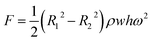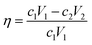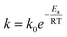DOI:
10.1039/D2SC06466A
(Edge Article)
Chem. Sci., 2023,
14, 3973-3981
A nanoparticle-coated microfluidic chip for automated, non-destructive extraction of encapsulated DNA in data storage†
Received
23rd November 2022
, Accepted 7th February 2023
First published on 11th February 2023
Abstract
DNA data storage based on tubes as physical storage carriers has been developed to solve the problem of the exponential growth of information. Compact disk (CD)-microfluidics that employs centrifugal forces for fluidic manipulation offers an attractive alternative that integrates complex assays onto a miniaturized platform to result in automation for DNA data storage. In this work, we develop a CD microfluidic chip modified with nanoparticles for accurate fluid flow control. The nanoparticle coating turns microchannels into valves or pumps, which reduces the error in fluidic control from 62% to 6%. Based on the nanoparticle coating, the chip integrates demineralization, nucleic acid amplification, and re-mineralization functions for automated, non-destructive information extraction. We prove the functionality of the chip with mineralized DNA data. Compared to purely manual operation and traditional amplification techniques, the microfluidic chip saves human power and time consumption, demonstrating an essential contribution to the development of DNA data storage.
Introduction
With the exponential growth of digital data production, the digital universe will grow to 175 zettabytes in 2025. 80% or more of the data after storage might not be needed for months, years, or even decades.1 DNA data storage has become an attractive alternative to existing cold data storage due to its high-density storage capacity and endurance.2 In recent years, the promise of DNA data storage has drawn attention from academia and the industry. For example, an automated end-to-end DNA data storage device has also been developed to explore the automation challenges, which includes a custom DNA synthesizer for writing data to a DNA oligonucleotide, a pool for liquid storage, and a Nanopore sequencer for reading.3 Among existing platforms, most rely on typical carriers such as tubes to store DNA samples because of convenience and immediate availability. However, these platforms are challenging to fulfill the requirements of data storage, including a high degree of miniaturization and integration.
Microfluidics has developed into a universal tool in the fields of synthesis, detection, and analysis because of its unique advantages, such as automation, miniaturization and integration.4–7 It offers a feasible solution for establishing automated devices integrated with multiplexed functional modules for DNA storage. Digital microfluidics has been used to retrieve dehydrated DNA spots on glass for scalable DNA data storage. This device demonstrates that 1 TB of data could be stored and retrieved.8 Another microfluidic platform composed of two-layer PDMS has been developed for automated storage and retrieval of data encoding DNA based on a combinatorial valve network.9 Compared to other platforms, compact disk (CD)-microfluidics stands out by the degree of maturity in which unit operations such as fluidic displacement, mixing, separation, and metering can be implemented, scaled, and standardized.10,11 Different strategies for fluid flow control in the microfluidic chip are emerging, such as photothermal pumping with high integratability,12–14 and an addressable electrowetting valve.15 The superhydrophobic surface has been used for self-cleaning,16 fog harvest,17 and droplet manipulation.18 Due to excellent physical properties, the superhydrophobic surface in microchannels provides more possibilities for accurate fluid flow control in microfluidic platforms. Two main strategies to promote superhydrophobicity are surface modification by a nanostructure and surface modification with surfactants. The former includes templating,19,20 microphase separation,21,22 nanoparticle coating,23–26 chemical etching,27,28 sol–gel process,29,30 electro-spinning,31,32 and so on. The latter is based on matching chemistry at the surface.33,34
In this work, we develop a nanoparticle modification method and first apply it to a CD microfluidic chip to yield superhydrophobic surfaces for accurate fluid flow control. The superhydrophobic coating composed of nanoparticles exhibited long-term stability and wide adaptability. The microchannels with the superhydrophobic surface could act as passive valves or pumps. We demonstrate applications of nanoparticle modification in the CD microfluidic chip, such as mixing liquids and accelerating reactions.
Furthermore, based on nanoparticle modification and DNA encapsulation methods, we design a CD microfluidic chip integrated with demineralization, nucleic acid amplification, and re-mineralization functions (Scheme 1). We adopt calcium carbonate nanoparticles with excellent biocompatibility for encapsulating DNA. In this way, information could be archived on DNA for over a 1000 years. The superhydrophobic nanoparticle coating drives the re-mineralized DNA sample to return to the sample hole against the direction of centrifugal force to realize non-destructive information extraction. As a proof-of-concept experiment, we encode the text information in DNA sequences and prove the functions of the chip with mineralized DNA data within 80 minutes, which is accurately restored.
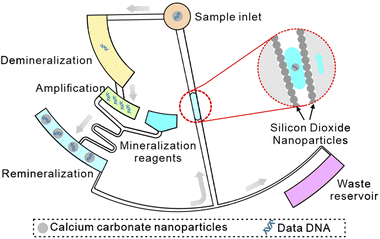 |
| | Scheme 1 Schematic of a CD microfluidic platform for non-destructive information extraction from mineralized DNA data. | |
Results and discussion
Test of superhydrophobic modification
To apply superhydrophobic modification in the CD microfluidic platform, we fabricated a superhydrophobic coating with PDMS and SiO2 nanoparticles. We characterized the superhydrophobic coating with ratios between PDMS and SiO2 nanoparticles ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 5
1 to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 by measuring the contact angles and the sliding angles (Fig. S2A and B†). The SEM images showed the nanostructure of superhydrophobic coatings decreased with the decreasing proportion of SiO2 nanoparticles (Fig. S3†). To keep the balance between the contact angle, the sliding angle, and the stability of the coating, we set the ratio of PDMS to SiO2 nanoparticles as 2
2 by measuring the contact angles and the sliding angles (Fig. S2A and B†). The SEM images showed the nanostructure of superhydrophobic coatings decreased with the decreasing proportion of SiO2 nanoparticles (Fig. S3†). To keep the balance between the contact angle, the sliding angle, and the stability of the coating, we set the ratio of PDMS to SiO2 nanoparticles as 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the following experiments. Furthermore, we tested the duration of contact and sliding angles after superhydrophobic modification. The result showed that the superhydrophobicity of the surface was stable over 100 days (Fig. S2C†). In addition, we repeatedly washed the microchannel modified by the superhydrophobic coating with water up to 200 times. There was no apparent change in the hydrophobicity of the coating. Given that ethyl acetate could react with some materials such as PC and PS, we used ethanol to extend the application range of the superhydrophobic coating, which also increased the hydrophobicity of the coating (Fig. S4†). Compared with ethyl acetate, ethanol generated a superhydrophobic coating with more nanostructures as a dispersant (Fig. S5†).
1 in the following experiments. Furthermore, we tested the duration of contact and sliding angles after superhydrophobic modification. The result showed that the superhydrophobicity of the surface was stable over 100 days (Fig. S2C†). In addition, we repeatedly washed the microchannel modified by the superhydrophobic coating with water up to 200 times. There was no apparent change in the hydrophobicity of the coating. Given that ethyl acetate could react with some materials such as PC and PS, we used ethanol to extend the application range of the superhydrophobic coating, which also increased the hydrophobicity of the coating (Fig. S4†). Compared with ethyl acetate, ethanol generated a superhydrophobic coating with more nanostructures as a dispersant (Fig. S5†).
Test of the superhydrophobic valve
To demonstrate the joint action of capillary force and major forces in the CD microfluidic chip (Fig. 1A), we designed and fabricated a CD microfluidic chip containing microchannels of different sizes as valves (Fig. 1B). We preloaded a solution with red dye into the chambers in the modified CD microfluidic chip and increased the rotation speed gradually. We recorded the rotation speed when the fluid was driven into the next chamber (Fig. 1C). Meanwhile, we set a microchannel without any treatment as the blank control. The maximum error of the superhydrophobic valve was no more than seven percent, while the maximum error of the blank control was about sixty percent (Table 1). The results showed that the superhydrophobic valve offered higher accuracy than the blank control because the effect of friction was minimized. Compared with existing approaches such as capillary valves and siphon valves, the superhydrophobic valve allows for integrating more reaction steps within a certain range of rotating speed because of high accuracy. The capillary force generated by the modified microchannel is related to the contact angle and channel size.35 The formula is given by:| | F = 2δ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ (w + h) θ (w + h) | (1) |
where δ is the surface tension coefficient of liquid, θ is the contact angle, and w and h are the width and height of the channel. The centrifugal force is related to the rotation speed, distance from the center of the circle, and channel size (Fig. S6†).36 The formula is given by:| | 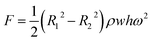 | (2) |
where R1 and R2 are the outer and inner radii of the liquid in the channel, ρ is the density of the liquid, w and h are the width and height of the channel, and ω is the rotation speed.
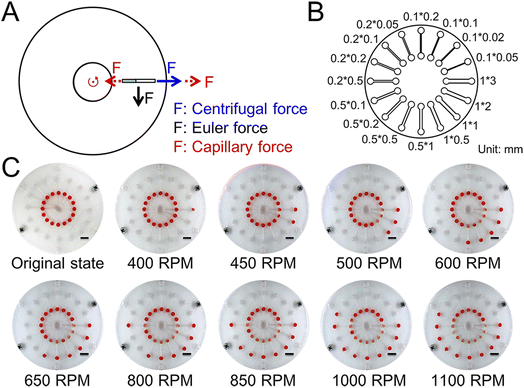 |
| | Fig. 1 Schematic and test of the superhydrophobic valve. (A) The force analysis of fluid in the CD microfluidic chip. (B) The structure of the CD microfluidic chip containing microchannels of different sizes. (C) Flow stage of the sample liquid in the chip with increasing rotation speed and time. The scale bar is 1 cm. | |
Table 1 The relationship between sizes of microchannels and thresholds of the hydrophobic valve
| Size/mm |
Blank control |
Superhydrophobic modification |
| Theoretical speed/rpm |
Experimental speed/rpm |
Error |
Theoretical speed/rpm |
Experimental speed/rpm |
Error |
| 1 × 3 |
233 |
350 |
−50.21% |
389 |
366 |
−5.74% |
| 1 × 2 |
247 |
400 |
−61.94% |
413 |
417 |
−0.97% |
| 1 × 1 |
285 |
450 |
−57.89% |
476 |
483 |
−1.54% |
| 1 × 0.5 |
349 |
500 |
−43.27% |
584 |
600 |
−2.74% |
| 0.5 × 1 |
349 |
467 |
−33.81% |
584 |
550 |
+5.82% |
| 0.5 × 0.5 |
403 |
533 |
−32.26% |
674 |
650 |
+3.56% |
| 0.5 × 0.2 |
533 |
617 |
−15.76% |
892 |
850 |
+4.71% |
| 0.5 × 0.1 |
698 |
883 |
−26.50% |
1168 |
1183 |
+1.28% |
| 0.2 × 0.5 |
533 |
650 |
−21.95% |
892 |
833 |
−6.61% |
| 0.2 × 0.2 |
637 |
767 |
−20.41% |
1066 |
1017 |
+4.59% |
Test of the superhydrophobic pump
To demonstrate the pumping function of microchannels modified with the superhydrophobic coating, we designed and fabricated another CD microfluidic chip containing chambers and winding microchannels of different sizes (Fig. 2A and B). The unit with a smaller size had a larger threshold value of the hydrophobic valve to prevent the liquid from entering the mixing microchannel. We set the unit as the control group. We increased the rotation speed of the chip gradually. Two fluids were driven into the modified channel from the mixing chamber under centrifugal force when the rotation speed reached the valve threshold. Fluids were driven back to the mixing chamber under capillary force after stopping the rotation of the chip. We repeated the process of reciprocating motion to mix the two liquids (Fig. 2C). We utilized two dyes to show the mixing process with the modified CD microfluidic chip. At a certain rotation speed, the mixing chamber marked with a red rectangle gradually became a uniform black as the number of cycles increased while the control group marked with a blue rectangle remained unchanged (Fig. 3A). We also utilized the synthesis reaction of Prussian blue to test the mixing effect of the chip. The mixing chamber marked with a red rectangle gradually turned blue as the cycles increased (Fig. 3B). It indicated that Prussian blue was synthesized in the mixing chamber as the reagents mixed. The colour change of the mixing unit was more evident than that in the control chamber marked with a blue rectangle. The result showed that microchannels modified with the superhydrophobic coating played the role of a pump as expected.
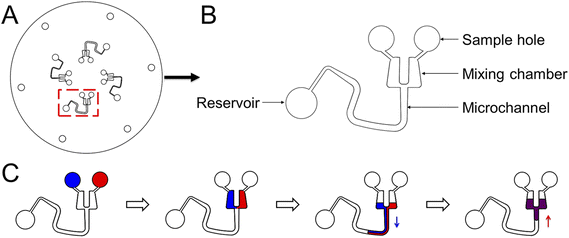 |
| | Fig. 2 The design of the CD microfluidic chip and illustration of the mixing process. (A) Illustration of the whole CD chip. (B) Illustration of one of the channels for mixing liquid. (C) The reciprocating motion of liquid in the modified microchannel. The blue arrow represents the direction of centrifugal force. The red arrow represents the direction of capillary force. | |
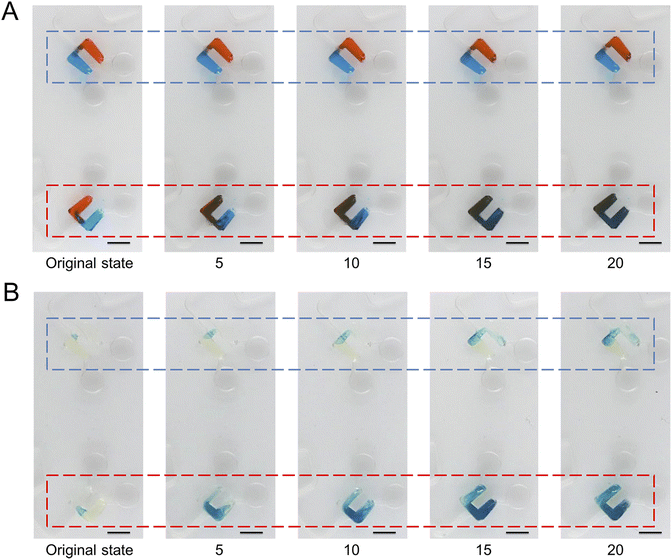 |
| | Fig. 3 Mixing of liquids under centrifugal force and capillary force. We took a picture every five rotations. (A) At a certain rotation speed, two dyes gradually mix under centrifugal force and capillary force in the mixing chamber marked with a red rectangle while the control group marked with a blue rectangle remains unchanged. (B) At a certain rotation speed, two reagents gradually mix and synthesize Prussian blue under centrifugal force and capillary force in the mixing chamber marked with a red rectangle, which is more evident than in the control group marked with a blue rectangle. The scale bar is 5 mm. | |
Test of DNA mineralization
DNA was encapsulated in calcium carbonate nanoparticles to reduce the decay rate and satisfy the requirement of DNA data storage (Fig. S7†). To test the effects of different components in the mineralization system on DNA encapsulation and release, we measured the concentration of free DNA in the solution with a fluorescent stain under different conditions. The concentration of protamine sulfate was set from 5 to 25 mg mL−1. As expected, the decreasing fluorescent signal showed that the combination of protamine sulfate and DNA increased with increasing concentration of protamine sulfate. The combination efficiency is saturated when the concentration is over 15 mg mL−1 (Fig. S8A†). We set the concentration of protamine sulfate as 15 mg mL−1 in the following experiments. The concentration of heparin sodium was optimized from 0.1 to 0.6 mg mL−1. As expected, the fluorescent signal increased, showing that the release of DNA increased with increasing concentration of heparin sodium. The release efficiency plateaued when the concentration was over 0.4 mg mL−1 (Fig. S8B†). We set the concentration of heparin sodium as 0.4 mg mL−1 in the following experiments. The concentrations of calcium chloride and sodium carbonate were optimized from 0.2 to 20 mM. The DNA loading in different samples was 20%, 2%, and 0.2%. The mineralization efficiency of DNA decreased with decreasing concentrations of calcium chloride and sodium carbonate (Fig. S8C†). The mineralization efficiency is given by:| | 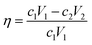 | (3) |
where η is the mineralization efficiency, c1 is the concentration of initial DNA, V1 is the volume of initial DNA, c2 is the concentration of free DNA, and V2 is the volume of free DNA. To increase the mineralization efficiency, we set the concentration of sodium carbonate as 0.2 mM and increased the concentration of calcium chloride from 0.2 to 20 mM. The mineralization efficiency increased to nearly one hundred percent with an increasing concentration of calcium chloride (Fig. S8D†). We set the concentration of calcium chloride as 20 mM in the following experiments.
To test the protective effect of mineralization under UV light and at high temperature, we made the standard curve between DNA concentrations and the PCR CT value in advance (Fig. S9†). The free and mineralized DNA samples were freeze-dried on a sheet of glass. We kept the sheet of glass in a UV light box with a power of 96 watts for 0, 2, 4, 6, and 8 hours. After collecting the samples left on the glass, we released DNA from the mineralization samples. The decay of the DNA under UV light could be monitored by qPCR analysis. We set the concentration of DNA without any treatment as C0. After being treated with UV light for 8 hours, the unprotected DNA decayed to about a ten-thousandth of C0, while the mineralized DNA decayed to about two percent of C0 (Fig. S10A†). We also heated the sheet of glass at 100, 120, and 140 °C for half an hour. When the temperature was up to 140 °C, the unprotected DNA decayed to about five percent of C0, while the mineralized DNA decayed to about twenty percent of C0 (Fig. S10B†). We also investigated the lifetime of DNA by accelerated aging tests in a climate box. The environmental conditions were set as a humidity of 50% and temperature ranging from 60 to 70 °C. On the sixth day, the concentrations of unprotected DNA samples decayed to eighteen percent, sixteen percent, and nine percent of C0 at temperatures of 60, 65, and 70 °C (Fig. S10C†). As expected, the higher the temperature, the faster the DNA decay. Meanwhile, the concentrations of mineralized DNA samples decayed to fifty-four percent, twenty-three percent, and sixteen percent of C0 at temperatures of 60, 65, and 70 °C (Fig. S10D†).
We further calculated the lifetime of DNA by assuming a first-order chemical degradation reaction. The temperature dependence of the reaction rate is given by the Arrhenius law.37 The decay rate equation is given by:
| | 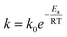 | (4) |
where
k is the reaction rate,
k0 is the pre-exponential factor,
Ea is the activation energy,
R is the gas constant, and
T is the temperature. We performed linear fits of the concentration data at different temperatures and obtained first-order decay rates of DNA (Table S1
†). Activation energies and pre-exponential factors of the individual reactions were obtained by fitting the data following the Arrhenius law (Table S2
†). The linear fit results of decay rates to temperatures showed that the decay rate of unprotected DNA was about 4.12 × 10
−7/s at 20 °C (Fig. S11A
†), and the decay rate of mineralized DNA was about 1.27 × 10
−8 s
−1 at 20 °C (Fig. S11B
†). The corresponding half-life of unprotected and mineralized DNA samples was 252 days and 791 years at minus 20 °C (
Table 2). The results showed that mineralization protection significantly inhibited the decay of DNA.
Table 2 The corresponding half-time of DNA under different conditions
|
|
20 °C |
0 °C |
−20 °C |
| Unprotected DNA |
19 days |
64 days |
252 days |
| DNA in calcium carbonate |
632 days |
30 years |
791 years |
Screening primers
To reduce the complexity of auxiliary equipment and time consumption, we screened primers of recombinase polymerase amplification (RPA) for DNA amplification. We randomly generated DNA sequences with an appropriate proportion of deoxynucleotides. After eliminating mismatch and hairpin structures, we selected two sets of DNA sequences with different lengths as primers (Table 3). After repeatedly amplifying the DNA amplicon five times, amplification products were analyzed by gel electrophoresis and high-throughput sequencing analysis. Gel electrophoresis images showed little difference between amplification products after purification, and some bonds with longer lengths appeared in both, indicating DNA spliced during isothermal amplification (Fig. S12A†). The high-throughput sequencing results showed that the target sequence read at a higher frequency with the shorter primers (Fig. S12B and C†). We selected primer set one for DNA amplification in the proof-of-concept experiment.
Table 3 Primer sequences
| Group |
Primer sequence |
| Set one |
F: TCTGCAAGTAGCCAAGGGTAAGCAAGGATC |
| R: TCTGCAAGTAGCCAAGGGTAAGCAAGGATC |
| Set two |
F: TCTGCAAGTAGCCAAGGGTAAGCAAGGATCGCATG |
| R: GAATCTGCACGAAGCACTTATGAACCCTTGGCACT |
Encoding data
To prove the concept of DNA data storage, we encoded the text information into seven DNA sequences with a length of 80 nt based on the principle of the DNA Fountain code38(Table 4). Each DNA molecule contained a seed region length of 16 nt, a data region length of 48 nt, an RS region length of 16 nt, and two adapter regions length of 30 nt for binding primers (Fig. S13†).
Table 4 The text information and encoded DNA sequences
| Text information |
DNA sequences |
| DNADataStorageBasedOnMicrofluidics |
AAAGAAAGGGAAGGAACGCTCGCCCAAGCGACCTATCGCCCGCACATTCGTGCATCCGGCCGATTGCTAGATGATGCTTT |
| AAGAAAGGGAAGGAAACACACATGCAACCACACGACCTCACGACCCATCTCACGTTCTAGCGACTTCGCAGAACCGGAGA |
| ACCCAACCAATACCAACACACATGCAACCACACGACCTCACGACCCATCTCACGTTCTAGCGACCGTATCGCCAAGGCGT |
| CCAACCAATACCTACCAGATAGGTAAATAGCCACAGACACAACCACTAACGGAGAGACGTAAAGCGTTTGACAGTAGTCT |
| CAACCAATACCTTCACCCACCACACGCCCAGCCGCTCTGACGACCTCCCTGCCCACACGTAAAGATAGATTTCAATGAGC |
| AAGGACGAGTTTAAAGATCGAGACAGCTAGGAACCAACTCAACCATGGACCTACTACTAGCGACCTAGCACTTGATCATT |
| GGACGAGTTTAAAGAAATCGAGACAGCTAGGAACCAACTCAACCATGGACCTACTACTAGCGACTGCTATCCGGTGGTAA |
DNA storage based on the microfluidic chip
To demonstrate the function of automated and non-destructive extraction of information from DNA data, we designed and fabricated a CD microfluidic platform integrated with demineralization, amplification, and re-mineralization of DNA (Fig. 4A). Modified microchannels in the platform acted as hydrophobic valves or passive pumps. We tested the fluid flow control function of the platform with three dyes. We added the mineralized DNA sample to the platform from the sample hole. The platform was preloaded with reagents for demineralization, amplification, and re-mineralization (Fig. 4B). We started to rotate the chip with a custom-made rotor and forced the mineralized DNA sample into the chamber preloaded with EDTA and heparin sodium solution (Fig. 4C). After demineralization for 15 minutes, we increased the rotation speed and forced the demineralized DNA sample and the amplification buffer into the next chamber preloaded with the isothermal amplification reagent (Fig. 4D). After heating the chip at 39 °C for one hour, we increased the rotation speed and forced the amplification product and mineralization reagents into the next chamber (Fig. 4E). The remineralized DNA sample was driven into the arc microchannel modified with a superhydrophobic coating, which was subjected to capillary forces in different directions and driven back to the center of the circle under the resultant force (Fig. S14†). We collected the remineralized DNA sample from the center of the circle (Fig. 4F). The remineralized DNA sample covered the loss of the original mineralized DNA sample and realized non-destructive extraction of information from DNA data. We carried out the control experiment off-chip following the same experimental procedure. We analyzed remineralized DNA samples with high-throughput sequencing to obtain DNA sequences encoding text information (Fig. 5A and B). We decoded DNA sequences to translate the original text information. The text information was successfully decoded from the high-throughput sequencing results (Fig. 5C and D).
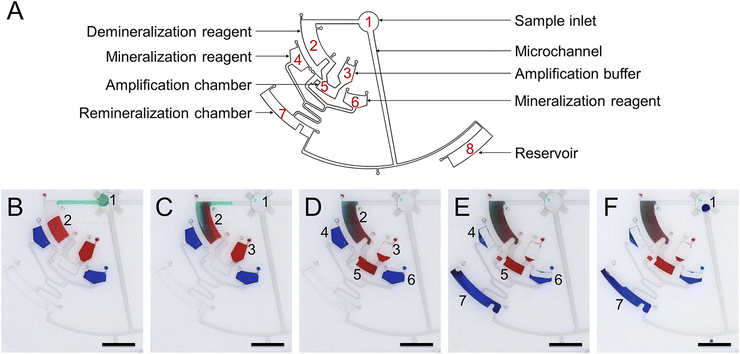 |
| | Fig. 4 Operation of the CD microfluidic chip. (A) Structure of the CD microfluidic chip and functional partitions. (B) Original state. Target DNA is mineralized with calcium carbonate nanoparticles in the original sample. (C) The sample in chamber one is driven into chamber two at 350 rpm. Mineralized target DNA is released with reagents for demineralization. (D) The demineralized sample in chamber two and amplification buffer in chamber three are driven into chamber five at 450 rpm. Free target DNA is amplified with RPA reagents. (E) The amplification product in chamber five and mineralization reagents in chambers four and six are driven into chamber seven at 550 rpm. The target DNA is remixed with reagents for mineralization. (F) The remineralized DNA sample in chamber seven is driven back to chamber one under capillary force. The scale bar is 1 cm. | |
 |
| | Fig. 5 Text information extraction from mineralized DNA data off-chip and on-chip. (A) The partial results of high-throughput sequencing (off-chip). (B) The partial results of high-throughput sequencing (on-chip). (C) The decoding text information from the results of high-throughput sequencing (off-chip). (D) The decoding text information from the results of high-throughput sequencing (on-chip). | |
Conclusions
In this paper, we develop a nanoparticle modification method with a wide range of applications and excellent hydrophobicity, and first apply it to the CD microfluidic chip. It improves liquid flow control in CD microfluidic chips without additional auxiliary equipment. Based on the nanoparticle modification method, we design a CD microfluidic chip integrating multiplexed functional modules. Calcium carbonate nanoparticles with excellent biocompatibility are adopted for encapsulating DNA data. The process of mineralization and demineralization does not require any organic reagent, which reduces the demand for materials of the microfluidic chip. It also provides a feasible idea for combining DNA data storage in vivo. We further utilize the passive capillary force generated by the superhydrophobic surface to drive the remineralized DNA return to the centre of the chip to cover the loss of the original sample. This structure design allows non-destructive extraction of encapsulated DNA, avoiding the positioning problem common to CD microfluidic chips and enabling us to manipulate it automatically more easily. We perform the conceptual experiment with DNA encoding the text information, proving a chip feasible for automated, non-destructive information extraction from mineralized DNA. The CD microfluidic chip integrated with the nanoparticle modification method as a passive liquid flow control strategy also has the potential to be used for multi-step reactions, including nanoparticle synthesis, sample preparation, immunoassays, nucleic acid analysis, cell lysis, separation, and trapping.
Data availability
The ESI† is available and contains experimental and analytical data.
Author contributions
X. J., S. L. and C. G. conceived the research. X. J. acquired funding. C. G. performed all the experiments and wrote the original draft of the manuscript. X. J. and S. L. co-supervised the project and edited the manuscript.
Conflicts of interest
The authors declare no competing financial interest.
Acknowledgements
We thank the National Key R&D Program of China (2018YFA0902600, 2021YFF1200801, 2021YFF1200802, 2020YFA0908900, and 2021YFF1200103), the Guangdong Provincial Key Laboratory of Advanced Biomaterials (2022B1212010003), the Shenzhen Bay Laboratory (SZBL2019062801004), the Shenzhen Science and Technology Program (KQTD20190929172743294 and JCYJ20200109141231365), the National Natural Science Foundation of China (21535001, 81730051, and 32071390), the Chinese Academy of Sciences (QYZDJ-SSW-SLH039 and 121D11KYSB20170026), the Shenzhen Key Laboratory of Smart Healthcare Engineering (ZDSYS20200811144003009), the Guangdong Innovative and Entrepreneurial Research Team Program (2019ZT08Y191), and the Tencent Foundation through the XPLORER PRIZE for financial support. The authors acknowledge the assistance of SUSTech Core Research Facilities.
Notes and references
- M. Dimopoulou, M. Antonini, P. Barbry and R. Appuswamy, Image Storage onto Synthetic DNA, Signal Process. Image Commun., 2021, 97, 116331 CrossRef.
- T. Zhou, Y. Luo and X. Jiang, DNA Data Storage: Preservation Approach and Data Encryption, Synth. Biol. J., 2021, 2, 371–383 Search PubMed.
- C. N. Takahashi, B. H. Nguyen, K. Strauss and L. Ceze, Demonstration of End-to-End Automation of DNA Data Storage, Sci. Rep., 2019, 9, 4998 CrossRef PubMed.
- Q. Hou, L. Wang, F. Xiao, L. Wang, X. Liu, L. Zhu, Y. Lu, W. Zheng and X. Jiang, Dual Targeting Nanoparticles for Epilepsy Therapy, Chem. Sci., 2022, 13, 12913–12920 RSC.
- S. Feng, S. Mao, J. Dou, W. Li, H. Li and J. M. Lin, An Open-space Microfluidic Chip with Fluid Walls for Online Detection of VEGF via Rolling Circle Amplification, Chem. Sci., 2019, 10, 8571–8576 RSC.
- M. Wu, D. Yuan, J. Xu and H. Chen, Electrochemiluminescence on Bipolar Electrodes for Visual Bioanalysis, Chem. Sci., 2013, 4, 1182–1188 RSC.
- W. Zhou, M. Dou, S. S. Timilsina, F. Xu and X. Li, Recent Innovations in Cost-effective Polymer and Paper Hybrid Microfluidic Devices, Lab Chip, 2021, 21, 2658–2683 RSC.
- S. Newman, A. P. Stephenson, M. Willsey, B. H. Nguyen, C. N. Takahashi, K. Strauss and L. Ceze, High Density DNA Data Storage Library via Dehydration with Digital Microfluidic Retrieval, Nat. Commun., 2019, 10, 1706 CrossRef CAS PubMed.
- Y. Luo, S. Wang, Z. Feng, J. Li, C. Mao, R. Wang and X. Jiang, Integrated Microfluidic DNA Storage Platform with Automated Sample Handling and Physical Data Partitioning, Anal. Chem., 2022, 94, 13153–13162 CrossRef CAS PubMed.
- Y. Chen, Y. Mei and X. Jiang, Universal and High-fidelity DNA Single Nucleotide Polymorphism Detection Based on a CRISPR/Cas12a Biochip, Chem. Sci., 2021, 12, 4455–4462 RSC.
- P. Arosio, T. Muller, L. Mahadevan and T. P. Knowles, Density-gradient-free Microfluidic Centrifugation for Analytical and Preparative Separation of Nanoparticles, Nano Lett., 2014, 14, 2365–2371 CrossRef CAS PubMed.
- G. Fu, Y. Zhu, W. Wang, M. Zhou and X. Li, Spatiotemporally Controlled Multiplexed Photothermal Microfluidic Pumping under Monitoring of On-Chip Thermal Imaging, ACS Sens., 2019, 4, 2481–2490 CrossRef CAS PubMed.
- G. Fu, W. Zhou and X. Li, Remotely Tunable Microfluidic Platform Driven by Nanomaterial-mediated on-demand Photothermal Pumping, Lab Chip, 2020, 20, 2218–2227 RSC.
- W. Zhou, G. Fu and X. Li, Detector-Free Photothermal Bar-Chart Microfluidic Chips (PT-Chips) for Visual Quantitative Detection of Biomarkers, Anal. Chem., 2021, 93, 7754–7762 CrossRef CAS PubMed.
- Y. Xia, C. Song, Y. Meng, P. Xue, A. J. deMello, Q. Gao, S. Stavrakis, S. Ma and X. Cao, An Addressable Electrowetting Valve for Centrifugal Microfluidics, Sens. Actuators, B, 2022, 369, 132276 CrossRef CAS.
- S. Roy, V. M. Suresh and T. K. Maji, Self-cleaning MOF: Realization of Extreme Water Repellence in Coordination Driven Self-assembled Nanostructures, Chem. Sci., 2016, 7, 2251–2256 RSC.
- J. Guo, W. Huang, Z. Guo and W. Liu, Design of a Venation-like Patterned Surface with Hybrid Wettability for Highly Efficient Fog Harvesting, Nano Lett., 2022, 22, 3104–3111 CrossRef CAS PubMed.
- S. Jiang, Y. Hu, H. Wu, R. Li, Y. Zhang, C. Chen, C. Xue, B. Xu, W. Zhu, J. Li, D. Wu and J. Chu, Three-Dimensional Multifunctional Magnetically Responsive Liquid Manipulator Fabricated by Femtosecond Laser Writing and Soft Transfer, Nano Lett., 2020, 20, 7519–7529 CrossRef CAS PubMed.
- B. Zhou, Y. Gao, Y. Mao and W. Wen, Facile Preparation of Superhydrophobic PDMS with Patternable and Controllable Water Adhesion Characteristics, J. Mater. Sci., 2017, 52, 11428–11441 CrossRef CAS.
- B. Chen, R. Zhang, H. Fu, J. Xu, Y. Jing, G. Xu, B. Wang and X. Hou, Efficient Oil-water Separation Coating with Robust Superhydrophobicity and High Transparency, Sci. Rep., 2022, 12, 2187 CrossRef CAS PubMed.
- S. Li, F. Zhao, Y. Bai, Z. Ye, Z. Feng, X. Liu, S. Gao, X. Pang, M. Sun, J. Zhang, A. Dong, W. Wang and P. Huang, Slippery Liquid-infused Microphase Separation Surface Enables Highly Robust Anti-fouling, Anti-corrosion, Anti-icing and Anti-scaling Coating on Diverse Substrates, Chem. Eng. J., 2022, 431, 133945 CrossRef CAS.
- Y. Wang, S. Tian, Q. Sun, W. Liu, R. Duan, H. Yang, X. Liu and J. Chen, Superhydrophobic Porous PLLA Sponges with Hierarchical Micro-/Nano-Structures for High-Efficiency Self-Cleaning, Macromol. Chem. Phys., 2019, 220, 1900338 CrossRef CAS.
- T. Ren, G. Tang, B. Yuan, Y. Yang, Z. Yan, L. Ma and X. Huang, Hexadecyltrimethoxysilane-Modified SiO2 Nanoparticle-Coated Halloysite Nanotubes Embedded in Silicone–Acrylic Polymer Films as Durable Fluorine-Free Superhydrophobic Coatings, ACS Appl. Nano Mater., 2020, 3, 5807–5815 CrossRef CAS.
- D. Li, H. Wang, Y. Liu, D. Wei and Z. Zhao, Large-scale Fabrication of Durable and Robust Super-hydrophobic Spray Coatings with Excellent Repairable and Anti-corrosion Performance, Chem. Eng. J., 2019, 367, 169–179 CrossRef CAS.
- L. Ejenstam, A. Swerin and P. M. Claesson, Toward Superhydrophobic Polydimethylsiloxane−Silica Particle Coatings, J. Dispersion Sci. Technol., 2015, 37, 1375–1383 CrossRef.
- G. Wu, D. Liu, J. Chen, G. Liu and Z. Kong, Preparation and Properties of Super Hydrophobic Films from Siloxane-modified Two-component Waterborne Polyurethane and Hydrophobic Nano SiO2, Prog. Org. Coat., 2019, 127, 80–87 CrossRef CAS.
- R. Sun, J. Zhao, Z. Li, N. Qin, J. Mo, Y. Pan and D. Luo, Robust Superhydrophobic Aluminum Alloy Surfaces with Anti-icing Ability, Thermostability, and Mechanical Durability, Prog. Org. Coat., 2020, 147, 105745 CrossRef CAS.
- W. Xu, P. Yi, J. Gao, Y. Deng, L. Peng and X. Lai, Large-Area Stable Superhydrophobic Poly(dimethylsiloxane) Films Fabricated by Thermal Curing via a Chemically Etched Template, ACS Appl. Mater. Interfaces, 2020, 12, 3042–3050 CrossRef CAS PubMed.
- S. Czyzyk, A. Dotan, H. Dodiuk and S. Kenig, Processing Effects on the Kinetics Morphology and Properties of Hybrid Sol–gel Superhydrophobic Coatings, Prog. Org. Coat., 2020, 140, 105501 CrossRef CAS.
- L. Liu, G. Kong, Y. Zhu and C. Che, Superhydrophobic Graphene Aerogel Beads for Adsorption of Oil and Organic Solvents via a Convenient in Situ Sol–gel Method, Colloid Interface Sci. Commun., 2021, 45, 100518 CrossRef CAS.
- T. Li, Y. Wang, H. Peng, X. Zhang, B. Shiu, J. Lin and C. Lou, Lightweight, Flexible and Superhydrophobic Composite Nanofiber Films Inspired by Nacre for Highly Electromagnetic Interference Shielding, Composites, Part A, 2020, 128, 105685 CrossRef CAS.
- X. Du, J. Xu, Q. Yan, B. Xin and C. Wang, Bio-inspired Hierarchically Porous Membrane with Superhydrophobic Antifouling Surface for Solar-driven Dehumidifying System, Colloids Surf., A, 2022, 636, 128168 CrossRef CAS.
- M. Ferrari, F. Cirisano and M. C. Morán, Mammalian Cell Behavior on Hydrophobic Substrates: Influence of Surface Properties, Colloids Interfaces, 2019, 3, 48 CrossRef CAS.
- X. Tian, V. Jokinen, J. Li, J. Sainio and R. H. Ras, Unusual Dual Superlyophobic Surfaces in Oil-Water Systems: The Design Principles, Adv. Mater., 2016, 28, 10652–10658 CrossRef CAS PubMed.
- N. Ichikawa, K. Hosokawa and R. Maeda, Interface Motion of Capillary-driven Flow in Rectangular Microchannel, J. Colloid Interface Sci., 2004, 280, 155–164 CrossRef CAS PubMed.
- M. Madou, J. Zoval, G. Jia, H. Kido, J. Kim and N. Kim, Lab on a CD, Annu. Rev. Biomed. Eng., 2006, 8, 601–628 CrossRef CAS PubMed.
- J. Koch, S. Gantenbein, K. Masania, W. J. Stark, Y. Erlich and R. N. Grass, A DNA-of-things Storage Architecture to Create Materials with Embedded Memory, Nat. Biotechnol., 2020, 38, 39–43 CrossRef CAS PubMed.
- Y. Erlich and D. Zielinsk, DNA Fountain Enables a Robust and Efficient Storage Architecture, Science, 2017, 355, 950–954 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a and
Xingyu
Jiang
*a and
Xingyu
Jiang
 *ab
*ab

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 5
1 to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 by measuring the contact angles and the sliding angles (Fig. S2A and B†). The SEM images showed the nanostructure of superhydrophobic coatings decreased with the decreasing proportion of SiO2 nanoparticles (Fig. S3†). To keep the balance between the contact angle, the sliding angle, and the stability of the coating, we set the ratio of PDMS to SiO2 nanoparticles as 2
2 by measuring the contact angles and the sliding angles (Fig. S2A and B†). The SEM images showed the nanostructure of superhydrophobic coatings decreased with the decreasing proportion of SiO2 nanoparticles (Fig. S3†). To keep the balance between the contact angle, the sliding angle, and the stability of the coating, we set the ratio of PDMS to SiO2 nanoparticles as 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the following experiments. Furthermore, we tested the duration of contact and sliding angles after superhydrophobic modification. The result showed that the superhydrophobicity of the surface was stable over 100 days (Fig. S2C†). In addition, we repeatedly washed the microchannel modified by the superhydrophobic coating with water up to 200 times. There was no apparent change in the hydrophobicity of the coating. Given that ethyl acetate could react with some materials such as PC and PS, we used ethanol to extend the application range of the superhydrophobic coating, which also increased the hydrophobicity of the coating (Fig. S4†). Compared with ethyl acetate, ethanol generated a superhydrophobic coating with more nanostructures as a dispersant (Fig. S5†).
1 in the following experiments. Furthermore, we tested the duration of contact and sliding angles after superhydrophobic modification. The result showed that the superhydrophobicity of the surface was stable over 100 days (Fig. S2C†). In addition, we repeatedly washed the microchannel modified by the superhydrophobic coating with water up to 200 times. There was no apparent change in the hydrophobicity of the coating. Given that ethyl acetate could react with some materials such as PC and PS, we used ethanol to extend the application range of the superhydrophobic coating, which also increased the hydrophobicity of the coating (Fig. S4†). Compared with ethyl acetate, ethanol generated a superhydrophobic coating with more nanostructures as a dispersant (Fig. S5†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ (w + h)
θ (w + h)