 68–71 CAS.
- Đ. Miodragović, A. Merlino, E. P. Swindell, A. Bogachkov, R. W. Ahn, S. Abuhadba, G. Ferraro, T. Marzo, A. P. Mazar, L. Messori and T. V. O'Halloran, Arsenoplatin-1 Is a Dual Pharmacophore Anticancer Agent, J. Am. Chem. Soc., 2019, 141, 6453–6457 Search PubMed.
- L. Messori, F. Scaletti, L. Massai, M. A. Cinellu, I. Russo Krauss, G. Di Martino, A. Vergara, L. Paduano and A. Merlino, Interactions of gold-based drugs with proteins: crystal structure of the adduct formed between ribonuclease A and a cytotoxic gold(III) compound, Metallomics, 2014, 6, 233–236 CAS.
- I. Russo Krauss, L. Messori, M. A. Cinellu, D. Marasco, R. Sirignano and A. Merlino, Interactions of gold-based drugs with proteins: the structure and stability of the adduct formed in the reaction between lysozyme and the cytotoxic gold(III) compound Auoxo3, Dalton Trans., 2014, 43, 17483–17488 CAS.
- A. Vergara, G. D'Errico, D. Montesarchio, G. Mangiapia, L. Paduano and A. Merlino, Interaction of Anticancer Ruthenium Compounds with Proteins: High-Resolution X-ray Structures and Raman Microscopy Studies of the Adduct between Hen Egg White Lysozyme and AziRu, Inorg. Chem., 2013, 52, 4157–4159 CAS.
- A. Casini, C. Temperini, C. Gabbiani, C. T. Supuran and L. Messori, The X–ray Structure of the Adduct between NAMI–A and Carbonic Anhydrase Provides Insights into the Reactivity of this Metallodrug with Proteins, ChemMedChem, 2010, 5, 1989–1994 CrossRef CAS PubMed.
- S. Ciambellotti, A. Pratesi, M. Severi, G. Ferraro, E. Alessio, A. Merlino and L. Messori, The NAMI A – human ferritin system: a biophysical characterization, Dalton Trans., 2018, 47, 11429–11437 RSC.
- L. Chiniadis, P. Giastas, I. Bratsos and A. Papakyriakou, Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points, Inorg. Chem., 2021, 60, 10729–10737 CrossRef CAS PubMed.
- M. Oszajca, M. Flejszar, A. Szura, P. Dróżdż, M. Brindell and K. Kurpiewska, Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies, Front. Chem., 2024, 12, 1371637 CrossRef CAS PubMed.
- A. Merlino, Interactions between proteins and Ru compounds of medicinal interest: A structural perspective, Coord. Chem. Rev., 2016, 326, 111–134 CrossRef CAS.
- M. P. Sullivan, M. Cziferszky, I. Tolbatov, D. Truong, D. Mercadante, N. Re, R. Gust, D. C. Goldstone and C. G. Hartinger, Probing the Paradigm of Promiscuity for N–Heterocyclic Carbene Complexes and their Protein Adduct Formation, Angew. Chem., Int. Ed., 2021, 60, 19928–19932 CrossRef CAS PubMed.
- G. Ferraro, L. Massai, L. Messori, M. A. Cinellu and A. Merlino, Structural evidences for a secondary gold binding site in the hydrophobic box of lysozyme, BioMetals, 2015, 28, 745–754 CrossRef CAS PubMed.
- A. Pratesi, D. Cirri, D. Fregona, G. Ferraro, A. Giorgio, A. Merlino and L. Messori, Structural Characterization of a Gold/Serum Albumin Complex, Inorg. Chem., 2019, 58, 10616–10619 CrossRef CAS PubMed.
- G. Ferraro, M. Paolillo, G. Sciortino, E. Garribba and A. Merlino, Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme, Inorg. Chem., 2022, 61, 16458–16467 Search PubMed.
- M. Paolillo, G. Ferraro, F. Pisanu, J. Maréchal, G. Sciortino, E. Garribba and A. Merlino, Protein–Protein Stabilization in V IV O/8−Hydroxyquinoline–Lysozyme Adducts, Chem. – Eur. J., 2024, e202401712 CrossRef CAS PubMed.
- G. Ferraro, M. Paolillo, G. Sciortino, F. Pisanu, E. Garribba and A. Merlino, Implications of Protein Interaction in the Speciation of Potential V IV O–Pyridinone Drugs, Inorg. Chem., 2023, 62, 8407–8417 CrossRef CAS PubMed.
- G. Ferraro, N. Demitri, L. Vitale, G. Sciortino, D. Sanna, V. Ugone, E. Garribba and A. Merlino, Spectroscopic/Computational Characterization and the X-ray Structure of the Adduct of the V IV O–Picolinato Complex with RNase A, Inorg. Chem., 2021, 60, 19098–19109 CrossRef CAS PubMed.
- F. Lanucara, S. W. Holman, C. J. Gray and C. E. Eyers, The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics, Nat. Chem., 2014, 6, 281–294 Search PubMed.
- M. P. Sullivan, M. Groessl, S. M. Meier, R. L. Kingston, D. C. Goldstone and C. G. Hartinger, The metalation of hen egg white lysozyme impacts protein stability as shown by ion mobility mass spectrometry, differential scanning calorimetry, and X-ray crystallography, Chem. Commun., 2017, 53, 4246–4249 Search PubMed.
- L. Eade, M. P. Sullivan, T. M. Allison, D. C. Goldstone and C. G. Hartinger, Not All Binding Sites Are Equal: Site Determination and Folding State Analysis of Gas–Phase Protein–Metallodrug Adducts, Chem. – Eur. J., 2024, 30, e202400268 CrossRef CAS PubMed.
- J. P. Williams, H. I. A. Phillips, I. Campuzano and P. J. Sadler, Shape changes induced by N-terminal platination of ubiquitin by cisplatin, J. Am. Soc. Mass Spectrom., 2010, 21, 1097–1106 CrossRef CAS PubMed.
- E. Escribano, S. Madurga, M. Vilaseca and V. Moreno, Ion mobility and Top-down MS complementary approaches for the structural analysis of protein models bound to anticancer metallodrugs, Inorg. Chim. Acta, 2014, 423, 60–69 Search PubMed.
- L. Cosottini, A. Geri, V. Ghini, M. Mannelli, S. Zineddu, G. Di Paco, A. Giachetti, L. Massai, M. Severi, T. Gamberi, A. Rosato, P. Turano and L. Messori, Unlocking the Power of Human Ferritin: Enhanced Drug Delivery of Aurothiomalate in A2780 Ovarian Cancer Cells, Angew. Chem., Int. Ed., 2024, e202410791 Search PubMed.
- R. Lucignano, A. Pratesi, P. Imbimbo, D. M. Monti, D. Picone, L. Messori, G. Ferraro and A. Merlino, Evaluation of Auranofin Loading within Ferritin Nanocages, Int. J. Mater. Sci., 2022, 23, 14162 CAS.
- D. M. Monti, G. Ferraro and A. Merlino, Ferritin-based anticancer metallodrug delivery: Crystallographic, analytical and cytotoxicity studies, Nanomedicine, 2019, 20, 101997 CrossRef CAS PubMed.
- C. G. Hartinger, Y. O. Tsybin, J. Fuchser and P. J. Dyson, Characterization of Platinum Anticancer Drug Protein-Binding Sites Using a Top-Down Mass Spectrometric Approach, Inorg. Chem., 2008, 47, 17–19 CrossRef CAS PubMed.
- J. Will, D. A. Wolters and W. S. Sheldrick, Characterisation of Cisplatin Binding Sites in Human Serum Proteins Using Hyphenated Multidimensional Liquid Chromatography and ESI Tandem Mass Spectrometry, ChemMedChem, 2008, 3, 1696–1707 CrossRef CAS PubMed.
- R. F. S. Lee, L. Menin, L. Patiny, D. Ortiz and P. J. Dyson, Versatile Tool for the Analysis of Metal–Protein Interactions Reveals the Promiscuity of Metallodrug–Protein Interactions, Anal. Chem., 2017, 89, 11985–11989 CrossRef CAS PubMed.
- J. Delafiori, G. Ring and A. Furey, Clinical applications of HPLC–ICP-MS element speciation: A review, Talanta, 2016, 153, 306–331 CrossRef CAS PubMed.
- G. Petruk, D. M. Monti, G. Ferraro, A. Pica, L. D'Elia, F. Pane, A. Amoresano, J. Furrer, K. Kowalski and A. Merlino, Encapsulation of the Dinuclear Trithiolato-Bridged Arene Ruthenium Complex Diruthenium-1 in an Apoferritin Nanocage: Structure and Cytotoxicity, ChemMedChem, 2019, 14, 594–602 CrossRef CAS PubMed.
- A. Bijelic, S. Theiner, B. K. Keppler and A. Rompel, X-ray Structure Analysis of Indazolium trans- [Tetrachlorobis(1 H -indazole)ruthenate(III)] (KP1019) Bound to Human Serum Albumin Reveals Two Ruthenium Binding Sites and Provides Insights into the Drug Binding Mechanism, J. Med. Chem., 2016, 59, 5894–5903 CrossRef CAS PubMed.
- K. Fujita, Y. Tanaka, T. Sho, S. Ozeki, S. Abe, T. Hikage, T. Kuchimaru, S. Kizaka-Kondoh and T. Ueno, Intracellular CO Release from Composite of Ferritin and Ruthenium Carbonyl Complexes, J. Am. Chem. Soc., 2014, 136, 16902–16908 CrossRef CAS PubMed.
- G. Ferraro, D. M. Monti, A. Amoresano, N. Pontillo, G. Petruk, F. Pane, M. A. Cinellu and A. Merlino, Gold-based drug encapsulation within a ferritin nanocage: X-ray structure and biological evaluation as a potential anticancer agent of the Auoxo3-loaded protein, Chem. Commun., 2016, 52, 9518–9521 RSC.
- D. M. Monti, G. Ferraro, G. Petruk, L. Maiore, F. Pane, A. Amoresano, M. A. Cinellu and A. Merlino, Ferritin nanocages loaded with gold ions induce oxidative stress and apoptosis in MCF-7 human breast cancer cells, Dalton Trans., 2017, 46, 15354–15362 RSC.
- G. Ferraro, A. Pica, G. Petruk, F. Pane, A. Amoresano, A. Cilibrizzi, R. Vilar, D. M. Monti and A. Merlino, Preparation, structure, Cytotoxicity and Mechanism of Action of ferritin-Pt(II) Terpyridine Compound Nanocomposites, Nanomedicine, 2018, 13, 2995–3007 CAS.
- G. Ferraro, A. Pratesi, D. Cirri, P. Imbimbo, D. Maria Monti, L. Messori and A. Merlino, Arsenoplatin-Ferritin Nanocage: Structure and Cytotoxicity, Int. J. Mater. Sci., 2021, 22, 1874 CAS.
- C. Lu, B. Maity, X. Peng, N. Ito, S. Abe, X. Sheng, T. Ueno and D. Lu, Design of a gold clustering site in an engineered apo-ferritin cage, Commun. Chem., 2022, 5, 39 CAS.
- B. Maity, S. Abe and T. Ueno, Observation of gold sub-nanocluster nucleation within a crystalline protein cage, Nat. Commun., 2017, 8, 14820 CAS.
- K. Fujita, Y. Tanaka, S. Abe and T. Ueno, A Photoactive Carbon–Monoxide–Releasing Protein Cage for Dose–Regulated Delivery in Living Cells, Angew. Chem., Int. Ed., 2016, 55, 1056–1060 CAS.
- A. R. Timerbaev, S. S. Aleksenko, K. Polec-Pawlak, R. Ruzik, O. Semenova, C. G. Hartinger, S. Oszwaldowski, M. Galanski, M. Jarosz and B. K. Keppler, Platinum metallodrug-protein binding studies by capillary electrophoresis-inductively coupled plasma-mass spectrometry: Characterization of interactions between Pt(II) complexes and human serum albumin, Electrophoresis, 2004, 25, 1988–1995 CAS.
- M. P. Sullivan, S. J. Morrow, D. C. Goldstone and C. G. Hartinger, Gel electrophoresis in combination with laser ablation–inductively coupled plasma mass spectrometry to quantify the interaction of cisplatin with human serum albumin, Electrophoresis, 2019, 40, 2329–2335 CAS.
- S. Abe, J. Niemeyer, M. Abe, Y. Takezawa, T. Ueno, T. Hikage, G. Erker and Y. Watanabe, Control of the Coordination Structure of Organometallic Palladium Complexes in an apo-Ferritin Cage, J. Am. Chem. Soc., 2008, 130, 10512–10514 CAS.
- T. Ueno, M. Abe, K. Hirata, S. Abe, M. Suzuki, N. Shimizu, M. Yamamoto, M. Takata and Y. Watanabe, Process of Accumulation of Metal Ions on the Interior Surface of apo-Ferritin: Crystal Structures of a Series of apo-Ferritins Containing Variable Quantities of Pd(II) Ions, J. Am. Chem. Soc., 2009, 131, 5094–5100 CAS.
- S. Abe, K. Hirata, T. Ueno, K. Morino, N. Shimizu, M. Yamamoto, M. Takata, E. Yashima and Y. Watanabe, Polymerization of Phenylacetylene by Rhodium Complexes within a Discrete Space of apo-Ferritin, J. Am. Chem. Soc., 2009, 131, 6958–6960 CAS.
- G. Ferraro, S. Ciambellotti, L. Messori and A. Merlino, Cisplatin Binding Sites in Human H-Chain Ferritin, Inorg. Chem., 2017, 56, 9064–9070 CAS.
- F. Hillenkamp, M. Karas, R. C. Beavis and B. T. Chait, Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Biopolymers, Anal. Chem., 1991, 63, 1193A–1203A CAS.
- M. Paolillo, G. Ferraro, I. Cipollone, E. Garribba, M. Monti and A. Merlino, Unexpected in crystallo reactivity of the potential drug bis(maltolato)oxidovanadium(IV) with lysozyme, Inorg. Chem. Front., 2024, 11, 6307–6315 CAS.
- E. Moreno-Gordaliza, B. Cañas, M. A. Palacios and M. M. Gómez-Gómez, Characterization of Pt-protein complexes by nHPLC–ESI-LTQ MS/MS using a gel-based bottom-up approach, Talanta, 2012, 88, 599–608 CAS.
- J. Wang, Y. Gou, Z. Zhang, P. Yu, J. Qi, Q. Qin, H. Sun, X. Wu, H. Liang and F. Yang, Developing an Anticancer Copper(II) Multitarget Pro-Drug Based on the His146 Residue in the IB Subdomain of Modified Human Serum Albumin, Mol. Pharmaceutics, 2018, 15, 2180–2193 CAS.
- P. Hao, H. Li, L. Zhou, H. Sun, J. Han and Z. Zhang, Serum Metal Ion-Induced Cross-Linking of Photoelectrochemical Peptides and Circulating Proteins for Evaluating Cardiac Ischemia/Reperfusion, ACS Sens., 2022, 7, 775–783 CrossRef CAS PubMed.
- X. Tan, H. Chen, C. Gu, J. Zang, T. Zhang, H. Wang and G. Zhao, Converting histidine-induced 3D protein arrays in crystals into their 3D analogues in solution by metal coordination cross-linking, Commun. Chem., 2020, 3, 151 CrossRef CAS PubMed.
- L. Zuily, N. Lahrach, R. Fassler, O. Genest, P. Faller, O. Sénèque, Y. Denis, M.-P. Castanié-Cornet, P. Genevaux, U. Jakob, D. Reichmann, M.-T. Giudici-Orticoni and M. Ilbert, Copper Induces Protein Aggregation, a Toxic Process Compensated by Molecular Chaperones, mBio, 2022, 13, e03251–e03221 CrossRef PubMed.
- M. Nie, Y. Luo and H. Li, Utilizing Platinum(II)-Based Cross-Linker and Two-Stage Data Analysis Strategy to Investigate the Allosteric in Glycogen Phosphorylase, Anal. Chem., 2025, 97, 3352–3360 CrossRef CAS PubMed.
- K. K. Raut, S. Pandey, G. Kharel and S. M. Pascal, Evidence of direct interaction between cisplatin and the caspase–cleaved prostate apoptosis response–4 tumor suppressor, Protein Sci., 2024, 33, e4867 CrossRef CAS PubMed.
- S. L. Binkley, C. J. Ziegler, R. S. Herrick and R. S. Rowlett, Specific derivatization of lysozyme in aqueous solution with Re(CO)3(H2O)3+, Chem. Commun., 2010, 46, 1203 RSC.
- F. J. F. Jacobs, J. R. Helliwell and A. Brink, Time-series analysis of rhenium(I) organometallic covalent binding to a model protein for drug development, IUCrJ, 2024, 11, 359–373 CrossRef CAS PubMed.
- B. Maity, M. Shoji, F. Luo, T. Nakane, S. Abe, S. Owada, J. Kang, K. Tono, R. Tanaka, T. T. Pham, M. Kojima, Y. Hishikawa, J. Tanaka, J. Tian, M. Nagama, T. Suzuki, H. Noya, Y. Nakasuji, A. Asanuma, X. Yao, S. Iwata, Y. Shigeta, E. Nango and T. Ueno, Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography, Nat. Commun., 2024, 15, 5518 CrossRef CAS PubMed.
- R. S. Das and Y. K. Agrawal, Raman spectroscopy: Recent advancements, techniques and applications, Vib. Spectrosc., 2011, 57, 163–176 CrossRef CAS.
- P. Carpentier, A. Royant, J. Ohana and D. Bourgeois, Advances in spectroscopic methods for biological crystals. 2. Raman spectroscopy, J. Appl. Crystallogr., 2007, 40, 1113–1122 CrossRef CAS.
- G. Kalosakas, A. R. Bishop and A. P. Shreve, Nonlinear disorder model for Raman profiles in naturally abundant PtCl, Phys. Rev. B: Condens. Matter Mater. Phys., 2002, 66, 094303 CrossRef.
- A. A. Petruk, A. Vergara, D. Marasco, D. Bikiel, F. Doctorovich, D. A. Estrin and A. Merlino, Interaction between Proteins and Ir Based CO Releasing Molecules: Mechanism of Adduct Formation and CO Release, Inorg. Chem., 2014, 53, 10456–10462 CAS.
- M. Caterino, A. A. Petruk, A. Vergara, G. Ferraro, D. Marasco, F. Doctorovich, D. A. Estrin and A. Merlino, Mapping the protein-binding sites for iridium(iii)-based CO-releasing molecules, Dalton Trans., 2016, 45, 12206–12214 CAS.
- T. M. Hunter, I. W. McNae, X. Liang, J. Bella, S. Parsons, M. D. Walkinshaw and P. J. Sadler, Protein recognition of macrocycles: Binding of anti-HIV metallocyclams to lysozyme, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 2288–2292 CAS.
- X. Liang, J. A. Parkinson, M. Weishäupl, R. O. Gould, S. J. Paisey, H. Park, T. M. Hunter, C. A. Blindauer, S. Parsons and P. J. Sadler, Structure and Dynamics of Metallomacrocycles: Recognition of Zinc Xylyl-Bicyclam by an HIV Coreceptor, J. Am. Chem. Soc., 2002, 124, 9105–9112 CAS.
- S. Ye, Probing electronic structures of transition metal complexes using electron paramagnetic resonance spectroscopy, Magn. Reson. Lett., 2023, 3, 43–60 CAS.
- K. E. Prosser and C. J. Walsby, Electron Paramagnetic Resonance as a Tool for Studying the Mechanisms of Paramagnetic Anticancer Metallodrugs, Eur. J. Inorg. Chem., 2017, 2017, 1573–1585 CAS.
- M. M. Roessler and E. Salvadori, Principles and applications of EPR spectroscopy in the chemical sciences, Chem. Soc. Rev., 2018, 47, 2534–2553 CAS.
- G. Sciortino, D. Sanna, V. Ugone, J.-D. Maréchal and E. Garribba, Integrated ESI-MS/EPR/computational characterization of the binding of metal species to proteins: vanadium drug–myoglobin application, Inorg. Chem. Front., 2019, 6, 1561–1578 CAS.
- J. C. Pessoa, M. F. A. Santos, I. Correia, D. Sanna, G. Sciortino and E. Garribba, Binding of vanadium ions and complexes to proteins and enzymes in aqueous solution, Coord. Chem. Rev., 2021, 449, 214192 CAS.
- M. F. A. Santos, G. Sciortino, I. Correia, A. C. P. Fernandes, T. Santos-Silva, F. Pisanu, E. Garribba and J. Costa Pessoa, Binding of V IV O 2+, V IV OL, V IV OL 2 and V V O 2 L Moieties to Proteins: X–ray/Theoretical Characterization and Biological Implications, Chem. A Eur. J., 2022, 28, e202200105 CAS.
- G. Ferraro, L. Vitale, G. Sciortino, F. Pisanu, E. Garribba and A. Merlino, Interaction of V IV O–8-hydroxyquinoline species with RNase A: the effect of metal ligands in the protein adduct stabilization, Inorg. Chem. Front., 2023, 10, 5186–5198 CAS.
- G. Ferraro, G. Tito, G. Sciortino, E. Garribba and A. Merlino, Stabilization and Binding of [V4O12]4− and Unprecedented [V20O54(NO3)]n− to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug VIVO(acetylacetonato)2, Angew. Chem., 2023, 135, e202310655 Search PubMed.
- G. Tito, G. Ferraro, F. Pisanu, E. Garribba and A. Merlino, Non–Covalent and Covalent Binding of New Mixed–Valence Cage–like Polyoxidovanadate Clusters to Lysozyme, Angew. Chem., Int. Ed., 2024, 63, e202406669 CAS.
- L. Vandebroek, E. De Zitter, H. G. T. Ly, D. Conić, T. Mihaylov, A. Sap, P. Proost, K. Pierloot, L. Van Meervelt and T. N. Parac-Vogt, Protein–Assisted Formation and Stabilization of Catalytically Active Polyoxometalate Species, Chem. – Eur. J., 2018, 24, 10099–10108 CAS.
- A. Sap, E. De Zitter, L. Van Meervelt and T. N. Parac-Vogt, Structural Characterization of the Complex between Hen Egg–White Lysozyme and Zr IV –Substituted Keggin Polyoxometalate as Artificial Protease, Chem. – Eur. J., 2015, 21, 11692–11695 CAS.
- A. Bijelic, C. Molitor, S. G. Mauracher, R. Al-Oweini, U. Kortz and A. Rompel, Hen Egg–White Lysozyme Crystallisation: Protein Stacking and Structure Stability Enhanced by a Tellurium(VI)–Centred Polyoxotungstate, ChemBioChem, 2015, 16, 233–241 CAS.
- S. Lentink, D. E. Salazar Marcano, M. A. Moussawi, L. Vandebroek, L. Van Meervelt and T. N. Parac-Vogt, Fine-tuning non-covalent interactions between hybrid metal-oxo clusters and proteins, Faraday Discuss., 2023, 244, 21–38 RSC.
- L. Vandebroek, Y. Mampaey, S. Antonyuk, L. Van Meervelt and T. N. Parac-Vogt, Noncovalent Complexes Formed between Metal–Substituted Polyoxometalates and Hen Egg White Lysozyme, Eur. J. Inorg. Chem., 2019, 2019, 506–511 CrossRef CAS.
- M. P. Sullivan, M. K. Nieuwoudt, G. A. Bowmaker, N. Y. S. Lam, D. Truong, D. C. Goldstone and C. G. Hartinger, Unexpected arene ligand exchange results in the oxidation of an organoruthenium anticancer agent: the first X-ray structure of a protein–Ru(carbene) adduct, Chem. Commun., 2018, 54, 6120–6123 CAS.
- N. Cetinbas, M. I. Webb, J. A. Dubland and C. J. Walsby, Serum-protein interactions with anticancer Ru(III) complexes KP1019 and KP418 characterized by EPR, J. Biol. Inorg. Chem., 2010, 15, 131–145 CAS.
- T. Furuya, D. Nakane, K. Kitanishi, N. Katsuumi, A. Tsaturyan, I. N. Shcherbakov, M. Unno and T. Akitsu, A novel hybrid protein composed of superoxide-dismutase-active Cu(II) complex and lysozyme, Sci. Rep., 2023, 13, 6892 CAS.
- H. Utkov, M. Livengood and M. Cafiero, Annual Reports in Computational Chemistry, Elsevier, 2010, vol. 6, pp. 96–112 Search PubMed.
- A. B. Rozhenko, in Application of Computational Techniques in Pharmacy and Medicine, ed. L. Gorb, V. Kuz'min and E. Muratov, Springer Netherlands, Dordrecht, 2014, vol. 17, pp. 207–240 Search PubMed.
- F. Himo, Recent Trends in Quantum Chemical Modeling of Enzymatic Reactions, J. Am. Chem. Soc., 2017, 139, 6780–6786 CAS.
- P. E. M. Siegbahn and F. Himo, The quantum chemical cluster approach for modeling enzyme reactions, Wiley Interdiscip. Rev. Comput. Mol. Sci., 2011, 1, 323–336 CAS.
- A. Parise, N. Russo and T. Marino, The platination mechanism of RNase A by arsenoplatin: insight from the theoretical study, Inorg. Chem. Front., 2021, 8, 1795–1803 CAS.
- F. Zobi and B. Spingler, Post-Protein-Binding Reactivity and Modifications of the fac -[Re(CO)3]+ Core, Inorg. Chem., 2012, 51, 1210–1212 CAS.
- D. Loreto, F. Fasulo, A. B. Muñoz-García, M. Pavone and A. Merlino, Unexpected Imidazole Coordination to the Dirhodium Center in a Protein Environment: Insights from X-ray Crystallography and Quantum Chemistry, Inorg. Chem., 2022, 61, 8402–8405 CAS.
- A. Terán, F. Fasulo, G. Ferraro, A. E. Sánchez-Peláez, S. Herrero, M. Pavone, A. B. Muñoz-García and A. Merlino, Exchange of equatorial ligands in protein-bound paddlewheel Ru 25+ complexes: new insights from X-ray crystallography and quantum chemistry, Inorg. Chem. Front., 2024, 11, 7803–7811 Search PubMed.
- Z. Ke, S. Abe, T. Ueno and K. Morokuma, Catalytic Mechanism in Artificial Metalloenzyme: QM/MM Study of Phenylacetylene Polymerization by Rhodium Complex Encapsulated in apo -Ferritin, J. Am. Chem. Soc., 2012, 134, 15418–15429 CAS.
- C. Pellegrini, The development of XFELs, Nat. Rev. Phys., 2020, 2, 330–331 Search PubMed.
- N. Pontillo, G. Ferraro, L. Messori, G. Tamasi and A. Merlino, Ru-Based CO releasing molecules with azole ligands: interaction with proteins and the CO release mechanism disclosed by X-ray crystallography, Dalton Trans., 2017, 46, 9621–9629 CAS.
- A. Warshel and M. Levitt, Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme, J. Mol. Biol., 1976, 103, 227–249 CAS.
- A. Roux, Z. Alsalman, T. Jiang, J. Mulatier, D. Pitrat, E. Dumont, F. Riobé, N. Gillet, E. Girard and O. Maury, Influence of Chemical Modifications of the Crystallophore on Protein Nucleating Properties and Supramolecular Interactions Network, Chem. – Eur. J., 2024, 30, e202400900 CAS.
- L. A. E. Batista De Carvalho, A. P. Mamede, A. L. M. Batista De Carvalho, J. Marques, G. Cinque, S. Rudić and M. P. M. Marques, Metallodrug-protein interaction probed by synchrotron terahertz and neutron scattering spectroscopy, Biophys. J., 2021, 120, 3070–3078 CAS.
- I. Khalaila, A. Bergamo, F. Bussy, G. Sava and P. J. Dyson, The role of cisplatin and NAMI-A plasma-protein interactions in relation to combination therapy, Int. J. Oncol., 2006, 29, 261–268 CAS.
- D. Loreto, G. Ferraro and A. Merlino, Protein-metallodrugs interactions: Effects on the overall protein structure and characterization of Au, Ru and Pt binding sites, Int. J. Biol. Macromol., 2020, 163, 970–976 CrossRef CAS PubMed.
- L. Skos, Y. Borutzki, C. Gerner and S. M. Meier-Menches, Methods to identify protein targets of metal-based drugs, Curr. Opin. Chem. Biol., 2023, 73, 102257 CrossRef CAS PubMed.
- H. U. Holtkamp and C. G. Hartinger, Advanced metallomics methods in anticancer metallodrug mode of action studies, TrAC, Trends Anal. Chem., 2018, 104, 110–117 CrossRef CAS.
- E. Michelucci, G. Pieraccini, G. Moneti, C. Gabbiani, A. Pratesi and L. Messori, Mass spectrometry and metallomics: A general protocol to assess stability of metallodrug-protein adducts in bottom-up MS experiments, Talanta, 2017, 167, 30–38 CrossRef CAS PubMed.
| This journal is © the Partner Organisations 2025 |