Effect of YOYO-1 on the mechanical properties of DNA
Binu
Kundukad
a,
Jie
Yan
abcd and
Patrick S.
Doyle
*ae
aBioSystems and Micromechanics (BioSym) IRG, Singapore MIT Alliance for Research and Technology (SMART), Singapore. E-mail: pdoyle@mit.edu
bDepartment of Physics, National University of Singapore, 117542, Singapore
cMechanobiology Institute, National University of Singapore, 117411, Republic of Singapore
dCentre for Bioimaging Sciences, National University of Singapore, 117546, Republic of Singapore
eDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA
First published on 28th October 2014
Abstract
YOYO-1 is a green fluorescent dye which is widely used to image single DNA molecules in solution for biophysical studies. However, the question of whether the intercalation of YOYO-1 affects the mechanical properties of DNA is still not clearly answered. Investigators have put forth contradicting data on the changes in persistence length of DNA. Here, we use atomic force microscopy to systematically study the changes in the mechanical properties of DNA due to the intercalation of YOYO-1. We first measured the persistence length, contour length and the bending angle distribution of the DNA–YOYO-1 complex. We find that the persistence length of DNA remains unaffected with the intercalation of YOYO-1. However the contour length increases linearly with about 38% increase at full saturation of 1 YOYO-1 per 4 base pairs of DNA. Next we measured the change in topology of relaxed closed circular DNA after the intercalation of YOYO-1. We find that YOYO-1 introduces supercoiling in closed circular DNA. Our observations indicate that the intercalation of YOYO-1 results in the underwinding of DNA duplex, but does not significantly change the persistence length.
1 Introduction
YOYO-1 (YOYO) is a green fluorescent dye which belongs to the family of cyanine dye and is a tetracationic homodimer of oxazole. Though YOYO is fairly non-fluorescent in solution, it forms a highly fluorescent complex with double stranded DNA, with more than 1000-fold increase in fluorescent intensity.1 Since the development of YOYO by Rye et al.,1 it has been widely used to visualize DNA molecules to measure self diffusion of DNA,2,3 study the hydrodynamic stretching of DNA4,5 and understand DNA dynamics and conformation in confinement.6,7 YOYO also enables visualizing the contour of elongated DNA for use in genomic mapping.8–10 In these and other studies, the DNA is usually modeled as a wormlike chain polymer – an elastic filament with a given bending rigidity that is embodied in the persistence length.11These studies use fluorescently labeled DNA to study the mechanical and structural properties of DNA under varying conditions like ionic strength, binding of organic compounds and biological agents. As YOYO-1 displays a strong fluorescent enhancement upon binding to DNA with increased signal to noise ratio, it enables an easy single molecule observation. YOYO binds to DNA by bis-intercalation of its two chromophore units into the DNA.1 The binding of YOYO to DNA changes the effective charge of DNA. Hence it is important to characterise whether YOYO binding changes the mechanical and structural properties of DNA.
Since the development of YOYO by Rye et al. in 1992, a variety of studies have been reported on the changes in persistence length and contour length of DNA due to the intercalation of YOYO. While there is consensus regarding the change in contour length upon YOYO binding to DNA, the results for changes in persistence length are conflicting. We summarize the results below – starting with the older studies and ending with the most recent findings.
In 1995 and 1997, Perkins et al. reported that the contour length of DNA increased by 35% when stained with TOTO and YOYO respectively at the saturation concentration.4,12 In 1997, Quake et al. cited unpublished work, according to which the intercalation of YOYO increased the contour length and persistence length of DNA by 22% and 32% respectively.13 In 1998, Bakajin et al. measured a 30% increase in contour length of DNA incubated with TOTO-1, an analogue of YOYO-1. They assumed that the persistence length also increased by the same factor as that of the contour length.5 These works were commonly referred to by many authors, until recently when various single molecule studies reported a decrease in persistence length up to 12 nm.14,15 A linear increase in contour length has also been reported by many authors.16,17
In more recent studies, the most common tools used for measuring the persistence length and contour length of DNA are magnetic/optical tweezers and atomic force microscopy (AFM). In 2002, atomic force microscopy studies by Berge et al. reported that the intercalation of a bis-intercalator, ditercalinium, increased the persistence length of DNA by two fold and the contour length by 28% that of the bare DNA.22
Tweezing experiments were reported in the subsequent years. In 2005, Sischka et al. showed that the persistence length of DNA decreased by 70% from 40 to 12 nm and the contour length increased by 36% when intercalated with YOYO.14 In 2010, Murade et al. reported a decrease in persistence length of DNA by about 71% from 52 to 15 nm when incubated with 500 nM YOYO.15 In tweezing experiments, a single DNA molecule in the desired buffer is stretched with a certain amount of force and the extension of the DNA molecule is measured. However, it has been observed that when a DNA molecule in a buffer containing YOYO is stretched, it exhibits a large hysteresis.23 Moreover this behaviour was strongly dependent on the speed at which the measurements are performed.16 This is due to the fact that YOYO exhibits different binding modes depending on the concentration of YOYO and time of incubation.24,25 The presence of free YOYO in the buffer has been shown to introduce time dependent changes in the mechanical properties of DNA.16 The studies mentioned above14,15,23 were done in non-equilibrium conditions as the DNA was stretched in a solution containing excess YOYO-1 molecules.
Taking into account the above mentioned limitations when using tweezers, Gunther et al., in 2010, measured the persistence length of DNA–YOYO complex using magnetic tweezers under equilibrium conditions.18 In their work, background DNA molecules were introduced so as to maintain a fixed staining ratio throughout the experiments. It was observed that under any staining ratio the persistence length remained unaffected and the contour length increased by 47% at a staining ratio of 1 YOYO per 3.2 base pairs.18 In 2010, Reuter et al. investigated the kinetics of binding, for single, hydrodynamically-stretched DNA molecules and found that the contour length increased by 36% at 1 YOYO per 3 base pairs.19
Recently, in 2013, Maaloum et al. found that the persistence length of DNA–YOYO complex decreased by 44% from 50 to 28 nm and the contour length increased by 46% at a staining ratio of 1 YOYO per 1 base pair.20 They used atomic force microscopy to measure the persistence length of DNA–YOYO complex from the mean square end to end distance and the contour length. A more recent work also reported a decrease in persistence length from 50 to 46 nm and an increase in contour length to 18%.21 A historical comparison of prior work on the effect of YOYO-1 on DNA properties is given in Table 1.
| Year | Reference | Technique | Dye | No. of dye/base pair | Persistence lengthsa | Contour length |
|---|---|---|---|---|---|---|
| a This column shows the persistence length of bare DNA and the YOYO stained DNA at the maximum concentration of YOYO used in each case. The value in the parentheses show the percentage increase or decrease. b The data is not reported and was never published. | ||||||
| 1997 | Perkins et al.12 | Hydrodynamic stretching | YOYO-1 | 1YOYO/4 bp | Not measured | 35% increase |
| 1997 | Quake et al.13 | Optical tweezers | YOYO-1 | Not controlled | 50–66 nm (32% increase)b | 22% increase |
| 2005 | Sischka et al.14 | Optical tweezers | YOYO-1 | Not controlled | 40–11.8 nm (70% decrease) | 36% increase |
| 2010 | Murade et al.15 | Optical tweezers | YOYO-1 | Not controlled | 52–14.9 nm (71% decrease) | 50% increase |
| 2010 | Gunther et al.18 | Magnetic tweezers | YOYO-1 | 1 YOYO/3.2 bp | Constant around 52 nm | 47% Increase |
| 2010 | Reuter et al.19 | Hydrodynamic stretching | YOYO-1 | 1 YOYO/3 bp | Not measured | 36% increase |
| 2013 | Maaloum et al.20 | Atomic force microscopy | YOYO-1 | 1 YOYO/1 bp | 50–28 nm (44% decrease) | 46% increase |
| 2014 | Shi et al.21 | Entropic force microscopy | YOYO-1 | 1 YOYO/0.8 bp | 51–46 nm (9% decrease) | 18% increase |
| 2014 | Present work | Atomic force microscopy | YOYO-1 | 1YOYO/4 bp | Constant around 57 nm | 38% increase |
Here we use atomic force microscope to systematically study the mechanical and the structural properties of DNA. Atomic force microscope allows easy visualization of the DNA–YOYO complex and is free from any artifacts related to aggregation, folding or bridging. Our study consist of two parts. In the first part we measure the persistence length, contour length and the bending angle distribution of the DNA–YOYO complex at different ratios of YOYO per base pair. In the second part, we measure the change in topology of relaxed closed circular DNA after incubating with YOYO. Eventually we rationalize our results in terms of unwinding of DNA by the interaction of YOYO.
2 Materials and methods
2.1 Sample preparation
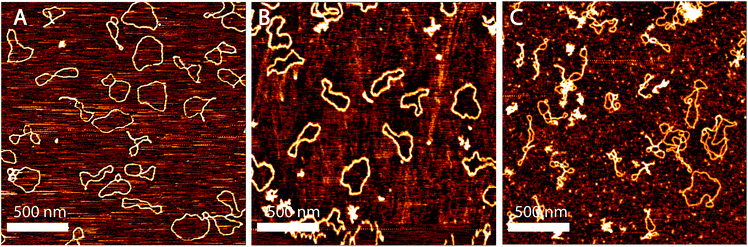
Monodisperse DNA fragments of 1000 base pairs and PUC 19 (2686 base pairs) was purchased from Thermoscientific (Vilnius, Lithuania). DNA was prepared with different concentrations of YOYO in a buffer containing 1× TE (10 mM Tris base, 1 mM EDTA, pH 8 titrated with HCl). The concentration of the dye was adjusted so that there is one YOYO dimer per 4, 8, 16 and 100 base pairs of DNA. The DNA–YOYO was incubated at 55 °C for 2 hours for homogenous distribution of the dye along the DNA. 20 μl of the sample was then deposited on a freshly cleaved SPI Grade V-4 mica and incubated for 30 min. This is then washed with DI water and blown dry using nitrogen gas. For imaging of bare DNA, 5 mM MgCl2 was used. This method allows the DNA molecules to equilibrate on the surface by diffusion. The interaction between DNA and mica mediated by Mg2+ is weak, so that the chain statistics is not affected. It has been shown previously that DNA deposited in this way equilibrates in a 2D conformation.26–29 For imaging DNA intercalated with YOYO, the sample was deposited on mica without MgCl2. YOYO being cationic facilitated the binding of the DNA–YOYO complex to mica. Here we assume that the binding of DNA–YOYO complex to the mica is 2D equilibrated due to the following reasons. (i) The size of the YOYO stained DNA molecules are similar to the 2D equilibrated DNA in the presence of Mg2+ as shown in panel A of Fig. 1. On the other hand, the kinetically trapped DNA molecules appear much smaller in size as shown in panel B of Fig. 1. The difference in size comes from considering a polymer equilibrated in 2D versus the projected image of a 3D polymer into a 2D surface. Rivetti et al. has shown that for a long DNA chain, the projection of the 3D image onto the 2D plane gives a mean-squared end-to-end size, 〈R2〉proj, which is 1/3 the value of the size of DNA equilibrated 2D, 〈R2〉2D.26 This difference in size can be used to distinguish between the 2D equilibrated and kinetically trapped DNA. (ii) We do not see any overlapping of the molecules as would be observed in the case of kinetically trapped molecules. | ||
| Fig. 1 (A) PUC 19 DNA (2686 base pairs) equilibrated on the surface of mica in the presence of Mg2+ (B) PUC 19 DNA kinetically trapped on the surface of polylysine treated mica. | ||
2.2 Atomic force microscopy
The imaging was done at room temperature in air with a Nanowizard II atomic force microscope (JPK Instruments, Berlin, Germany). Images were acquired in the tapping mode with Nanosensor silicon (Si) cantilevers (spring constant of 10–130 N m−1) and operated below their resonance frequency (typically 200–500 kHz).2.3 Data analysis
There are two ways to determine the persistence length of a polymer: (i) from the bending angle or (ii) from the end to end distance of the DNA molecule. For both cases, the first step is to obtain the centerline of the DNA molecules. This was done by moving along the DNA molecules in a small step size of half the cross sectional diameter and locating the point with maximum height. For a DNA molecule equilibrated in 2 dimensions, the energy needed to bend a segment of length, L through angle θ is given by | (1) |
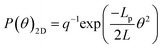 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θs,s+L〉 which can be written as:
θs,s+L〉 which can be written as:〈cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θs,s+L〉2D = exp(−L/2Lp) θs,s+L〉2D = exp(−L/2Lp) | (3) |
We can also find the mean-squared end-to-end distance by taking the integral of the tangent–tangent correlation function and it is written as follows:
 | (4) |
3 Results
3.1 Persistence length
We first determined the persistence length of bare DNA. For this purpose, a DNA concentration of 1.37 mg l−1 in 5 mM MgCl2 and an incubation time of 5 min was used for acquiring the images. Here we used short DNA fragments of 1000 base pairs. DNA molecules of this size allows easy visualisation of structural changes in DNA with minimal effect due to aggregation and folding. Moreover, DNA molecules of size less than 1000 nm are shown to be unaffected by excluded volume interactions26 and hence consistent with the use of eqn (1)–(4). For DNA–YOYO complex, a concentration of 2.1 mg l−1 and an incubated time of 30 min was used. Fig. 2 show the representative images of bare DNA and the DNA–YOYO complex and also their corresponding contours.From the DNA contours, the tangent autocorrelation function, 〈cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θs,s+L〉 versus the separation, L was plotted. The curve thus obtained was fitted with eqn (3), from which the persistence length was obtained (Fig. 3). We obtained a persistence length of 56 nm for bare DNA which is in good agreement with previously reported values.27,29
θs,s+L〉 versus the separation, L was plotted. The curve thus obtained was fitted with eqn (3), from which the persistence length was obtained (Fig. 3). We obtained a persistence length of 56 nm for bare DNA which is in good agreement with previously reported values.27,29
 | ||
Fig. 3 The correlation function 〈cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θs,s+L〉 versus separation, L for DNA with (A) no YOYO (B) 1 YOYO per 100 base pairs (C) 1 YOYO per 16 base pairs (D) 1 YOYO per 8 base pairs (E) 1 YOYO per 4 base pairs. The filled circles are the experimental data obtained by averaging 30 molecules. The line is the fit to the experimental data using eqn (3). θs,s+L〉 versus separation, L for DNA with (A) no YOYO (B) 1 YOYO per 100 base pairs (C) 1 YOYO per 16 base pairs (D) 1 YOYO per 8 base pairs (E) 1 YOYO per 4 base pairs. The filled circles are the experimental data obtained by averaging 30 molecules. The line is the fit to the experimental data using eqn (3). | ||
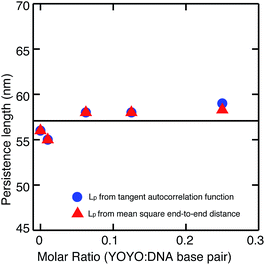
We then measured the persistence length of DNA–YOYO complex at various YOYO per base pair concentrations. We find that the persistence length of DNA–YOYO complex does not deviate much from that of bare DNA and stays within the error limits with an average value of 57 nm. We also measured the mean-squared separation, 〈(Rs,s+L)2〉 between pairs of points located at s and s + L and averaging for all values of s along the contour. Fig. 4 shows the plot of the mean-square end-to-end distance versus the separation, L for the same conditions as in Fig. 3. The closed circles are the experimental data. This curve is fitted with eqn (4), from which the persistence length is obtained. The persistence length obtained from the two methods are in good agreement (Fig. 5).
 | ||
| Fig. 4 The mean square end to end distance versus the separation, L for DNA with (A) no YOYO (B) 1 YOYO per 100 base pairs (C) 1 YOYO per 16 base pairs (D) 1 YOYO per 8 base pairs (E) 1 YOYO per 4 base pairs. The filled circles are the experimental data obtained by averaging 30 molecules. The line is the fit to the experimental data using eqn (4). | ||
3.2 Bending angle distribution
One might expect the persistence length of the DNA–YOYO complex to be reduced due to the charge neutralization of DNA by YOYO. However, persistence length is the bending rigidity of DNA which is derived from the bending angle distribution of DNA. The persistence length from Fig. 3 and 4 is obtained by averaging the bending rigidity along the contour of the DNA molecules. YOYO binding could cause structural changes in DNA which could lead to the local changes in the bending rigidity. The observed persistence length could be the result of larger bending angles due to kinks counterbalancing the smaller bending angle due to stiffening. This could lead to the overall persistence length being unaffected. To investigate this, we plotted the probability distribution of the bending angles obtained from all the molecules in case of bare DNA and DNA–YOYO complex. Fig. 6 shows the probability distribution of the bending angles for angle, θ between the tangents separated by a contour length of 20 nm. This length scale was used as it has been shown earlier that the DNA is well described by the wormlike chain model for length scales more than three helical turns of DNA (10.5 nm).31 Here we have used 20 nm to accommodate for any extension of the helical turns resulting from the YOYO intercalation. We find that the probability distribution of the bending angle is unaffected by the intercalation of YOYO and fits the curve which describes the wormlike chain model. Thus we conclude that the persistence length of DNA–YOYO complex is unaffected by the intercalation of YOYO.3.3 Contour length
Next, we measured the contour length of the DNA–YOYO complex. From the AFM images, we find that the contour length of bare DNA is 328 ± 8 nm. With the staining ratio of 1 YOYO per 100 base pairs, there is almost no change in contour length. However, the contour length of the DNA–YOYO complex increases linearly with increase in staining ratio as shown in Fig. 7. At a staining ratio of 1 YOYO per 4 base pairs, the contour length of DNA increases by 38% from 328 ± 8 nm to 453 ± 17 nm. From these values, we find that each bound YOYO extends the DNA molecule by 0.5 nm. | ||
| Fig. 7 Contour length of DNA–YOYO complex increases linearly with increase in staining ratio. The circles represent the average contour length of DNA–YOYO complex averaged over 30 molecules. The data points correspond to the DNA–YOYO ratios mentioned in Fig. 2. The error bars are the standard deviations and the line is a linear fit to the data points. | ||
3.4 YOYO changes the helical twist of DNA
We find that YOYO increases the contour length of DNA–YOYO complex without affecting the persistence length. We propose that this elongation of DNA–YOYO complex is due to the under twisting of DNA. YOYO induced under twisting of DNA has been reported earlier, however there has not been a direct evidence for this process. Earlier NMR studies have shown that YOYO under twists the DNA by 106°.32 Magnetic tweezers experiments have shown that, a torsionally constrained DNA–YOYO complex becomes negatively supercoiled when destained. The authors measured an untwisting of 24° per YOYO molecule.18In order to investigate the supercoiling of DNA by the intercalation of YOYO, we used relaxed closed circular DNA. For this purpose, PUC 19 DNA was treated with Topo 1 which relaxes both the left and the right handed supercoiling. Topo 1 is a class of DNA topoisomerase that cuts a single strand of the DNA helix and allows the complimentary strand to pass through the nick and reseals the nick, thereby reducing the linking number and relaxing the DNA.33 The closed circular DNA thus obtained was then incubated with YOYO in the ratio 1 YOYO per 100 bp and 1 YOYO per 4 base pair at 55 °C for 2 hours. Panel A of Fig. 8 shows the relaxed closed circular PUC 19 DNA. When incubated with 1 YOYO per 100 bp, we find that there is no change in the topology of DNA (panel B of Fig. 8). However the DNA topology is fundamentally different when incubated with 1 YOYO per 4 base pairs as shown in panel C of Fig. 8. We find that the majority of the DNA population is plectonemically supercoiled. Less that 5% of the DNA is not supercoiled as the presence of nicks in one of the strands in DNA helix prevents the DNA from supercoiling. Some of the molecules appear compacted due to the increased helical twist in the DNA. We measure the length of the superhelical axis of the discernible plectonemes and found that the ratio of the average length of the superhelical axis of the plectonemes and the contour length of the circular DNA is 0.42 which is in agreement with the value reported in the literature.34 Our observations show that the YOYO induced supercoiling in the relaxed closed circular DNA is due to the under twisting of DNA by the intercalated YOYO.
4 Discussion
Here we used atomic force microscopy to study the mechanical and structural properties of the DNA–YOYO complex. We find that the persistence length of the DNA–YOYO complex does not deviate from that of the bare DNA for YOYO concentrations ranging from very low staining ratio of 1 YOYO per 100 base pairs to the saturation level of 1 YOYO per 4 base pairs. It should be noted that our methodology is able to measure an increase or decrease in persistence length precisely as reported in our previous work.28 Our results are at odds with most of the previously reported results14,15,19–21 which showed that the persistence length of DNA decreases with increasing concentration of YOYO. Our results do however agree with the recent study by Gunther et al.18 These discrepancies in the literature can be due to the following reasons. (i) Most of the single molecule experiments were not done at equilibrium conditions. Single molecules experiments are usually done by stretching a single DNA molecule in a buffer containing excess of YOYO. This would mean that the stretching of DNA molecules would result in the association and dissociation of the YOYO-1 and hence affect the persistence length. This issue was taken into consideration by Gunther et al. who designed their experiments such that the number of YOYO per base pair remains fixed.18 (ii) YOYO binding has been shown to depend on the amount of force applied and the stretching speed.16Using atomic force microcopy, we are able to visualize the molecules under equilibrium conditions and unperturbed by external force. However our results on persistence length do not agree with the previous atomic force microscopy results20 due to the following reasons. The prior authors determined the persistence length from the end-to-end distance and the contour length of DNA.20 They used polydispersed DNA molecules with sizes ranging from about 200 to 1500 nm. However it has been shown that for DNA molecules equilibrated in 2D, the excluded volume effects significantly alters the chain dimensions of DNA molecules with contour length more than 20 times the persistence length. Taking these factors into consideration, we have plotted the tangent auto correlation function and the mean square end-to-end distance for short length scales of 4–100 nm. We have used monodisperse DNA molecules of 1000 bp (340 nm) to avoid any effects due to the excluded volume interactions and other artifacts. It should be noted that our results in air well represent the results in solution due to our sample preparation methods. Zuccheri et al.35 has shown earlier that while imaging DNA in solution, the DNA is slight mobile on the mica surface in the presence of buffer. However when deionized water is injected to replace the buffer, the DNA molecules freezes in the same conformation as they had been immediately before the injection of water.35 In our sample preparation we freeze the conformation with deionized water before drying the sample. This enables us to capture the conformation of DNA in the desired buffer conditions.
Another important factor that affects the binding of YOYO to DNA is the time and temperature of incubation. We find that for the equilibrium distribution of YOYO along the DNA molecules, the sample has to be incubated at 55 °C for at least 2 hours. Previous electrophoresis studies have also shown this as a necessary condition for equilibration of the DNA–YOYO complex.25 Most of the previously reported experiments were performed with samples incubated at room temperature.
Another factor which may be the cause of the inconsistency in the reported values of persistence length and contour length is the ionic strength. Firstly, the persistence length of DNA has been shown to be dependent on the ionic strength. At ionic strength more than 7.44 mM, the persistence length of DNA remains constant between 50 to 60 nm.36 The ionic strength of 1× TE (10 mM Tris base, 1 mM EDTA, pH 8 titrated with HCl) is about 8.4 mM. At this ionic strength we measured the persistence length of DNA to be 56 nm. This agrees well with the previously reported values.27,36 The binding of YOYO has also been shown to be dependent on the ionic strength. It has been shown by Gunther et al. that the binding of YOYO is stable at ionic strengths less than 100 mM.18 The choice of our buffer conditions was made to ensure that it is high enough to provide a stable condition for bare DNA and at the same time low enough for the stable binding of YOYO.
The contour length measurements show that the length of DNA increases by 38% at the maximum staining ratio of 1 YOYO per 4 base pairs. Thus each bound YOYO extends the DNA molecule by 0.5 nm which is in agreement with previously reported results.18 The measurement of the persistence length and the contour length of DNA shows that the intercalation of YOYO changes the contour length of DNA without affecting the persistence length. However one would expect the persistence length of DNA–YOYO complex to be reduced due to the charge neutralisation of DNA by YOYO. The bending angle distribution shows that the intercalation of YOYO does not introduce local changes in the bending angles along the DNA helix. The linear increase in contour length of DNA with increasing concentration of YOYO should be due to the underwinding of the DNA helix. In the case of closed circular DNA, such a change in helical twist should result in a change in the linking number deficit and hence the superhelical density. Hence the relaxed closed circular DNA should become supercoiled with the change in the helical twist.33 Our experiments on closed circular DNA intercalated with YOYO have confirmed the fact YOYO changes the helical twist of DNA and hence leading to the supercoiling of DNA.
5 Conclusion
AFM is an effective tool which can be used to determine the mechanical and structural properties of DNA. Given the fact that AFM allows the measurement of persistence length and contour length of 2D equilibrated DNA under zero force, it is complementary to other methods such as magnetic and optical tweezers. Here we have used atomic force microscopy to measure the persistence length and the contour length of DNA and YOYO intercalated DNA by carefully considering the experimental factors that affect these measurements. Taking into consideration the experimental factors such as incubation time and temperature for equilibrium distribution of the YOYO on DNA, the ionic strength of the buffer and using short DNA fragments of DNA, we find that the our results agree with those reported by Gunther et al. Moreover, we have confirmed a possible mechanism due to which the mechanical and structural properties of DNA is affected when stained with YOYO. We have shown that YOYO binding leads to under winding of DNA which leads to the change in topology in the case of closed circular DNA.Acknowledgements
This research was supported by the National Research Foundation Singapore through the Singapore MIT Alliance for Research and Technology's BioSystems and Micromechanics (BioSym) IRG research program and the National Science Foundation (CBET-1335938). Piewen Cong is thanked for the contribution of matlab program to determine the centreline of DNA.References
- H. S. Rye, S. Yue, D. E. Wemmer, M. A. Quesada, R. P. Haugland, R. A. Mathies and A. N. Glazer, Nucleic Acids Res., 1992, 20, 2803–2812 CrossRef CAS PubMed.
- D. Wirtz, Phys. Rev. Lett., 1995, 75, 2436–2439 CrossRef CAS.
- B. Maier and J. O. Rädler, Phys. Rev. Lett., 1999, 82, 1911–1914 CrossRef CAS.
- T. T. Perkins, D. E. Smith, R. G. Larson and S. Chu, Science, 1995, 268, 83–87 CAS.
- O. Bakajin, T. Duke, C. Chou, S. Chan, R. Austin and E. Cox, Phys. Rev. Lett., 1998, 80, 2737–2740 CrossRef CAS.
- J. O. Tegenfeldt, C. Prinz, H. Cao, S. Chou, W. W. Reisner, R. Riehn, Y. M. Wang, E. C. Cox, J. C. Sturm and P. Silberzan, et al. , Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10979–10983 CrossRef CAS PubMed.
- C.-C. Hsieh and P. S. Doyle, Korea Aust Rheol J, 2008, 20, 127–142 Search PubMed.
- D. C. Schwartz, X. Li, L. I. Hernandez, S. P. Ramnarain, E. J. Huff and Y.-K. Wang, Science, 1993, 262, 110–114 CAS.
- R. H. Meltzer, J. R. Krogmeier, L. W. Kwok, R. Allen, B. Crane, J. W. Griffis, L. Knaian, N. Kojanian, G. Malkin and M. K. Nahas, et al. , Lab Chip, 2011, 11, 863–873 RSC.
- K. D. Dorfman, AIChE J., 2013, 59, 346–354 CrossRef CAS PubMed.
- J. F. Marko and E. D. Siggia, Macromolecules, 1995, 28, 8759–8770 CrossRef CAS.
- T. T. Perkins, D. E. Smith and S. Chu, Science, 1997, 276, 2016–2021 CrossRef CAS.
- S. R. Quake, H. Babcock and S. Chu, Nature, 1997, 388, 151–154 CrossRef CAS PubMed.
- A. Sischka, K. Toensing, R. Eckel, S. D. Wilking, N. Sewald, R. Ros and D. Anselmetti, Biophys. J., 2005, 88, 404–411 CrossRef CAS PubMed.
- C. U. Murade, V. Subramaniam, C. Otto and M. L. Bennink, Nucleic Acids Res., 2010, 38, 3423–3431 CrossRef CAS PubMed.
- M. L. Bennink, O. D. Schärer, R. Kanaar, K. Sakata-Sogawa, J. M. Schins, J. S. Kanger, B. G. de Grooth and J. Greve, Cytometry, 1999, 36, 200–208 CrossRef CAS.
- P. S. Doyle, B. Ladoux and J.-L. Viovy, Phys. Rev. Lett., 2000, 84, 4769–4772 CrossRef CAS.
- K. Günther, M. Mertig and R. Seidel, Nucleic Acids Res., 2010, 38, 6526–6532 CrossRef PubMed.
- M. Reuter and D. T. Dryden, Biochem. Biophys. Res. Commun., 2010, 403, 225–229 CrossRef CAS PubMed.
- M. Maaloum, P. Muller and S. Harlepp, Soft Matter, 2013, 9, 11233–11240 RSC.
- N. Shi and V. M. Ugaz, Small, 2014, 10, 2553–2557 CrossRef CAS PubMed.
- T. Berge, N. S. Jenkins, R. B. Hopkirk, M. J. Waring, J. M. Edwardson and R. M. Henderson, Nucleic Acids Res., 2002, 30, 2980–2986 CrossRef CAS PubMed.
- C. Murade, V. Subramaniam, C. Otto and M. L. Bennink, Biophys. J., 2009, 97, 835–843 CrossRef CAS PubMed.
- A. Larsson, C. Carlsson, M. Jonsson and B. Albinsson, J. Am. Chem. Soc., 1994, 116, 8459–8465 CrossRef CAS.
- C. Carisson, M. Johnson and B. Åkerman, Nucleic Acids Res., 1995, 23, 2413–2420 CrossRef PubMed.
- C. Rivetti, M. Guthold and C. Bustamante, J. Mol. Biol., 1996, 264, 919–932 CrossRef CAS PubMed.
- P. A. Wiggins, T. Van Der Heijden, F. Moreno-Herrero, A. Spakowitz, R. Phillips, J. Widom, C. Dekker and P. C. Nelson, Nat. Nanotechnol., 2006, 1, 137–141 CrossRef CAS PubMed.
- B. Kundukad, P. Cong, J. R. van der Maarel and P. S. Doyle, Nucleic Acids Res., 2013, 41, 8280–8288 CrossRef CAS PubMed.
- P. J. Hagerman, Annu. Rev. Biophys. Biophys. Chem., 1988, 17, 265–286 CrossRef CAS PubMed.
- J. Abels, F. Moreno-Herrero, T. van der Heijden, C. Dekker and N. Dekker, Biophys. J., 2005, 88, 2737–2744 CrossRef CAS PubMed.
- A. K. Mazur and M. Maaloum, Phys. Rev. Lett., 2014, 112, 068–104 CrossRef.
- F. Johansen and J. P. Jacobsen, J. Biomol. Struct. Dyn., 1998, 16, 205–222 CAS.
- A. D. Bates and A. Maxwell, DNA topology, Oxford University press, 2005 Search PubMed.
- T. C. Boles, J. H. White and N. R. Cozzarelli, J. Mol. Biol., 1990, 213, 931–951 CrossRef CAS.
- G. Zuccheri, R. T. Dame, M. Aquila, I. Muzzalupo and B. Samori, Appl. Phys. A: Mater. Sci. Process., 1998, 66, S585–S589 CrossRef CAS.
- C. G. Baumann, S. B. Smith, V. A. Bloomfield and C. Bustamante, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 6185–6190 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2014 |