DOI:
10.1039/D4RA04038D
(Review Article)
RSC Adv., 2024,
14, 28487-28515
Bioreceptor modified electrochemical biosensors for the detection of life threating pathogenic bacteria: a review
Received
1st June 2024
, Accepted 23rd August 2024
First published on 6th September 2024
Abstract
The lack of reliable and efficient techniques for early monitoring to stop long-term effects on human health is an increasing problem as the pathogenesis effect of infectious bacteria is growing continuously. Therefore, developing an effective early detection technique coupled with efficient and continuous monitoring of pathogenic bacteria is increasingly becoming a global public health prime target. Electrochemical biosensors are among the strategies that can be utilized for accomplishing that goal with promising potential. In recent years, identifying target biological analytes by interacting with bioreceptors modified electrodes is among the most commonly used detection techniques in electrochemical biosensing strategies. The commonly employed bioreceptors are nucleic acid molecules (DNA or RNA), proteins, antibodies, enzymes, organisms, tissues, and biomimetic components such as molecularly imprinted polymers. Despite the advancement in electrochemical biosensing, developing a reliable and effective biosensor for detecting pathogenic bacteria is still in the infancy stage with so much room for growth. A major milestone in addressing some of the issues and improving the detection pathway is the investigation of specific bacterial detection techniques. The present study covers the fundamental concepts of electrochemical biosensors, human PB illnesses, and the latest electrochemical biosensors based on bioreceptor elements that are designed to detect specific pathogenic bacteria. This study aims to assist researchers with the most up-to-date research work in the field of bio-electrochemical pathogenic bacteria detection and monitoring.
1. Introduction
Pathogenic bacteria (PB) can cause life-threatening infections and are one of the main causes of mortality worldwide because PB poses an urgent threat to global health. Due to the shield created by PB, essential antibiotics are insufficient to fight against PB,1 thus, antibiotics are becoming ineffective against PB.2 Paralysis, tetanus, fever, cramping in the stomach, respiratory and urinary tract infections, cancer, diarrhea, cholera, and fever are the deadliest illnesses and diseases caused by PB.3 The World Health Organization (WHO) reported that antibiotic resistance poses a serious worldwide problem with far-reaching implications for public health and the economy.4 Intestinal infectious disorders are caused by bacteria that are associated with water, food, and physiological fluids. These bacteria also propagate infectious diseases among humans and animals.5 The primary cause of foodborne illness outbreaks is undercooked or processed ready-to-eat (RTE) meat, dairy products, fruit, and vegetables.6–8 The primary reservoirs for many foodborne diseases include the environment (soil and water), foods produced from animals (meat, milk, and eggs), and agricultural goods (fruit and vegetables).5,9 The most common foods that can harbor bacteria such as Salmonella spp., Staphylococcus aureus, Campylobacter, Listeria, Shigella, or Escherichia coli O157:H7 are fruit, vegetables, fish, meat, eggs, and dairy products. Due to the severe pandemic brought on by the life-threatening infection of PB, preventive measures should be taken.10 Nonetheless, the development of diagnosis and prevention tools and routes can address the intense outbreak of PBs.
Electrochemical biosensors have garnered increasing attention from researchers in recent years because of their extremely sensitive and selective method of detecting PB. The primary function of a biorecognition element on a biosensor is to give target analyte specificity. The bioreceptors are immobilized on a sensor surface to capture the target analyte. Antibodies, enzymes, cells, aptamers, DNAs, biomimetics, and phages are the most frequent types of bioreceptors or biorecognition elements, also known as molecular probes. Bioreceptor-modified electrochemical biosensors are a potent tool for the identification of pathogenic bacteria, providing quick, accurate, and targeted diagnostics in a range of applications. Table 1 summarizes the advantages of electrochemical biosensing over other methods of detecting PBs is presented below.
Table 1 Advantages of electrochemical biosensor over other methods for PBs detectiona
| Specification |
Electrochemical method |
Other methods |
| Abbreviations: ELISA: enzyme-linked immunoassay, CFU: colony forming unit, PCR: polymerase chain reaction. |
| Sensitivity |
Electrochemical biosensors are highly sensitive to specific analytes for the lowest limit of detection.11 |
Most of the conventional methods are low sensitive, e.g., ELISA requires long incubation time (24–48 hours) with low sensitivity (≥105 CFU mL−1) yield12 |
| Selectivity and viability |
Electrochemical biosensors are highly selective, bind and detect specific analyte, discriminate between live and dead bacteria.13,14 |
All methods are less selective, PCR techniques cannot distinguish between nonviable and viable bacteria cells due to false positive cross-amplification and false negative DNA polymerase inhibition.15 |
| Stability |
Retained their efficiency for a long time, it shows high efficiency.16 |
Insufficient stability, low efficiency of signal amplification.17 |
| Linearity and limit of detection |
Wide dynamic linear range of biosensor response and lowest limit of detection.18 |
Lack of linear dynamic range and poor limit of detection.17 |
| Detection method |
This method is rapid, real-time with portable device.16 |
Most of the conventional methods are time consuming, and long working protocol requires heavy machinery.19,20 |
In recent years, electrochemical biosensors, especially portable electrochemical biosensors, have been the preferred bioanalytical method for detecting PBs. Numerous biosensors were previously reported such as mass-based (piezoelectric21 and surface acoustic22), optical (light scattering,22 fiber optics,23 and SPR24), and electrochemical (amperometry,25 potentiometric,26 and impedance27). However, they all face the challenge of identifying pathogens in real samples (such as food) at low analyte concentrations with excellent sensitivity and selectivity. Nonspecific adsorption of biomolecules in the sample (either from the matrix or microbes) can severely impede the biosensor surface.28 This increases background “noise”, reduces the selectivity of the biosensor, and weakens the signal. Recently, several studies have been performed on novel and portable biosensors that can outperform conventional detection technologies. More research is needed on biosensors that can quantitatively identify and screen infections in clinical, environmental, and dietary samples.29 Electrochemical platforms are the commonly used biosensor due to their high analyte selectivity and ability to perform multiplex analyses. Furthermore, they can achieve high analytical precession even in complex food matrices with varying densities, compositions, and pH levels. As a diagnostic tool for clinical applications, electrochemical-based biosensors have shown considerable promise. However, they still face some obstacles that must be overcome. Among these challenges, preserving the sensor's stability and repeatability in intricate real matrices, generating a low limit of detection (LOD), and preventing non-specific adsorption of interfering species are the three main obstacles faced in the development of electrochemical biosensing platforms.30 Pathogens are electrochemically detected using a working electrode that has been modified with recognition components (such as an antibody, aptamer, or DNA probe) to guarantee measurements with high selectivity, sensitivity, and specificity. Employing a working electrode modified with recognition elements (e.g., an antibody, aptamer, or DNA probe) allows for the electrochemical detection of pathogens while maintaining measurement selectivity, sensitivity, and specificity. For this reason, the most widely used biosensors are electrochemical-based platforms.
Incorporating biorecognition elements into the electrode surface of the electrochemical biosensing to selectively detect and identify biologically significant microorganisms such as pathogens can significantly enhance the accuracy and specificity of biosensing recognition platforms. Therefore, bioreceptors incorporated transducers-based bio-electrochemical sensors increase the overall platform viability for the diagnostic and detecting applications. This study covers the fundamentals of electrochemical biosensors, human PBs illnesses, and the most recent advancements in bioreceptor element-based electrochemical biosensors for the detection of specific pathogenic bacteria. To the best of our knowledge, there is no published survey that comprehensively review electrochemical biosensors based on bioreceptors surface modification for the detection of pathogenic bacteria with a great length of details on the commonly used biorecognition elements and a large selection of life threating pathogenic bacteria along with the most frequent diseases caused by these infectious microorganisms. This work can serve as a reference for researchers with interest in developing reliable and highly sensitive electrochemical based biosensors to selectively detect deadly pathogens for point of care applications in the health care sector.
2. Principle of an electrochemical biosensor
A combined receptor-transducer device that employs a biological recognition element for selected quantitative or semi-quantitative analytical data is called an electrochemical biosensor.31 In 1962, Clark and Lyons introduced the term “biosensor”. Their outstanding achievements has made them the original founders of biosensing concept.31 To measure the amount of dissolved oxygen in blood using the amperometry technique, they developed an oxygen electrode and a glucose oxidase enzyme.32 The electrochemical biosensors are the most extensively used biosensing platform and are successfully commercialized to monitor glucose for diabetic patients. For biosensing detection, pathogenic bacteria are collected from host body, cultured, and screened. The desired biochemical conversion is performed to bring detectable condition. Thereafter, pathogenic, or nonpathogenic bacteria can be detected using a group of bioreceptors (enzyme, antibody, cell, bacteriophage, nucleic acid) fabricated electrode.33,34 Both live and dead bacteria can be detected by electrochemical biosensors. The interaction of bioreceptors with bacterial cellular components-which can be found in both living and dead bacteria is frequently the basis for detection mechanisms.14,35 Additionally, the bacteriophage base biosensor cannot detect dead bacteria.36 The receptor–analyte interaction is measured using electrochemical techniques such as amperometry, potentiometry, impedimetric, and conductometry. The most used signal measurement techniques are cyclic voltammetry (CV), differential pulse voltammetry (DPV), stripping voltammetry, alternating current voltammetry (ACV), polarography, square wave voltammetry (SWV), and linear sweep voltammetry (LSV). Different electrochemical signal measurement techniques are suitable for various biosensing applications, and each method has its own merits. The type of analyte, the desired level of sensitivity and specificity, and the practical considerations all play a role in the measurement systems selection. These approaches can be classified into many types based on which characteristics of the electrode are regulated and which are measured. For incidence, potentiometry is used to quantify the difference in electrode potentials, amperometry analyses electric current, and coulometry records charge passed during a given time period.37 Technological developments in these areas keep improving the functionality and performance of electrochemical biosensors across a range of applications.
Recent wearable electrochemical sensors have attracted a great deal of attention due to their wide range of applications in the human body. These electrochemical devices are used numerous forms such as contact lenses, Google glass, skin-patch, mouth gourds, smartwatches, underwear, and wristbands.38 Additionally, electrochemical biosensor has been trailed for bacteria detection in the human body. Mannoor et al., developed a wireless and wearable biosensor based on graphene-electrode-silk hybrid structure for Staphylococcus aureus bacteria detection on tooth enamel.39 The developed sensor displayed a high sensitivity and with detection limits down to a single bacterium. Another flexible, wearable, wireless and battery-free DNA hydrogel-based biosensor was developed by Xiong and his colleagues. The introduced biosensor can identify wound infections caused by pathogenic bacteria (such as Staphylococcus aureus.) prior to any evident signs of wound infection. The fully integrated wound infections sensor integrates biologically responsive DNA hydrogel and Near-field communication (NFC) module to support a smartphone-based readout for updating the wound infection condition.40 Based on the previously reported finding that in infected wounds, the pH becomes alkaline which have been linked predominantly to the presence of bacteria.41 Manjakkal et al. introduced graphite-polyurethane thick electrode for potentiometric electrochemical pH-based wearable sensor for pH level detection, thus determining the wound healing stage.42
Overall, graphical representation of pathogenic bacteria detection strategy is represented in Fig. 1. The three main parts of a biosensor are (i) a bioreceptor, also known as a biorecognition element, which identifies the desired analyte; (ii) a transducer that changes biological (electrochemical) impulses into electrical signals, (iii) a signal processing system presents these electrical signals in a recognizable format. Furthermore, biosensors are classified according to the type of transducer used, which includes electrochemical, calorimetric (thermometric), mass (piezoelectric or surface acoustic wave devices), and optical.43 Electrochemical biosensors are becoming increasingly popular for applications in biotechnology, food safety, environmental monitoring, and clinical diagnostics due to their rapid reaction, ease of use, and low cost.
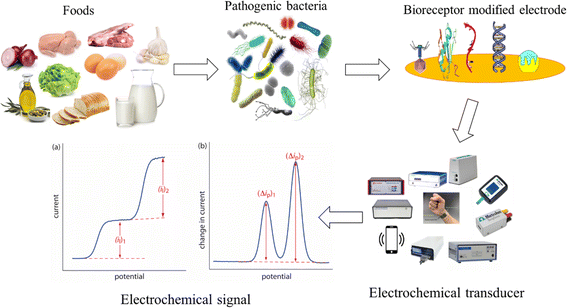 |
| | Fig. 1 Schematic diagram showing the typical electrochemical biosensing process for the detection of pathogenic bacteria. It consists of PB, bioreceptor modified electrode, electrochemical workstation measuring the receptor–analyte interaction signal. | |
2.1. Bioreceptors
A molecule known as a bioreceptor binds to analytes through biological processes. Bioreceptors are categorized into affinity (such as nucleic acid and antibodies) and catalytic (such as enzymes, cells, and tissues) types. Biosensors are further classed according to the type of bioreceptor they use enzymes, cells, antibodies, or nucleic acid based. Immobilization techniques that can be utilized to attach bioreceptors on the surface of sensors include membrane and matrix entrapment, covalent and ionic binding, physical adsorption, and intermolecular crosslinking.44 The physical adsorption of bioreceptors is mediated by Van der Waals forces, hydrophobic interactions, hydrogen bonds, and ionic forces. Covalent bonding between bioreceptors and sensor surfaces involves functional groups such as amino, sulfhydryl, hydroxyl, phenolic, and thiol groups, among others. It is worth noting that covalent binding is commonly utilized for enzyme immobilization. Polymeric materials such as polyacrylamide, cellulose acetate, starch, alginate, pectate, polyvinyl alcohol, polyvinyl chloride, polycarbonate, and silica gel are employed to entrap matrices or pathogens. Glutaraldehyde, hexamethylene diisocyanate, 1,5-difluoro 2,4-dinitrobenzene, and bisdiazobenzidine-2,2′-disulphonic acid are examples of bi- or multi-functional cross-linking chemicals used in intermolecular cross-linking.45 Furthermore, successful immobilization techniques include covalent attachment to a functionalized substrate, affinity immobilization (attachment of biotinylated probes to streptavidin-coated surfaces), and self-assembling (chemisorption of thiol-modified probes onto gold surfaces). A short summary of bioreceptor bonding on transducer surface is presented in Fig. 2. Bioreceptor can be also classified into two main categories: catalytic biochemical receptors such as enzyme, cells, and DNA zymes; and affinity biochemical receptors such as antibody, aptamer, DNA/RNA oligonucleotides, engineered and protein.
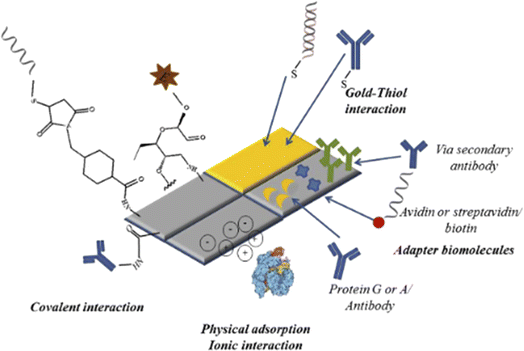 |
| | Fig. 2 Schematic diagram illustrating examples of matrix-analyte bonding formation techniques. The figure was adopted from ref. 46 with permissions, copyright @ Elsevier. | |
2.1.1 Enzymes. Proteins called enzymes function as biological catalysts in organisms to quicken chemical reactions. Because they are composed of polypeptide chains of amino acids, the three-dimensional structure of the enzyme is determined by the arrangement of these amino acids.47 Enzymes were the first molecular recognition components to be incorporated in biosensors.48 Enzyme-based biosensors use a stable source of enzyme material (primarily through bio-renewable sources) and have a high possibility of modifying the catalytic properties or substrate specificity of the enzymes by means of genetic engineering. Finally, catalytic amplification of the biosensor response can be achieved by the modulation of the enzyme activity with respect to the target analyte.46 The capacity to speed biochemical activities.49To improve detection stability and repeatability, enzymes can be immobilized on the transducer matrix surface. Therefore, the choice of inert, stable, and resistant support material is essential for improving enzyme activity.50 The enzymes can be immobilized on support material via adsorption, covalent bonding, crosslinking, encapsulation, and entrapment as illustrated in Fig. 3. Literature revealed that enzyme activity varies depending on the type of bonding with the surface of the sensor. Peroxidases and oxidoreductases have received a great deal of attention within the biosensor scientific community because they are the most stable enzymes for catalyzing oxide reduction reaction.54 Listeria monocytogenes,55 Escherichia coli56, and campylobacter jejuni57 pathogenic bacteria detection was reported by enzymes modified electrochemical biosensor. Even though enzyme application takes up a lot of space on the catalytic biosensor, the enzyme quickly loses its ability to function after two to four weeks after application.58
2.1.2 Cells. A wide range of applications, including the detection of diverse analytes from the environment, food, clinical settings, and other sources, can be fulfilled by creating biosensors through live cell-based assays.59 It has previously been documented that several cells, including bacteria, yeast, and higher eukaryotic cells like vertebrata or mammals, were used as bioreceptors within bio-electrochemical sensors. Moreover, earlier research revealed that mammalian tissue slices or cells might be successfully employed as biorecognition components in studies utilizing biosensors.36 The use of living cells as identification elements can result in low detection limits and functional stability due to their great sensitivity. Banerjee et al.,60 investigated a cell-based sensing mechanism based on collagen-encapsulated mammalian cells for rapid detection of pathogenic bacteria and toxins. Zhao et al.,61 discussed the synthesis of silica nanocomposite doped with liposome to be exploited as an artificial cell-based biosensor. The synthesized artificial cell can bind and detect hemolysin and Listeriolysin O, secreted by pathogen L. monocytogenes bacterium. One of the disadvantages of the proposed cell-based sensor is that the presence of undesired enzymes can cause a complicated response and result in a lack of selectivity.62
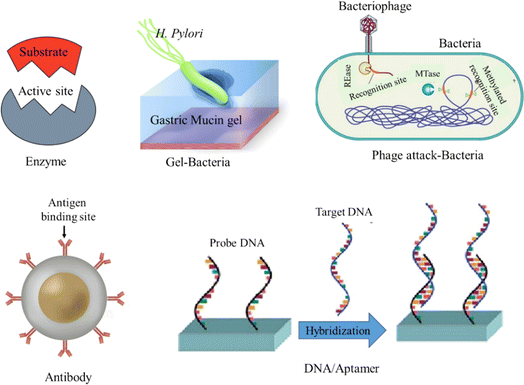 |
| | Fig. 3 Bioreceptor–analyte interaction for electrochemical biosensor detection. Figure was created by authors based on ideas by ref. 51–53. | |
2.1.3 Antibodies. Antibodies (Ab) or immunoglobulins (Ig) are large Y-shaped glycoproteins that have a high specificity for recognizing antigens, which are substances that might trigger an immune response.63 Each antigen contains discrete areas called epitopes to which the antibody can bind. While monoclonal antibodies are selective for only one epitope within an antigen, polyclonal antibodies can bind many epitopes inside an antigen. To analyze surface plasmon resonance (SPR) immunosensors using optical transduction, biosensor chips were coated with various secreted antibodies, including monoclonal, polyclonal, and recombinant.64 To facilitate the conjugation of the transducer and the antibody, the transducer surface had to be modified by adding functional groups such as carboxyl, amino, aldehyde, or sulfhydryl groups using a polymer or monomer. The antigen-specific antibody binding acts as a lock and key mechanism, and it is very simple in SPR, thus making it more accurate and faster than traditional assay.65 The results, however, were highly variable because of the limits of the antibody-decorated immunosensors with irreversibility, binding affinity, temperature, and the pH level of the reaction.66
2.1.4 Bacteriophages. Viruses known as bacteriophages, which range in size from 20 to 200 nm, are extracted from host cell lysis and can be employed to decorate sensitive and specific electrochemical biosensors. While most phages have a net negative charge due to their positively charged tail fibers and negatively charged capsid (head), researchers have suggested several methods to immobilize phages on electrochemical biosensor platforms, including covalent and ionic techniques. The dipole attraction force mechanism was preserved in ionic bonding to immobilize the phage on the electrode surface.65 Furthermore, the presence of a particular receptor on their tail protein makes the lysis bacteriophage more selective to the host strain. In addition, bacteriophages have distinct morphologies, binding affinities, and variable temperature, pH, and ionic strength compared to immunosensors, which makes them uniquely distinctive.67 Phage function as a biorecognition component for pathogen exposure has been declared by researchers for a variety of pathogens, including Escherichia coli, Staphylococcus aureus, and Bacillus anthracis spores.68
2.1.5 Aptamers and nucleic acids. Small single-stranded RNA or DNA oligonucleotides, known as aptamers, typically have a length of 20 to 60 nucleotides and are highly selective and affinity-bound to target molecules. Presently produced aptamers have a broad spectrum of binding sites, ranging from simple inorganic molecules to whole cells and huge protein complexes. The aptamers are the nucleotide counterparts of antibodies; yet the synthesis of aptamers is significantly less costly and complex than that of antibodies.69,70 Furthermore, aptamers are not poisonous or immunogenic.69 These nucleic acid segments were bioreceptor components with broad use for transducer manufacturing. Phosphate and sugar (deoxyribose) groups alternate to form nucleic acid strands. One of the nitrogen bases; adenine (A), cytosine (C), guanine (G), or thymine (T) is held by each sugar group. As in the cases of adenine:thymine (A:T) and cytosine:guanine (C:G), one strand nitrogen base is joined to another strand by hydrogen bonds.71 Signal amplification strategies were also studied by attaching reporter probe with suitable condition. The transducer surface is properly functionalized by a monomer, polymer, or composite to immobilize the nucleic acid receptor.11 The ssDNA immobilized biosensor has a broad range of applications in both experimental and real sample medium. Literature from various studies reporting on nucleic acid-based electrochemical biosensors was discussed in this study.
3. Pathogenic bacteria respective diseases
PBs cause a great deal of human disorders by interfering with normal bodily functions. The intestinal mucosa serves as the body's largest interface for the colonization of both pathogenic and nonpathogenic bacterial species in most cases. Some of which combat the pathogenesis infections by fighting against infectious bacteria.72 Beneficial bacteria may prevent the cluster of pathogenic bacteria by forming colonization and invasion. Nevertheless, pathogenesis begins with the transmission of the bacterial infectious agent to the host. Then bacteria remain at the site for colonization in the host system. The sickness is finally caused by the host immune system. The most common pathogenic bacteria and the resulting infectious diseases in humans are listed in Table 2. Hazardous infections can be caused by a variety of bacteria, viruses, and fungi, particularly in environments that support their growth and survival. Pneumonia and diarrhea together are the third cause of death among children under 5 years of age, accounting for 2 million deaths per year.100 Food and waterborne PB can cause acute or chronic infections in most individuals. A list of 26 human microbial species studied in electrochemical biosensor detection review on 13 PBs are summarized in Table 2. Here, the focus is on literature studies and reporting on electrode fabrication material, testing performance parameters, and concluding remarks on discussing the PBs.
Table 2 The most common pathogenic bacteria along with the resulting infectious diseases in humans
| Pathogenic bacteria |
Human diseases |
Pathogenic bacteria |
Human diseases |
| (1) Salmonella spp. |
Diarrhea, fever, abdominal cramps73 |
(14) Pseudomonas aeruginosa |
Urinary tract, respiratory, wound infections74 |
| (2) Escherichia spp. |
Urinary tract infection, kidney failure75 |
(15) Klebsiella pneumoniae |
Pneumonia, urinary tract infection, bloodstream infections76,77 |
| (3) Vibrio spp. |
Cholera, diarrhea, and dehydration78 |
(16) Listeria monocytogenes |
Fever, muscle aches, meningitis79 |
| (4) Shigella spp. |
Dysentery, diarrhea, fever, stomach cramps80 |
(17) Yersinia pestis |
Bubonic, septicemic, and pneumonic plague81 |
| (5) Bacillus spp. |
Anthrax (skin, lungs, gastrointestinal tract)82 |
(18) Enterococcus faecalis |
Urinary tract infections, endocarditis (inflammation of heart lining)83 |
| (6) Clostridium spp. |
Tetanus, botulism, paralysis, colitis84 |
(19) Francisella tularensis |
Tularensis illness85,86 |
| (7) Neisseria spp. |
Gonorrhea, meningitis87 |
(20) Haemophilus influenzae |
Respiratory tract infection (pneumonia, sinusitis, and ear infections)88 |
| (8) Mycobacterium spp. |
Tuberculosis, leprosy, skin, peripheral nerves, and mucosa of respiratory tract89 |
(21) Corynebacterium diphtheriae |
Diphtheria (pharyngitis, fever, swelling of the neck)90 |
| (9) Staphylococcus spp. |
Skin, respiratory, bloodstream, bone and joint, heart infection91 |
(22) Bordetella parapertussis |
Pertussis, whooping cough92 |
| (10) Streptococcus spp. |
Pneumonia, skin infection93 |
(23) Borrelia burgdorferi |
Lyme diseases (joint, heart, nervous infections)94 |
| (11) Brucella spp. |
Brucellosis, zoonotic diseases (fever, and joint pain, infections), flu-like infections91 |
(24) Chlamydia trachomatis |
Lung carcinoma, trachoma, detrimental effects on female reproductive health95 |
| (12) Legionella spp. |
Legionnaires (pneumonia, Pontiac fever)96 |
(25) Campylobacter spp. |
Campylobacteriosis (diarrhea, cramping, abdominal pain, and fever)97 |
| (13) Helicobacter pylori |
Gastritis, peptic ulcers, stomach cancer98 |
(26) Serratia marcescens |
Urinary tract infections, ocular lens infections99 |
4. Electrochemical biosensors for pathogenic bacteria detection
The electrode is a critical component of an electrochemical sensor that serves as a substrate for attaching the bioreceptor and target analyte while also converting the biological signal into an analogue electric signal. The performance of electrochemical sensors is affected differentially by various electrode materials. When developing high-performance electrochemical sensing platforms that use different analytical techniques to identify target molecules, electrode material is a crucial component in determining the sensitivity of electrochemical sensors.101 That is the sensitivity of electrochemical sensors can be maximized for a variety of applications by carefully choosing the electrode materials.
4.1. Salmonella spp.
One of the main culprits behind bacterial foodborne illnesses worldwide is Salmonella spp., which is a Gram-negative bacterium. Many studies reported on the detection of Salmonilla typhi by ssDNA modified electrochemical biosensor. Bacchu et al. fabricated a highly sensitive gold nanoparticles (AuNPs) and polycysteine (P-Cys) modified screen-printed electrode (SPE), SPE/P-Cys@AuNPs, electrochemical biosensor for the detection of Salmonella typhi in anthraquinone-2-sulfonic acid monohydrate sodium salt (AQMS) and characterized by Differential pulse voltammetry (DPV).102 Target DNA was detected by the developed electrochemical biosensor with a detection range of 1 × 10−6 to 1 × 10−22 mol L−1 and a limit of detection (LOD) of 6.8 × 10−25 mol L−1. The modified biosensor has excellent discrimination ability and is reusable up to 6 to 7 times. The developed sensor displayed an excellent detection performance in real samples such as blood, poultry feces, egg, and milk. The detection summary is represented in Fig. 4. Recently all reported electrochemical biosensors research work for Salmonilla typhi is shown in Table 3. Nanoporous gold decorated glassy carbon electrode (GCE) aptasensor was also utilized for Salmonella typhi detection.
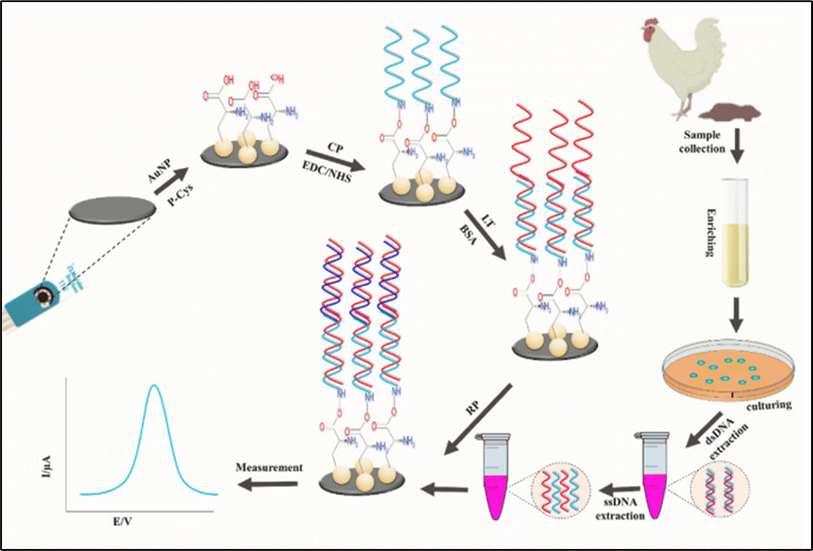 |
| | Fig. 4 Schematic illustration for the fabrication of DNA-biosensor and the detection of Salmonilla Typhi from real samples. The figure was adapted from ref. 102 and reproduced with permission, copyright @ Elsevier. | |
Table 3 Examples of the most recent reported electrochemical biosensors for the detection of pathogenic Salmonella spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| ITO: indium tin oxide, CHI: chitosan, GO: graphene oxide, MPTS: organosilane 3-mercaptopropyltrimethoxy silane, GNPs: gold nanoparticles, CFU: colony forming unit, GNAs: gold nanoaggregates, Glu: glutaraldehydes, NPG: nanoporous gold; AQMS: anthraquinone-2-sulfonic acid monohydrate sodium salt, N/A: not/absent. |
| ITO/GO-CHI |
NH2-ssDNA |
Salmonella typhi |
DNA biosensor |
DPV |
5.0 × 10−8 to 1.0 × 10−14 mol L−1 |
1.0 × 10−14 mol L−1 |
PBS |
N/A |
103 |
| SPE/MPTS-AuNPs |
SH-ssDNA |
Salmonella typhi |
DNA biosensor |
DPV |
5.0 × 10−8 to 1.0 × 10−10 mol L−1 |
5.0 × 10−11 mol L−1 |
PBS |
Serum |
104 |
| GCE/GO/GNPs |
SH-ssDNA |
Salmonella |
DNA biosensor |
EIS |
2.4–2.4 × 103 CFU mL−1 |
3.0 CFU mL−1 |
[Fe(CN)6]3−/4− |
Pork |
105 |
| ITO/GNAs |
NH2-ssDNA |
Salmonella typhi |
DNA biosensor |
EIS |
2.4 × 10−14 to 4.0 × 10−18 mol L−1 |
4.0 × 10−18 mol L−1 |
[Fe(CN)6]3−/4− |
Urine and blood |
106 |
| Au/Cys/Glu |
HO-ssDNA |
Salmonella typhi |
DNA biosensor |
EIS |
N/A |
1.91 mg mL−1 |
[Fe(CN)6]3−/4− |
N/A |
107 |
| GCE/Au/NPG |
SH-ssDNA |
Salmonella typhi |
Aptasensor |
EIS |
6.5 × 102 to 6.5 × 108 CFU mL−1 |
1 CFU mL−1 |
PBS |
Egg |
108 |
| SPE/P-Cys@AuNPs |
HN2-ssDNA |
Salmonella typhi |
DNA biosensor |
DPV |
1 × 10−6 to 1 × 10−10; 1 × 10−10 to 1 × 10−22 mol L−1 |
6.8 × 10−23 mol L−1 |
AQMS |
Blood, poultry feces, egg, milk |
102 |
Electrochemical impedance spectroscopy (EIS) was applied to characterize and evaluate the results of the electrochemical detection.108 It was concluded that the developed sensor performed well in real samples with the ability to detect and distinguish between live and dead Salmonella bacterial cells in egg samples. The main distinguishing mechanism between live and dead bacteria was based on the value of the charge transfer resistance (Rct); it increases after binding with live bacteria and significantly decreases in the presence of dead bacterial cells. Additionally, in another research gold nanoaggregates modified surface was prepared to covalently bond with 5′NH2 modified ssDNA probes.106 The target DNA was hybridized at 35 °C for 60 seconds. The binding efficiency was analyzed by standard EIS techniques in [5 mM K3Fe(CN)6] in 0.1 mol L−1 KCl solution. For the detection of Salmonilla typhi, DNA probe modified electrode displayed the most superior performance among the reported electrochemical biosensors.
4.2. Escherichia spp.
Among the family of Escherichia species, E. coli bacterial cells are frequently found in the digestive tracts of healthy individuals; yet a small number of clones are accountable for severe diarrhea and infections beyond the intestines. Escherichia spp. was detected by biosensors modified with different bioreceptors such as antibody, DNA and bacteriophage. M. Barreiros et. al. reported on the detection of E. coli O157:H7 using Anti-E. Coli onto an epoxy silane modified indium tin oxide (ITO) electrode (Barreiross dos Santos et al., 2015).109 In this study, EIS technique was applied with a linear range of 10 to 106 CFU mL−1 and limit of detection of 1 CFU mL−1. The developed biosensor was found to be highly selective in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 Salmonella typhimurium/E. coli O157:H7 medium. Many of these impedimetric immunosensors displayed high sensitivity and selectivity. Other antibody modified immunosensors were developed and reported for the detection of Escherichia spp., and they are listed in Table 4. Furthermore, Xu et.al. reported on NH2-ssDNA modified ssDNA/GO/CS/GCE electrode for the electrochemical detection of E. coli O157:H7 using EIS and DPV. In this study, the developed sensor showed an excellent performance on the detection of E. coli O157:H7.118 A T4 phagosensor was decorated on micro-electrochemical sensor with a 3-mercaptoreopionic acid modified gold electrode.122 Utilizing DPV, the modified sensor showed a limit of detection of 14 ± 5 CFU mL−1 and a wide dynamic range of 1.9 × 101 to 1.9 × 108 CFU mL−1. This phagosensor was successfully able to selectively distinguish between viable and dead bacteria cells. The schematic representation of the AuE/Cys/PDCIT/T4 phage electrode fabrication aided with applied chemistry is shown in Fig. 5.
500 Salmonella typhimurium/E. coli O157:H7 medium. Many of these impedimetric immunosensors displayed high sensitivity and selectivity. Other antibody modified immunosensors were developed and reported for the detection of Escherichia spp., and they are listed in Table 4. Furthermore, Xu et.al. reported on NH2-ssDNA modified ssDNA/GO/CS/GCE electrode for the electrochemical detection of E. coli O157:H7 using EIS and DPV. In this study, the developed sensor showed an excellent performance on the detection of E. coli O157:H7.118 A T4 phagosensor was decorated on micro-electrochemical sensor with a 3-mercaptoreopionic acid modified gold electrode.122 Utilizing DPV, the modified sensor showed a limit of detection of 14 ± 5 CFU mL−1 and a wide dynamic range of 1.9 × 101 to 1.9 × 108 CFU mL−1. This phagosensor was successfully able to selectively distinguish between viable and dead bacteria cells. The schematic representation of the AuE/Cys/PDCIT/T4 phage electrode fabrication aided with applied chemistry is shown in Fig. 5.
Table 4 Examples of reported electrochemical biosensors for the detection of pathogenic Escherichia spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| EDC: 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; BSA: bovine serum albumin, NHS: N-hydroxysuccinimide, MWCNT: multiwall carbon nanotube, PGE: pencil graphite electrode; PPy: polypyrrole; GOx:glucose oxidase; PDA: polydopamine; SPCE: screen printed carbon electrode, CDs: carbon dots, PANI: polyaniline, MCE: mercaptoethanol, GNS: graphene nano sheets; IDE: Interdigitated electrode; APTES: (3-Aminopropyl) triethoxysilane; CS: chitosan, PCR: olymerase chain reaction, MCH: 6-mercapto-1-hexanol, DNR: daunomycin; PEI: polyethylenenimine; CNT: carbon nanotube; CNFs: carbon nanofibers; AuE: gold electrode; EIS: electrochemical impedance spectroscopy; DPV: differential pulse voltammetry; 3-MPA: 3-mercaptoreopionic acid; EI: electrostatic interaction; CL: covalent linkage; Cys: cysteamine; PDCIT: 1,4-dithiocyanate; GOx: glucose oxidase; HRP: horseradish peroxide; AuNPs: gold nanoparticles; Thi: thionine; AMP: Amperometric; AP: alkaline phosphatase; β-gal: β-galactosidase; PAPG: 4-aminophenyl-β-galactopyranoside. |
| AuE/Ag@BSA/EDC |
Anti-E. coli |
E. coli O157:H7 |
Immunosensor |
DPV |
3.0 × 102 to 3.0 × 108 CFU mL−1 |
N/A |
[Fe(CN)6]3−/4− |
N/A |
110 |
| PGE/PPy/AuNP/MWCNT@Chi |
Anti-E. coli |
E. coli O157:H7 |
Immunosensor |
Amperometry |
3 × 101 to 3 × 107 CFU mL−1 |
30 CFU mL−1 |
[Fe(CN)6]3−/4− |
Food |
111 |
| SPE/PDA@GOx-MBs/AuNPs/GOx |
Anti-E. coli |
E. coli O157:H7 |
Immunosensor |
Amperometry |
102 to 106 CFU mL−1 |
102 CFU mL−1 |
Glucose |
Beef |
112 |
| ITO/Epoxysilane |
Anti-E. coli |
E. coli O157:H7 |
Immunosensor |
EIS |
10 to 106 CFU mL−1 |
1 CFU mL−1 |
[Fe(CN)6]3−/4− |
N/A |
113 |
| SPCE/CDs/ZnO nanorod/PANI |
pDNA |
E. coli O157:H7 |
Aptasensor |
DPV |
1.3 × 10−18 to 10 × 10−12 mol L−1 |
1.3 × 10−18 mol L−1 |
[Fe(CN)6]3−/4− |
Water |
113 |
| AuE/MCE/pDNA/GNSs |
pDNA |
E. coli O157:H7 |
Genosensor |
SWVs |
7.3 to 1.0 × 10−17 μmol L−1 |
1.0 × 10−17 μmol L−1 |
(Fe(CN)63−/4−–KCl) |
Water |
114 |
| GCE/CDs-Fe3O4 |
DNA |
E. coli O157:H7 |
DNA biosensor |
DPV |
10 to 108 CFU mL−1 |
6.88 CFU mL−1 |
[Fe(CN)6]3−/4− |
Milk, water |
115 |
| Au IDE/APTES |
COOH-ssDNA |
E. coli O157:H7 |
DNA biosensor |
Amperometry |
1 × 10−16 to 10 × 10−6 mol L−1 |
0.8 fmol L−1 |
[Fe(CN)6]3−/4− |
N/A |
116 |
| Co-aliginic acid |
NH2-ssDNA |
E. coli |
DNA biosensor |
DPV |
1.0 × 102 to 2.0 × 103 cells per mL |
50 cells mL−1 |
PBS |
Water |
117 |
| GCE/GO/CS/EDC/NHS |
NH2-ssDNA |
E. coli O157:H7 |
DNA biosensor |
EIS |
1.0 × 10−14 to 1.0 × 10−8 mol L−1 |
3.584 × 10−15 mol L−1 |
[Fe(CN)6]4−/3− |
N/A |
118 |
| PGE/MWCNT/ssDNA |
NH2-ssDNA |
E. coli |
DNA biosensor |
DPV |
NA |
17 nmol L−1 |
Guanine |
PCR sample |
119 |
| AuE/ECA I or ECA II or ECA–MCH |
SH-ECA |
E. coli OMPs |
Aptasensor |
FIS |
1 × 10−7 to 2 × 10−6 mol L−1 |
NA |
[Fe(CN)6]3−/4− |
Water |
120 |
| GCE/PEI-CNT |
T2-phage |
Escherichia coli B |
Phagosensor |
EIS |
103 to 107 CFU mL−1 |
102 CFU mL−1 |
[Fe(CN)6]3−/4− |
N/A |
65 |
| SPE/CNFs |
T4-phage |
Escherichia coli |
Phagosensor |
EIS |
102 to 106 CFU mL−1 |
36 CFU mL−1 |
[Fe(CN)6]3−/4− |
Apple juice |
121 |
| AuE/3-MPA |
T4-phage |
Escherichia coli B |
Phagosensor |
DPV |
19–1.9 × 109 CFU mL−1 |
14 ± 5 CFU mL−1 |
[Fe(CN)6]3−/4− |
N/A |
122 |
| SPE/EDC |
T4-phage |
E. coli K12 cells |
Phagosensor |
EIS |
102 to 108 CFU mL−1 |
103 CFU mL−1 |
[Fe(CN)6]3−/4− |
Milk |
123 |
| AuE/Cys/PDCIT |
T4-phage |
Escherichia coli |
Phagosensor |
EIS |
102 to 107 CFU mL−1 |
8.0 × 102 CFU mL−1 |
[Fe(CN)6]3−/4− |
N/A |
124 |
| GOx/HRP-Cu3(PO4)/AuNPs/Thi |
T4-phage |
Escherichia coli |
Phagosensor |
EIS |
1.5 × 101–1.5 × 108 CFU mL−1 |
1 CFU mL−1 |
PBS + [Fe(CN)6]3−/4− |
Urine |
67 |
| PtE/β-gal |
T7 |
Escherichia coli |
Phagosensor |
DPV |
N/A |
102 CFU mL−1 |
PAPG |
Water |
125 |
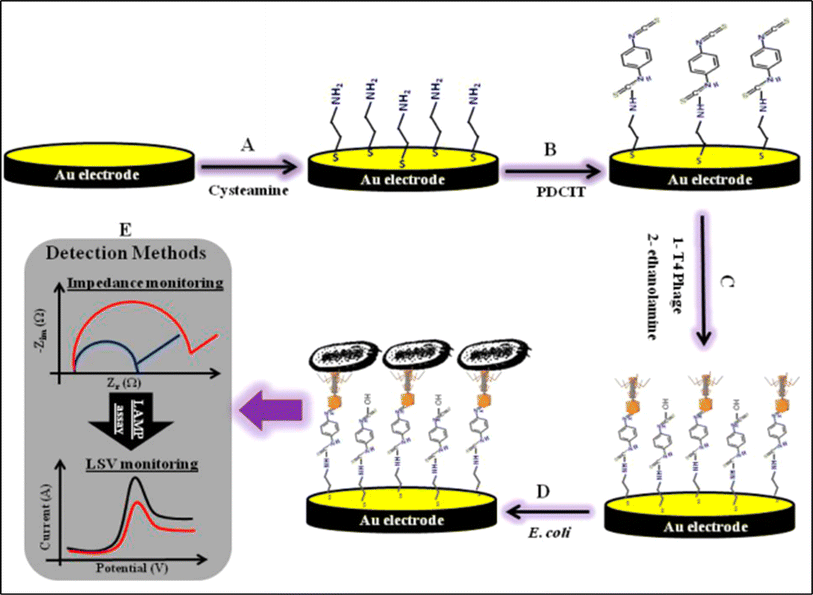 |
| | Fig. 5 Schematic illustration showing the fabrication of AuE/Cys/PDCIT/T4 phage electrode. The figure was adapted from ref. 122 reproduced with permission, copyright @ Elsevier. | |
Furthermore, T4 bacteriophage modified electrochemical biosensor was developed for rapid detection of live pathogenic bacteria in urine medium with GOx/HRP-Cu3(PO4)/AuNPs/Thi composite modified electrode. This modified electrode showed a linear range of 15–1.5 × 108 CFU mL−1 with limit of detection of 1 CFU mL−1 in clinical settings.67 It was concluded that bacteriophage based electrochemical biosensor detection strategy is simple, quick, highly selective, and sensitive.
4.3. Vibrio spp.
Most diseases in humans connected to the natural microbiota of aquatic environments and seafood are caused by Gram-negative bacteria called Vibrio spp. Vibrio cholerae are among the most common species of Vibrio spp. Ali et al. designed and fabricated gold nanotube and 3-aminopropyltriethoxysilane/N-hydroxysuccinimide modified glassy carbon electrode (GCE/AuNC/APTES/NHS) for DNA modified biosensor for the detection of pathogenic Vibrio cholera in real sample.126 In this work, stepwise GCE modification was performed with analytical confirmation and characterization with SEM and FTIR. The developed biosensor had a dynamic linear range of 10−8 to 10−14 mol L−1 to detect the target DNA. The established electrochemical biosensor was highly sensitive as shown in results summarized in Table 5. Poly (propylene imine) dendrimer (PPI) and gold nanoparticles (AuNP) composite modified electrode was chosen as a platform for antibody immobilization in the development of immunosensor.133 Anti-cholera toxin antibody was attached to the developed PPI-AuNP composite for final electrode fabrication. The fabricated electrode was able to detect vibrio cholera toxin with a dynamic range of 10−7 g mL−1 to 10−12 g mL−1 with two different limits of detection 7.2 × 10−13 and 4.2 × 10−13 g mL−1. It is worth mentioning that the LOD is the lowest reported limit of detection for the detection of vibrio cholera. In another research work, a label-free immunosensor was developed by antibody binding with amino eater composite on cerium oxide nanowire. The graphical representation of this immunosensor is shown in Fig. 6. In immunosensor assay, label free immunosensor was developed for the detection of Vibrio cholerae O. The immobilization of anti-V. cholerae O1 onto CeO2 nanowire-deposited sensor was performed via an amino ester. The electrochemical response of an immunosensor modified electrode with an immobilized Vibrio cholerae O was measured in [Fe(CN)6]3−/4− using standard EIS method. In the author's opinion, immunity receptor modified electrochemical biosensor detection mechanism have occupied great attention in research field except DNA probe modified detection strategy.
Table 5 Examples of reported electrochemical biosensors for the detection of pathogenic Vibrio spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| npcRNA: non-protein coding RNA; PSA: polystyrene-co-acrylic acid; ncDNA: non complementary DNA; ME: microelectrode; PPI: poly(propylene imine) dendrimer, PANnf: polyacrylonitrile nanofiber, Anti-CT: anti cholera toxin, PPy-NTA: polypyrrole-nitrilotriacetic acid. |
| GCE/AuNC/APTES/NHS |
HN2-ssDNA |
Vibrio cholerae |
DNA biosensor |
DPV |
10−8 to 10−14 10−14 to 10−27 mol L−1 |
7.41 × 10−30 mol L−1 |
AQMS |
Poultry feces |
126 |
| ITO/ZrO2 |
ssDNA |
Vibrio cholerae |
DNA biosensor |
DPV |
10−10 to 1017 mol L−1 |
10−17 mol L−1 |
Methylene blue |
N/A |
127 |
| AuE |
SH-ssDNA |
Vibrio cholerae |
DNA biosensor |
CV |
500–100 ng μL−1 |
100 ng μL−1 |
Methylene blue |
N/A |
128 |
| Carbon electrode |
npcRNA |
Vibrio cholera |
DNA biosensor |
SWV |
NA |
6 × 10−17 mol L−1 |
Acetate buffer |
PCR sample |
129 |
| SPE/AuNPs/PSA |
ncDNA |
Vibrio cholera |
DNA biosensor |
DPV |
10−8 to 10−17 mol L−1 |
10−21 mol L−1 |
AQMS |
N/A |
130 |
| ME/CeO2@APTES/NHS/EDC |
Anti-V. cholerae O1 |
Vibrio cholerae O1 |
Immunosensor |
EIS |
1.0 × 102 to 1.0 × 104 CFU mL−1 |
1.0 × 102 CFU mL−1 |
[Fe(CN)6]3−/4− |
N/A |
131 |
| GCE/CNF |
Anti-V. cholera toxin |
Vibrio cholerae toxin |
Immunosensor |
EIS |
1.3 × 10−13 to 4.56 × 10−5 g mL−1 |
1.2 × 10−13 g mL−1 |
[Fe(CN)6]3−/4− |
Water |
132 |
| GCE/AuNPs-PPI |
Anti-cholera toxin |
Cholera Toxin |
Immunosensor |
SWV, EIS |
10−7 to 10−12 g mL−1 |
7.2 × 10−13 (SWV), 4.2 × 10−13 (EIS) g mL−1 |
[Fe(CN)6]3−/4− |
N/A |
133 |
| ITO/ZnO |
Anti-cholera toxin B |
Vibrio cholerae toxin B |
Immunosensor |
DPV |
12.5–500 g mL−1 |
0.16 g mL−1 |
[Fe(CN)6]3−/4− |
Selective medium |
134 |
| ITO/PANnf |
Anti-cholera toxin B |
Vibrio cholerae toxin B |
Immunosensor |
DPV |
6.25 × 10−9 to 5.00 × 10−7 g mL−1 |
22 × 10−11 g mL−1 |
[Fe(CN)6]3−/4− |
Selective medium |
135 |
| GCE/CNT/PPy-NTA |
Anti-CT |
Anti-cholera toxin antibody |
Immunosensor |
EIS |
10−13 to 10−5 g mL−1 |
∼10−13 g mL−1 |
[Fe(CN)6]3−/4− |
N/A |
136 |
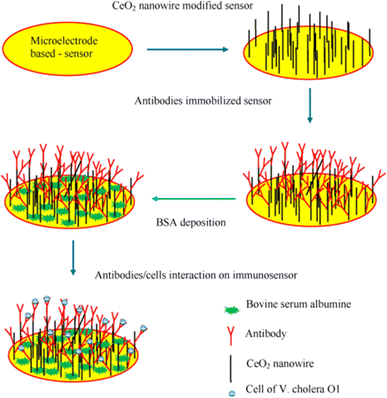 |
| | Fig. 6 Schematic illustration of the immunosensor fabrication for the detection of V. cholera O1 detection. The figure was adapted from ref. 133 and reproduced with permission, copyright @ Elsevier. | |
4.4. Shigella spp. and Bacillus spp.
Shigella spp. are Gram negative bacteria that are most known for their intestinal infection (shigellosis). Ali et al. demonstrated a label free electrochemical detection of Shigella flexneri in real food samples by applying a series of chemical modification to establish a highly efficient immobilization of ssDNA capture probe.137 For this purpose, poly-melamine, poly-glutamic acid, and disuccinimidyl suberate functionalized indium tin oxide electrode was prepared to immobilize NH2-ssDNA. Then, using one or two base pair mismatches, a reporter probe, a modified electrode was bound with the linear target. Lastly, the DPV response was measured in anthraquinone-2-sulfonic acid monohydrate sodium salt (AQMS) acting as a redox probe mediator. They concluded that the proposed biosensor could serve as a model methodology for the detection of other pathogens. The ITO/P-Mel/PGA/DSS electrode preparation of the above-mentioned sensor is illustrated in Fig. 7. Shigella dysenteriae was detected via thiolated aptamer modified electrodeposited GCE.138 The assay has a linear dynamic range that extends from 101 to 106 CFU mL−1 and a limit of detection of 10° CFU mL−1. The amino functionalized ssDNA, and aptamer-based sensor have extensive applications for the detection of Shigella spp. compared with other reported research work.
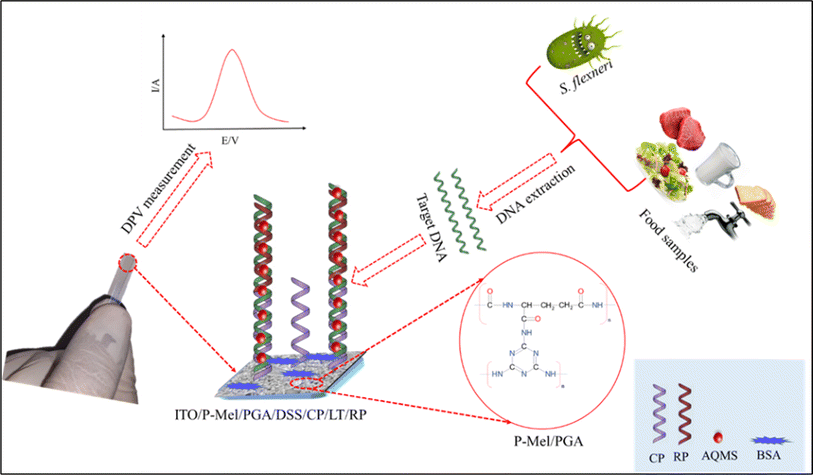 |
| | Fig. 7 Schematic diagram illustrating the fabrication process of the ITO/P-Mel/PGA/DSS modified electrode for the detection Shigella flexneri. The figure was adapted from ref. 137 and reproduced with permission, copyright @ Elsevier. | |
Bacillus subtilis are among the bacterial family of Bacillus spp., Gram positive bacterial species famous for causing foodborne illness. It was previously reported that SH-ssDNA modified gold electrode was used to detect Bacillus subtilis bacteria utilizing conventional DPV technique.139 The developed sensor successfully detected the target pathogenic species (Bacillus subtilis) with a linear range of 0.1–20 fmol L−1 and a limit of detection down to 0.08 fmol L−1. They concluded that the results were in strong agreement with that of quantitative polymerase chain reaction (QPCR) detecting system. Table 6 summaries the reported electrochemical biosensors for the detection of Shigella spp. and Bacillus spp. In another detection strategy, protein-based Bacillus licheniformis detection strategy was developed by H. Wu et al.145 In this assay, the biosensor was constructed using a D-amine acid containing substrate peptide via self-assembly of cysteine residual at the C-terminal. A biotin modifier was labelled at the N-terminal of the substrate peptide. This enabled the sensitive electrochemical detection of the intact substrate peptide using a streptavidin-conjugated alkaline phosphatase, which catalyzes the conversion of electrochemically inactive 1-naphthyl phosphate into electrochemically active phenol. Under optimized conditions, the protease can be determined in concentration range from 0.5 to 100 mg mL−1 with a detection limit to 0.16 mg mL−1. From Table 6 and the performance parameter analysis, it is evident that DNA probe and peptide based electrochemical biosensor reported the best result for the detection of Bacillus spp.
Table 6 Examples of reported electrochemical biosensors for the detection of pathogenic Shigella spp. and Bacillus spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| P-Mel: poly-melamine, PGA: poly-glutamic acid, DSS: disuccinimidyl suberate, HOOCA: 4-amino benzoic acid tetrafluoroborate, SA: sodium aliginate, GE: gold electrode; GNPs: gold nanoparticles, MCH: mecatohexanol; GNPs: gold nanoparticles; PBSE: pyrenebutanoic acid succinimidyl ester. |
| Shigella spp. |
| ITO/P-Mel/PGA/DSS |
NH2-ssDNA |
Shigella flexneri |
DNA biosensor |
DPV |
1 × 10−21 to 1 × 10−6 CFU mL−1 |
10 cells per ml |
AQMS |
Food |
137 |
| 80 to 8 × 1010 cells per mL |
| GCE/AuNPs |
Aptamer |
Shigella dysenteriae |
Aptasensor |
EIS |
101 to 106 CFU mL−1 |
1.0 CFU mL−1 |
[Fe(CN)6]3−/4− |
Mik, water |
138 |
| SPE/HOOCA |
Aminated-DNA aptamer |
S. typhimurium |
Aptasensor |
EIS |
101 to 108 CFU mL−1 |
6 CFU mL−1 |
[Fe(CN)6]3−/4− |
Apple juice |
140 |
| SPE/MWCNT/SA |
Anti-S. flexneri |
Shigella flexneri |
Immunosensor |
CV |
104 to 1010 CFU mL−1 |
3.1 × 103 CFU mL−1 |
HAc–NaAc buffer |
N/A |
141 |
| SPE/MWCNT/CHI |
Anti-S. flexneri |
Shigella flexneri |
Immunosensor |
CV |
104 to 1010 CFU mL−1 |
2.3 × 103 CFU mL−1 |
HAc–NaAc buffer |
N/A |
142 |
| Carbon electrode |
npcRNA |
Shigella flexneri |
DNA biosensor |
SWV |
N/A |
6 × 10−17 mol L−1 |
Acetate buffer |
PCR product |
129 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| Bacillus spp. |
| GE |
SH-ssDNA |
Bacillus subtilis |
DNA biosensor |
DPV |
0.1 fmol L−1 to 20 fmol L−1 |
0.08 fmol L−1 |
N.BstNBI and NEBuffer 3 |
PCR product |
139 |
| PGE/GNPs-SH |
SH-ssDNA |
Bacillus cereus |
DNA biosensor |
EIS |
0.02–10 nmol L−1 |
100 CFU mL−1 |
[Fe(CN)6]4−/3− |
Food |
143 |
| 0.01–10 μmol L−1 |
9.4 × 10−12 mol L−1 |
| AuE/SWCNTs/1-PBSE |
Anti-B. subtilis |
Bacillus subtilis |
Immunosensor |
Potentiostatic |
102 to 1010 CFU mL−1 |
102 CFU mL−1 |
PBS |
Dust |
144 |
| AuE/Peptide/MCH |
Peptide |
Bacillus licheniformis |
Peptide biosensor |
DPV |
0.5–100 μg mL−1 |
0.16 μg mL−1 |
PBS |
Serum, saliva |
145 |
| GCE/Chit/GNPs |
Anti-Bacillus cereus |
Bacillus cereus |
Immunosensor |
Amperometric |
5.0 × 101 to 5.0 × 104 CFU mL−1 |
10.0 CFU mL−1 |
MYP medium |
Milk |
146 |
| SPCE/immuno-c/sNP |
Anti-Bacillus cereus |
Bacillus cereus |
Immunosensor |
CV |
N/A |
40 CFU mL−1 |
HCl solution |
Food, water |
147 |
4.5. Clostridium spp. and Neisseria spp.
Clostridium spp. are Gram positive bacteria, cause bacterial infection diseases such as botulism and tetanus. Qian et al.,148 developed a nanocomposite modified ssDNA/CeO2/CHIT/GCE biosensor for the detection of Clostridium perfringens extracted from dairy products. With favorable selectivity, they were able to achieve a linear dynamic range of 1.0 × 10−14 to 1.0 × 10−7 mol L−1 and a limit of detection of 7.06 × 10−15 mol L−1. Neisseria spp. are Gram negative bacterial microorganisms that are incriminated for causing human diseases such as gonorrhea, leprosy, and Mycobacterium leprae. In another study, antitoxin B single domain antibody receptor was modified to decorate polyurethane (PU) nanospiked gold electrode-based label-free electrochemical immunosensor for Clostridium difficile toxin B detection.149 This electrochemical immunosensor can detect within a concentration range of 1–130 pg mL−1 and a limit of detection of 0.5 pg mL−1. Therefore, it can be concluded that immunosensor reported the lowest detection capability amongs different bioreceptor modified electrochemical biosensors.
Singh et al. developed DNA-biosensor for the detection of a sexually transmitted disease, gonorrhoeae, caused by Neisseria gonorrhoeae, pathogenic bacteria.150 In this study, thiolated capture probe was immobilized on gold electrode and 6-Mercapto-1-hexanol (MCH) was used to block nonspecific agents to facilitate oligos “stand”. A complete graphical representation of the electrode preparation and DPV analysis in methylene blue (MB) medium is shown in Fig. 8. The most updated reported research is summarized in Table 7. 5′ Aminated capture probes and super sandwiched detector probes were utilized for the detection of Neisseria gonorrhoeae. The capture probe modified surface hybridize with gonorrheal DNAs and after sandwich type detector probe amplified the detection signal. Further, the biosensing assay displayed a wide linear range of 100 aM to 100 nM (109 orders of magnitude) with an excellent sensitivity of 22.6 kΩ·(log[concentration])−1. This type of strategic detection occupies a prizeworthy position in the research community.
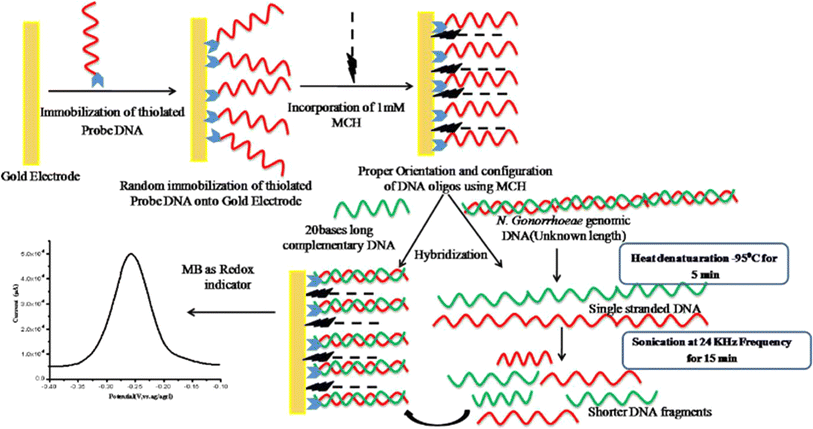 |
| | Fig. 8 Schematic diagram showing the fabrication process of the ssDNA-Au electrode for the detection of Neisseria gonorrhoeae. The figure was adapted from ref. 150 and reproduced with permission, copyright @ Elsevier. | |
Table 7 Examples of reported electrochemical biosensors for the detection of pathogenic Clostridium spp. and Neisseria spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| cMWCNT: carboxylated MWCNT; ZNF: zinc oxide nanoflower; PU: polyurethane, SA: streptavidin. |
| Clostridium spp. |
| GCE/CHIT/CeO2 |
ssDNA |
Clostridium perfringens |
DNA biosensor |
EIS |
10−14 to 10−7 mol L−1 |
7.06 × 10−15 mol L−1 |
[Fe(CN)6]3−/4− |
Milk |
148 |
| GCE/MWCNT/AuNP |
ssDNA |
Clostridium tetani |
DNA biosensor |
DPV |
NA |
1.0 × 10−16 mol L−1 |
MB |
N/A |
151 |
| AuE/PU |
Anti-C. Difficile toxin B |
Clostridium difficile |
Immunosensor |
DPV |
1–130 pg mL−1 |
0.5 pg mL−1 |
[Fe(CN)6]3−/4− |
Stool |
149 |
| SPE/AuNPs |
SA aptamer |
Clostridium perfringens |
Aptasensor |
DPV |
10−12 to 10−6 mol L−1 |
10−12 mol L−1 |
PBS |
N/A |
152 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| Neisseria spp. |
| C5@paper/cMWCNT |
NH2-ssDNA |
Neisseria gonorrhoeae |
DNA biosensor |
EIS |
(5 zmol–5 pmol) |
(45 aM) |
PBS |
Selective medium |
153 |
| SPAuE |
SH-ssDNA |
Neisseria gonorrhoeae |
DNA biosensor |
DPV |
1 × 10−15 to 1 × 10−22 mol L−1 |
NA |
PBS |
N/A |
154 |
| AuE |
SH-ssDNA |
Neisseria gonorrhoeae |
DNA biosensor |
DPV |
1.0 × 10−6–0.5 × 10−18 mol L−1 |
1.0 × 10−18 mol L−1 |
PBS + MB |
Selective medium |
150 |
| Glass subs/Cr/AuNPs |
SH-ssDNA |
Neisseria gonorrhoeae |
DNA biosensor |
CV |
10–60 ng μl−1 |
NA |
PBS + MB |
N/A |
155 |
| Si/Pt/ZNF |
SH-ssDNA |
Neisseria meningitidis |
DNA biosensor |
DPV, EIS |
5–240 ng μl−1 |
5 ng μl−1 |
[Fe(CN)6]3−/4− |
N/A |
156 |
4.6. Mycobacterium spp.
Mycobacterium spp. are Gram positive bacteria causing several infectious human diseases such as Tuberculosis, leprosy, Buruli ulcer and tuberculous mycobacterium. Among Mycobacterium spp., Mycobacterium tuberculosis-based DNA detection mechanism was reported based on gold nanotube array (AuNTsA) electrode platform with a 1.5 μm in length and 200 nm in diameter.157 For the biosensor fabrication, AuNTsA was vertically aliened on Au thick film during nanotubes synthesis process and a DNA probe was immobilized using Tris–EDTA for 12 hours. Next, different complementary DNA was immobilized on captured DNA probe in an incubated environment at 37 °C for 45 minutes. The schematic representation for the biosensor decoration is presented in Fig. 9. Recently, another research work was carried out for the early diagnosis of tuberculosis caused by ESAT-6 antibody.158 A Ni-rGO-PANI composite film modified electrode was fabricated for the immobilization of anti-ESAT-6 antibody. They reported a linear range of 1–100 ng mL−1 and a detection limit of 1.042 ng mL−1. Table 8 summarizes the previously reported electrochemical biosensors for the detection of Mycobacterium spp. A sensitive electrochemical DNA biosensor based on functionalized iron oxide with mercaptopropionic acid (MPA-Fe3O4) nanoparticle for the detection of Mycobacterium tuberculosis was reported.163 A DNA probe was immobilized on MPA-Fe3O4/NCC/CTAB electrode and sequentially bond with the target DNA and signal amplification, ruthenium bipyridyl Ru(bpy)32+. The sensing mechanism offered a wide detection range of 1.0 × 10−6 to 1.0 × 10−12 M and limit of detection of 7.96 × 10−13 M.163 Therefore, it can be concluded that with some limitations DNA biosensor, immunosensor, and aptasensor bioreceptor modified biosensor showed good result for the detection of Mycobacterium spp.
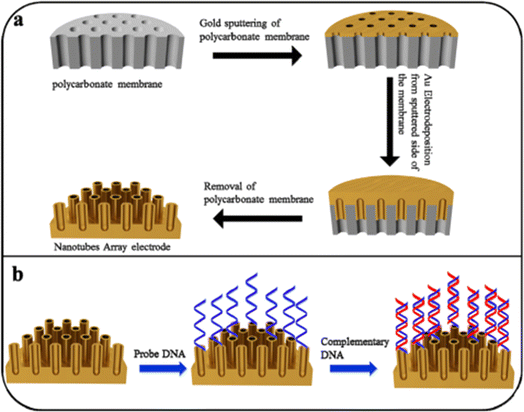 |
| | Fig. 9 Graphical representation of (a) AuNTsA synthesis; (b) decoration of DNA based biosensor on AuNTsA electrodes. The figure was adapted from ref. 157 and reproduced with permission, copyright @ Elsevier. | |
Table 8 Examples the previously reported electrochemical biosensors for the detection of pathogenic Mycobacterium spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| FTO: tin-doped fluorine, PDITC: phenylene diisothiocyanate, MPA: mercaptopropionic acid, NCC/CTAB: nanocellulose crystalline functionalized cetyltrimethyl ammonium bromide, SPGE: screen-printed gold electrode. |
| FTO/APTES/PDITC |
NH3-ssDNA |
Mycobacterium tuberculosis |
DNA biosensor |
CV |
1–100 nmol L−1 |
1.5 ng μL−1 |
[Fe(CN)6]3−/4− |
Sputum, urine |
159 |
| SPAuE |
SH-ssDNA |
Mycobacterium tuberculosis |
DNA biosensor |
DPV |
0.5–10 nmol L−1 |
1.90 nmol L−1 |
[Fe(CN)6]3−/4− |
Sputum |
160 |
| AuNTsA |
SH-ssDNA |
Mycobacterium tuberculosis |
DNA biosensor |
DPV |
0.01 ng μL−1 to 100 ng μL−1 |
0.05 ng μL−1 |
[Fe(CN)6]3−/4− |
N/A |
157 |
| GCE/AuNPs-rGO |
SH-ssDNA |
Mycobacterium tuberculosis |
DNA biosensor |
DPV |
1.0 × 10−15 to 1.0 × 10−9 mol L−1 |
NA |
PBS |
Selective medium |
161 |
| ITO/APTES/AuNPs |
SH-ssDNA |
Mycobacterium sp. genomic DNA. |
DNA biosensor |
DPV |
1.25 to 50 ng mL−1 |
1.25 ng mL−1 |
Tris–HCl |
Sputum |
162 |
| SPCE/MPA-Fe3O4/NCC/CTAB/EDC/NHS |
ssDNA |
Mycobacterium tuberculosis |
DNA biosensor |
DPV |
1.0 × 10−6 to 1.0 × 10−12 mol L−1 |
7.96 × 10−13 mol L−1 |
Ru(bpy)3 2+ |
Selective medium |
163 |
| SPCE/PANI/GP/EDC-NHS |
ssDNA |
Mycobacterium tuberculosis |
DNA biosensor |
DPV |
10−6 to 10−9 mol L−1 |
7.853 × 10−7 mol L−1 |
PBS + MB |
N/A |
164 |
| PANI-rGO-Ni-DSP |
Anti-ESAT-6 |
ESAT-6 |
Immunosensor |
CV |
1–100 ng mL−1 |
1.042 ng mL−1 |
PBS |
Blood |
158 |
| SPGE/GP/PANI/EDC-NHS |
Anti-CFP10-ESAT6 |
CFP10-ESAT6 |
Immunosensor |
DPV |
10–500 ng mL−1 |
1.5 ng mL−1 |
[Fe(CN)6]3−/4− |
Sputum |
165 |
| SPGE/GP/PANI |
Anti-CFP10 |
CFP10 antigen |
Immunosensor |
DPV |
20–100 ng mL−1 |
15 ng mL−1 |
[Fe(CN)6]3−/4− |
Sputum |
166 |
| SPGE/PANI/GP |
CapAptamer |
CFP10-ESAT6 |
Aptasensor |
DPV |
5–500 ng mL−1 |
1.5 ng mL−1 |
PBS |
Sputum |
167 |
4.7. Staphylococcus spp.
Gram-positive Staphylococcus spp. species can cause suppuration and other infectious illnesses in both people and animals. In 2020, Roushani et al.168 described the use of chicken IgY antibody as an immunosensing agent that was covalently bound to AuNPs as a biosensor based on AuNPs modified GCE for the detection of Staphylococcus aureus. The antibody was bonded with G-producing Staphylococcus and the interference from EIS study was reported for the detection of Staphylococcus aureus. This biosensor reported by this work displayed a wide linear dynamic range from 10 to 107 CFU mL−1 and a limit of detection of 3.3 CFU mL−1 with a 3.0% of relative standard deviation (RSD). This sensor successfully identified the pathogenic species in both milk and human blood serum. In another study, nuc and mecA genes of methicillin-resistant Staphylococcus aureus (MRSA) was detected for the quantification of Staphylococcus aureus.169 For this purpose, methylene blue (MB) and epirubicin (EP) were encapsulated in UiO-66-NH2 and locked by hybrid double-stranded DNA. Based on an electroactive dye release approach, the target DNA hybridizes with the displacement DNA (DEP and DMB) from metal–organic frameworks (MOF). The detection performance was significantly enhanced by Co–Zn bimetallic zeolitic imidazolate framework-derived N-doped porous carbon (BMZIF) nanocomposite modified electrode compared with intrinsic electrode. Table 9 presents examples of the previously reported electrochemical biosensors for the detection of Staphylococcus spp.
Table 9 Examples of reported electrochemical biosensors for the detection of pathogenic Staphylococcus spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| IgY: anti-protein antibody, SAM: self-assembled molecular monolayer, TTF: tetrathiafulvalene, MB: magnetic bead, BMZIF: Co–Zn bimetallic ZIF, GGP: guinea grass leaves. |
| GCE@AuNP |
IgY |
Staphylococcus aureus |
Immunosensor |
EIS |
10 to 107 CFU mL−1 |
3.3 CFU mL−1 |
[Fe(CN)6]4−/3− |
Milk, blood |
168 |
| AuE/SAMs |
Anti-S. aureus |
Staphylococcus aureus |
Immunosensor |
EIS |
101 to 107 CFU mL−1 |
10 CFU mL−1 |
[Fe(CN)6]4−/3− |
N/A |
170 |
| TTF-MPA-AuE |
RbIgG |
Staphylococcus aureus |
Immunosensor |
Amperometric |
2.2 × 105 to 9.2 × 105 CFU mL−1 |
1.6 × 105 CFU mL−1 |
NaAc/NaCl buffer |
N/A |
171 |
| PtE/GA-PEI |
Anti-S. aureus |
Staphylococcus aureus |
Immunosensor |
Amperometric |
101 to 108 CFU mL−1 |
10 CFU mL−1 |
[Fe(CN)6]4−/3− |
Milk, cheese meat |
172 |
| GCE/CNFs/CNPs/AuNPs |
SH-anti-S. aureus |
Staphylococcus aureus |
Aptasensor |
EIS |
1.2 × 101 to 1.2 × 108 CFU mL−1 |
1 CFU mL−1 |
Fe(CN)63−/4− |
Serum |
173 |
| MBs |
Anti-S. aureus |
Staphylococcus aureus |
Aptasensor |
DPV |
10−1 × 106 CFU mL−1 |
1.0 CFU mL−1 |
Buffer C |
Water |
174 |
| GCE/MOFs/AuNPs |
SH-DNA |
Staphylococcus aureus |
Aptasensor |
DPV |
7–7 × 106 CFU mL−1 |
1.9 and 5.2 CFU mL−1 |
[Fe(CN)6]4−/3− |
Urine |
175 |
| AuE |
DNA nanoflowers |
Staphylococcus aureus |
Aptasensor |
DPV |
60–6 × 107 CFU mL−1 |
9.0 CFU mL−1 |
MB |
Water, honey |
176 |
| GCE/AuNP-ssDNA-rGO |
ssDNA |
Staphylococcus aureus |
Aptasensor |
EIS |
10 to 106 CFU mL−1 |
10 CFU mL−1 |
[Fe(CN)6]4−/3− |
Food |
177 |
| SPGE |
Aptamer |
Staphylococcus aureus |
Aptasensor |
CV |
N/A |
39 and 414 CFU mL−1 |
[Fe(CN)6]4−/3− |
Water |
178 |
| GCE/BMZIF-derived NPCs |
dsDNA |
Staphylococcus aureus |
DNA biosensor |
DPV |
N/A |
3.7 fmol L−1 and 1.6 fmol L−1 |
PBS |
Water |
169 |
| SPGE-cys-GGP |
GGP |
Staphylococcus aureus |
Biosensor |
CV |
3 × 102 and 3 × 108 CFU mL−1 |
102 CFU mL−1 |
[Fe(CN)6]4−/3− |
Food |
179 |
| BC/c-MWCNTs-PEI |
Phage |
Staphylococcus aureus |
Phagosensor |
DPV |
3 × 100 to 3 ×107 CFU mL−1 |
3 CFU mL−1 |
PBS |
Milk, food |
14 |
| SPE/CNT-f-PEI |
Phage |
Staphylococcus aureus |
Phagosensor |
EIS |
102 to 107 |
1.23 × 102 |
Fe(CN)63−/4− |
Blood plasma |
180 |
| 102 to 105 CFU mL−1 |
1.29 × 102 CFU mL−1 |
| SPE-COOH |
Phage |
Staphylococcus aureus |
Phagosensor |
EIS |
2.0–2.0 × 106 CFU mL −1 |
N/A |
Fe(CN)63−/4− |
Apple juice, water |
181 |
Additionally, a sensitive and specific Staphylococcus aureus detection system was developed using a gold nanoparticle/carbon nanoparticle/cellulose nanofiber nanocomposite (AuNPs/CNPs/CNFs) synthesized on the surface of GCE. This combination worked as a sensing element, immobilizing a specific S. aureus aptamer. With a LOD of 1 CFU mL−1, the fabricated aptasensor demonstrated a broad linear dynamic range (1.2 × 101 to 1.2 × 108) CFU mL−1 and was able to precisely identify and quantify Staphylococcus aureus in human blood serum, a clinical sample with a complicated matrix. The published research on an electrochemical biosensor for the detection of pathogenic Staphylococcus species showed good performance, with a few limitations.173 The working procedures are summarized and presented in Fig. 10.
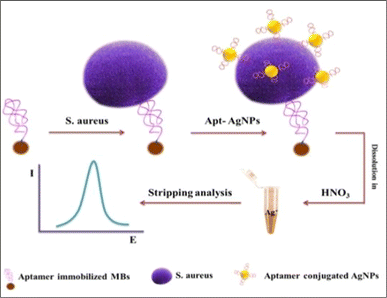 |
| | Fig. 10 Aptamer-based electrochemical determination of S. aureus. The figure was adapted from ref. 173 and reproduced with permission, copyright @ Elsevier. | |
4.8. Streptococcus spp. and Legionella spp.
A Gram-positive bacterium called Streptococcus spp. is linked to bacterial intra-mammary infections in bovine that cause mastitis. Recently, Yaghoobi et al.182 demonstrated a research work on the detection and quantification of Streptococcus Pneumoniae bacteria (SPB) with lead nanoparticles (Pb NPs) and DNA-FAM-S modified GCE. After binding with the complementary targeted DNA, the electrode performance was measured by standard EIS technique. The systematic representation is shown in Fig. 11. They concluded that the produced biosensor's selectivity is on par with that of the standard NanoDrop technique. In a different study, an electrochemical immunosensor was created by anchoring DNA tetrahedrons (DNA TH) with hollow structures to gold electrodes. This allowed for the quick detection of pneumococcal surface protein A (PspA) peptide and Streptococcus pneumoniae lysate from both synthetic and real human samples. Furthermore, with a LOD of 0.093 CFU mL−1, the developed DNA-TH-based immunosensor demonstrates strong sensing efficacy against Streptococcus pneumoniae lysate in a therapeutically relevant linear range from 5 to 100 CFU mL−1. This leads to the conclusion that Streptococcus spp. can be effectively detected in immunological and biosensors using electrochemical sensing technology.
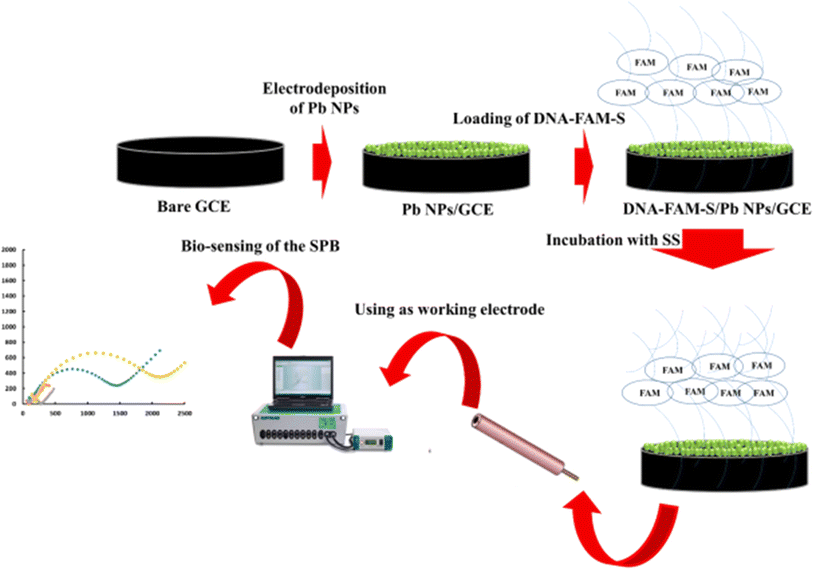 |
| | Fig. 11 Schematic diagram showing the preparation procedure for AuE/AuNPs/CysA/pDNA electrode and the detection of SPB. The figure was adapted from ref. 182 [open access]. | |
Literature revealed that Legionella pneumophila was detected by DNA based bioassay of square wave voltammetry technique.183 In this study, mip gen of Legionella pneumophila was detected by AuE/AuNPs/Cys A/pDNA biosensor. A linear dynamic calibration line with a range of 1 μmol L−1 to 1 zmol L−1 and low limit of detection were reported. These results are among the highest for the detection of Legionella pneumophila. Table 10 shows some examples of the previously reported electrochemical biosensors for the detection of Streptococcus spp. and Legionella spp. Additionally, complementary DNA was hybridized to provide a DNA probe-based detection technique.189 For the quantification of the ss 21mer DNA sequence, the developed biosensor showed a wide linear range over seven orders of magnitude with an ultrasensitive detection limit of 3.1 × 10−13 M. It also selectively distinguished the complementary sequence from target sequences that had single base mismatches (MM1) and triple base mismatches (MM3) of different strains of Legionella spp. In contrast, other immunosensor showed unsatisfactory results.186
Table 10 Examples of previously reported electrochemical biosensors for the detection of pathogenic Streptococcus spp. and Legionella spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| 6-MHA: 6-mercaptohexanoic acid, TMB: 3,3′,5,5′-tetramethylbenzidine, 16-AHT: 16-amino-1-hexadecanethiol. |
| Streptococcus spp. |
| AuE |
DNA tetrahedron |
Streptococcus pneumonia lysate |
Immunosensor |
SWV |
5–100 ng mL−1 |
0.093 ng mL−1 |
PBS |
Axilla, nasal cavity, mouth |
184 |
| SPGE/GA |
Anti-Streptococcus pyogenes |
Anti-Streptococcus pyogenes |
Immunosensor |
EIS |
4.2 × 102 to 4.2 × 106 CFU mL−1 |
9.3 CFU mL−1 |
Fe(CN)63−/4− |
N/A |
185 |
| GCE/Pb NPs |
DNA-FAM-S |
Streptococcus Pneumoniae |
DNA biosensor |
EIS |
0.0031 ng mL−1- 0.4 ng mL−1 |
0.0022 ng mL−1 |
Fe(CN)63−/4− |
N/A |
182 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| Legionella spp. |
| AuE/6-MHA |
Anti-L. pneumophila |
Lp02-GFP-L. pneumophila |
Immunosensor |
EIS |
N/A |
2.0 × 102 cells per mL |
PBS |
N/A |
186 |
| SPE/TMB |
LP3IIG2 anti-Legionella |
Legionella pneumophila |
Immunosensor |
Chronoamperometry |
101 and 104 CFU mL−1 |
4 CFU mL−1 |
PBS |
N/A |
187 |
| SPGE/16-AHT |
Anti-Legionella |
Legionella pneumophila |
Immunosensor |
SWV |
10 to 108 CFU mL−1 |
10 CFU mL−1 |
Fe(CN)63−/4− |
Artificially contaminated samples |
188 |
| PtE/GA |
ssDNA |
Legionella pneumophila |
DNA biosensor |
DPV |
10−13 to 10−6 mol L−1 |
3.1 × 10−13 mol L−1 |
Fe(CN)63−/4− |
N/A |
189 |
| AuE/AuNPs/Cys a |
ssDNA |
Legionella pneumophila |
DNA biosensor |
SWV |
1 μmol L−1 to 1 zmol L−1 |
1 zepto-molar |
Tris–HCl |
N/A |
183 |
4.9. Brucella spp.
The Gram-negative bacterium Brucella spp. is the primary cause of the infectious, communicative, and contagious disease brucellosis, which mostly affects cattle, bison, and pigs. The development of novel composite materials and effective detection approach could address these issues. Fe3O4@Au nanocomposite has been widely used to fabricate electrochemical biosensor. A poly(ethylene glycol) (PEG) and hyaluronic acid (HA) modified Fe3O4@Au NPs electrochemical immunosensor was proposed by Lv et al. to detect disease markers that are specific to brucellosis antibodies.190 The graphical explanation of the devolped sensor is shown in Fig. 12. It's interesting to note that this immunosensor can test in 100% serum without biological interference and has a linear response range of 10−15 to 10−11 g mL−1 towards brucellosis antibodies. Rahi et al.191 created a three-dimensional nanostructure with gold nanoribbons encased in gold nanoblooms using the sonoelectrodeposition technique. The suggested nanostructure was employed as a transducer to create a genosensor and to immobilize a probe unique to Brucella. The zepto-molar electrochemical detection of Brucella in blood samples from brucellosis patients was carried out using this technology. Table 11 summarizes examples of previously reported electrochemical biosensors for the detection of Brucella spp. Additionally, a probe specific to Brucella was immobilized by fabricating 6-Mercapto-1-hexanol (MCH) on the nPd electrode surface.194 The created genosensor was assessed for the purpose of testing the bacteria in human and cultured samples both with and without PCR. With a sensitivity of 0.02 μA dm3 mol−1, a linear concentration range of 1.0 × 10−12 to 1.0 × 10−19 mol dm−3, and a detection limit of 2.7 × 10−20 mol dm−3, the genosensor was able to identify the complimentary sequence. This sensor exhibited the lowest limit of detection among the surveyed literature.
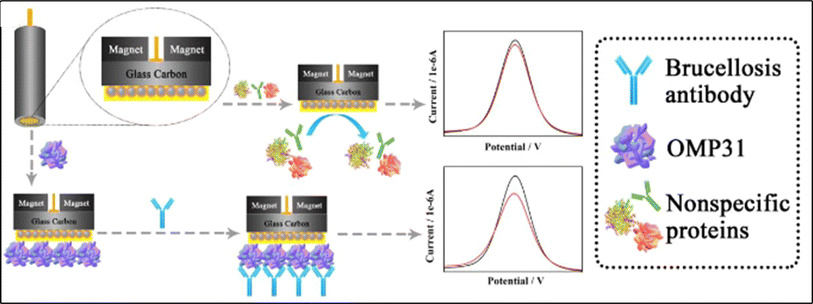 |
| | Fig. 12 Schematic diagram explanting the fabrication of Fe3O4@Au@PEG@HA NPs electrochemical immunosensor. The figure was adapted from ref. 190 and reproduced with permission, copyright @ Elsevier. | |
Table 11 Examples of reported electrochemical biosensors for the detection of pathogenic Brucella spp.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| PEG-MA: poly(ethyleneglycol)-methacrylate, GMA: glycidylmethacrylate, PEG: polyethylene glycol, HA: hyaluronic acid, GSH: glutathione. |
| Fe3O4@SiO2@p(PEG-MA-GMA) |
Aptamer |
Brucella melitensis |
Aptasensor |
EIS |
102 to 107 CFU mL−1 |
N/A |
PBS |
Milk, milk products |
192 |
| AuE/nAu |
p-ssDNA |
Brucella genome |
DNA biosensor |
DPV |
10 zmol dm−3 to 10 pmol dm−3 |
1.71 zmol dm−3 |
MB |
N/A |
191 |
| AuE |
ssDNA |
Cas12/crRNA/target DNA |
DNA biosensor |
SWV |
4.5 × 108 to 4.5 × 101 copies per reaction |
2 copies per reaction |
MB |
Blood, milk |
193 |
| AuE/nPd |
p-ssDNA |
t-ssDNA |
Genosensor |
DPV |
1.0 × 10−12 to 1.0 × 10−19 mol dm−3 |
2.7 × 10−20 mol dm−3 |
Fe(CN)63−/4− |
Serum |
194 |
| (GO–NW@Au-NF)FTO/Au-NF@NW-GO |
OMP31 antigen |
Br-antibody |
Immunosensor |
DPV |
N/A |
9.3 fg mL−1 |
PBS |
Serum |
190 |
| GCE/Fe3O4@Au@PEG@HA NPs |
OMP31 antigen |
Brucellosis antibody |
Immunosensor |
DPV |
10−15 g mL−1 to 10−11 g mL−1 |
0.36 fg mL−1 |
PBS |
Serum |
190 |
| GCE/AuNPs/GSH/HA |
OMP31 antigen |
Antibody |
Immunosensor |
DPV |
2.08 × 10−15 to 1.04 × 10−12 g mL−1 |
0.50 fg mL−1 |
Fe(CN)63−/4− |
Serum |
195 |
| SPCE/GNP |
Anti-Brucella melitensis |
Brucella melitensis |
Immunosensor |
EIS |
4 × 104 to 4 × 106 CFU mL−1 |
1 × 104 CFU mL−1 |
Fe(CN)63−/4− |
Milk |
196 |
4.10. Helicobacter pylori
Gram-negative Helicobacter pylori bacteria are well known for damaging the stomach lining and negatively impacting human health. Recently, Jaradat et al. developed HopQ (protein) biomarker grafted screen-printed carbon electrodes with MWCNT-COOH decorated with gold nanoparticles (AuNP), SPCE/MWCNT/AuNP, immunosensor for the detection of Helicobacter pylori (H. pylori) pathogenic bacterium.197 It is important to emphasize that the linearity was discovered to be within the range of 10 pg mL−1 to 100 ng mL−1. The platform showed limit of detection (LOD) and limit of quantification (LOQ) of 2.0 pg mL−1 and 8.6 pg mL−1, respectively. Another study found that the AuNPs-based electrochemical biosensor may be used to detect H. pylori bacterium iDNA sequences by introducing initiator DNA (iDNA) triggered hybridization chain reaction (HCR).198 Gel electrophoresis image confirmed that the HCR occurs with the free DNA at the DNA-modified AuNPs. Authors highlighted that HCR converts the iDNA to long dsDNA concatemer. Furthermore, it was observed that electrochemical active molecule [Ru(NH3)5L]2+ intercalated into dsDNA. Table 12 sums up examples of some previously reported electrochemical biosensors for the detection of Helicobacter pylori. Furthermore, it was revealed that an electrochemical biosensor based on SiO2 nanoparticle decorated molecularly imprinted polymer (MIP) was created to detect Helicobacter pylori on a screen-printed electrode (SPE), which is thought to function as a receptor by using template VacA antigen.201 Schematic illustration showing the fabrication process of the devoloped biosensor is presented in Fig. 13. The developed VacA-MIP/SiO2 @SPE sensor shows excellent sensitivity (0.304 mA ng ml−1) and a very low detection limit (0.01 ng mL−1) in a linear range of 0.01–100 ng mL−1 under optimal testing conditions. Consequently, the effective detection approach was occupied by MIP electrochemical detection techniques, DNA biosensors, and immunosensors with no restrictions.
Table 12 Examples of some previously reported electrochemical biosensors for the detection of pathogenic Helicobacter pylori.a
| Electrode |
Bioreceptor |
Analyte |
Biosensor type |
Electrochemical method |
Linear range |
Limit of detection |
Redox probe |
Real sample |
Ref. |
| PEDOT: poly-3,4 ethylene dioxythiophene, g-C3N4: graphitic carbon nitrite, Pin5COOH: polyindole carboxylic acid, ZnO-T: zinc oxide tetrapods. |
| AuE |
SH-ssDNA |
Helicobacter pylori |
DNA biosensor |
DPV |
20.0–410.0 nmol L−1 |
7.2 nmol L−1 |
Chlorogenic acid |
N/A |
199 |
| SPE@rGO/Au |
MIP-CagA |
Helicobacter pylori |
Biosensor |
DPV |
0.05–50 ng mL−1 |
0.05 ng mL−1 |
[Fe(CN)6]3−/4− |
Blood |
200 |
| SPE@SiO2 |
MIP-VacA |
Helicobacter pylori |
Biosensor |
DPV, EIS |
0.01–100 ng mL− 1 |
0.01 ng mL− 1 |
[Fe(CN)6]3−/4− |
Serum |
201 |
| GCE/Ti3C2Tx/AuNPs |
SH-cpDNA |
Helicobacter pylori |
DNA biosensor |
DPV |
10−11 to 10−14 mol L−1 |
1.6 × 10−16 mol L−1 |
PBS |
Milk, serum |
202 |
| AuE |
SH-ssDNA |
Helicobacter pylori |
DNA biosensor |
DPV |
0.3–240 nmol L−1 |
0.15 nmol L−1 |
β-cyclodextrin |
N/A |
203 |
| GCE/GO/AuNP |
ssDNA |
Helicobacter pylori |
DNA biosensor |
DPV |
60.0–600.0 p mol L−1 |
27.0 p mol L−1 |
Oracet blue |
SDS, ethanol, glucose, CTAB |
204 |
| AuE |
SH-cpDNA |
Helicobacter pylori |
DNA biosensor |
DPV |
0.01 fmol L−1 to 0.5 fmol L−1, 1 fmol L−1 to 100 fmol L−1 |
0.68 amol L−1 |
[Ru(NH3)5L]2+ |
N/A |
198 |
| AuE/PEDOT/rGO/Pdnano |
BabA antibody |
Helicobacter pylori |
Immunosensor |
CV |
0.2–20 ng mL−1 |
0.2 ng mL−1 |
[Fe(CN)6]3−/4− |
Stool |
205 |
| AuET/ZnO/g-C3N4 |
VacA antigen |
Helicobacter pylori |
Immunosensor |
DPV |
0.1–12.8 ng mL−1 |
0.1 ng mL−1 |
[Fe(CN)6]3−/4− |
Serum |
200 |
| AuE/red-GOx/PEDOT/Ptnano |
CagA antibody |
Helicobacter pylori |
Immunosensor |
DPV |
0.1 ng mL−1 to 30 ng mL−1 |
0.1 ng mL−1 |
[Fe(CN)6]3−/4− |
Serum |
206 |
| SPAuE@ZnO-T |
CagA antigen |
Helicobacter pylori |
Immunosensor |
CV, SWV |
0.2 ng mL−1 to 50 ng mL−1 |
0.2 ng mL−1 |
[Fe(CN)6]3−/4− |
Serum |
207 |
| AuE/Pin5COOH/c-MWCNT/TiO2NPs |
Cag aantibody |
Helicobacter pylori |
Immunosensor |
SWV |
0.1–8.0 ng mL−1 |
0.1 ng mL−1 |
[Fe(CN)6]3−/4− |
Stool |
208 |
| SPCE/MWCNT/AuNPs |
HopQ antibody |
Helicobacter pylori |
Immunosensor |
SWV |
10 pg mL−1 to 100 ng mL−1 |
2.0 pg mL−1 |
[Fe(CN)6]3−/4− |
Artificial saliva |
197 |
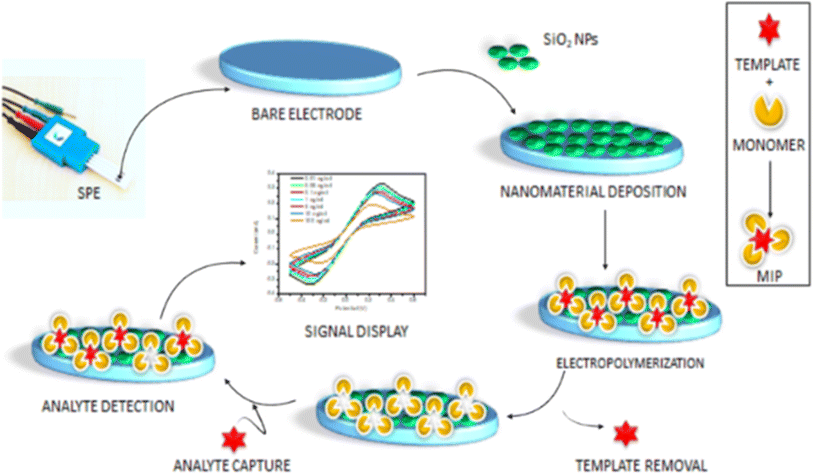 |
| | Fig. 13 Molecularly imprinted polymer (MIP) based biosensor. The figure was adapted from ref. 201 and reproduced with permission, copyright @ Elsevier. | |
5. Conclusions
This review of the literature concentrates on the bioreceptor theme for electrochemical biosensor modification, which offers a wide range of applications as a platform for pathogen detection. The performance metrics and a summary of the electrode construction techniques for identifying pathogenic bacteria are provided. Additionally, new developments on various bioreceptors, detection techniques, assay plans, redox probes, and material kinds are summarized and tallied. Thus, combining electrochemical-based biosensors with electrode materials and bioreceptors makes it easier to determine the optimal approach for identifying specific infections. This work also includes a comprehensive analysis of various electrochemical biosensor production methods for the detection of pathogenic bacteria and the process by which they are modified. All bioreceptors have a specific role in PB detection, however the detection limit of DNA and antibody-modified electrochemical biosensors is lower than that of other bioreceptors. Furthermore, phagosensors, or phage base sensors, are more straightforward than others. Thus, this review focuses on biosensing techniques that have led to notable advancements in the survivability, response time, selectivity, and sensitivity of bacterial detection. To prevent negative effects as early as possible, a great deal of research has recently been conducted with the goal of sensitively detecting infectious microorganisms. Additionally, combining the bioreceptor immobilized electrochemical biosensor into a single, reliable, and integrated platform for the management of healthcare is becoming a global aim for the scientific community. For scientists conducting research on electrochemically based bioreceptor modified sensors, this review can therefore be used as a starting point and a reference.
6. Present challenges and future perspectives
Water, food, hospitals, and human fluids can harbor or be the source of a variety of pathogenic bacteria that can cause potentially fatal illnesses and become resistant to antibiotics. Due to the drawbacks of bioreceptors, such as limited stability for antibodies, difficulties binding to DNA targets for nucleic acids. That decreased sensitivity to nuclease for aptamers, electrochemical biosensors have several limitations when it comes to practical applications. However, the combination of clustered regularly interspaced short palindromic repeats (CRISPR) associated technology209 and electrochemical DNA sensors can improve the sensitivity and precision. One of the major challenges in electrochemical biosensing of pathogenic bacteria in real samples is the need for multiple sample preparation stages. Furthermore, despite the potential for extreme sensitivity and robustness, selective biorecognition element-based bioanalytical techniques necessitate the addition of reagents to the sample and laborious sample preparation procedures, which lengthen the time-to-results (TTR). Additionally, furthermore, a wide range of molecules, including proteins, lipids, nucleic acids, and other cellular debris, are commonly present in bacterial samples and can obstruct the response of biosensors. It can be challenging to preserve the viability of bacteria during sample preparation, particularly when live bacteria is required for the analytical phases of the detection process. The wide fluctuation in bacterial concentrations in samples makes it difficult to consistently and reliably detect bacteria. False detection or a drop in signal to noise ratio may arise due to environmental conditions or contamination from other microorganisms such as intercellular bacterial leakage. Literature revealed that electrochemical biosensing still suffer from low yield during simultaneous detection. Furthermore, it is difficult to design compact, portable electronics without sacrificing effectiveness. For practical applications, biosensors must be integrated with electronic systems for data processing and wireless communication; however, this requires sophisticated engineering solutions.
Future research could benefit from adding nanomaterials to electrochemically based biosensors to increase their performance even further. The design of biosensors has been significantly impacted by the swift advancement of nanomaterials research. Moreover, a range of organic groups, including conductive polymers, thiols, and silanes, can be functionalized on the surfaces to effectively immobilize bioreceptors.210 The development of point-of-need diagnostic instruments that incorporate nanomaterials, microfluidics, and electrochemical biosensors can benefit from cooperation between several disciplines, including nanotechnology, food science, material science, electrochemistry, microbiology, and system design and integration. At every stage of the process, including production, packaging, distribution, storage, and consumption, these devices will offer food safety evaluations and food screening capabilities. Academic and industrial researchers are collaborating to collectively commercialize a few fully integrated biosensing detection technologies. However, further research is required to precisely identify infectious bacteria.211 Electrochemical biosensors can be used as a wearable detection tool in the near future,16 collecting data wirelessly near the field to quickly identify pathogenic bacteria that pose a threat to human life. Such wearable and wireless integrated systems should be also commercially attractive. For the PBs detection, further bacterial culture protocol improvements is currently being aggressively sought. Achieving efficient bacterial disruption might require a combination of chemical (such as detergents), mechanical (e.g., bead-beating), and enzymatic (for incident lysozyme) lysis techniques. Immuno-magnetic beads or selective growth media is often employed to extract the desired bacterial population from the sample. Removal of cellular debris through filtration and centrifugation after lysis is part sample preparation procedure. Setting up and following standardized protocols for the collection, storage, and preparation of bacterial samples are recommended steps to ensure high yield and reproducibility. By implementing efficient and reliable strategies to tackle the above challenges, electrochemical biosensors can greatly enhance the accuracy and dependability of bacterial sample analysis.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.
Author contributions
Md. Abdul Khaleque: conceptualized the main idea, structured, and wrote the original manuscript; S. I. Hossain: contributed to the writing and revision of the manuscript; Md. Romzan Ali: contributed to the writing and revision of the manuscript; Hala S. Abuelmakarem: wrote sections of the manuscript; Mohamed Aly Saad Aly: updated the main concepts and ideas, wrote, reviewed, and edited the manuscript, supervised and evaluated the overall work; Muhammad Shamim Al Mamun: designed, wrote, reviewed and edited sections of the manuscript; Md. Zaved Hossain Khan: contributed to the main concept, supervised and evaluated the overall concepts.
Conflicts of interest
The authors declare that there is no conflict of interest.
Acknowledgements
This research received specific funding facilities from Georgia Tech Shenzhen Institute (GTSI).
References
- T. M. Uddin, A. J. Chakraborty, A. Khusro, B. R. M. Zidan, S. Mitra, T. Bin Emran, K. Dhama, M. K. H. Ripon, M. Gajdács, M. U. K. Sahibzada, M. J. Hossain and N. Koirala, J. Infect. Public Health, 2021, 14, 1750–1766 CrossRef PubMed.
- E. Peterson and P. Kaur, Front. Microbiol., 2018, 9, 1–21 CrossRef PubMed.
- S. W. Smith, Approach to Internal Medicine, 2016 Search PubMed.
- S. Ajulo and B. Awosile, PLoS One, 2024, e0297921, DOI:10.1371/journal.pone.0297921.
- B. Wang, H. Wang, X. Lu, X. Zheng and Z. Yang, Foods, 2023, 2795, DOI:10.3390/foods12142795.
- B. Wang, H. Wang, X. Lu, X. Zheng and Z. Yang, Foods, 2023, 2795, DOI:10.3390/foods12142795.
- L. K. Strawn, E. D. Fortes, E. A. Bihn, K. K. Nightingale, Y. T. Gröhn, R. W. Worobo, M. Wiedmann and P. W. Bergholz, Appl. Environ. Microbiol., 2013, 79, 588–600 CrossRef CAS PubMed.
- M. A. Khaleque, M. R. Ali, M. S. Bacchu, M. R. A. Mamun, M. I. Hossain, M. S. Hossain, M. Aly Saad Aly and M. Z. H. Khan, Heliyon, 2022, e20676, DOI:10.1016/j.heliyon.2023.e20676.
- M. Aijuka and E. M. Buys, Food Microbiol., 2019, 82, 363–370 CrossRef PubMed.
- D. P. Kafetzopoulos, E. L. Psomas and P. D. Kafetzopoulos, Food Control, 2013, 33, 505–513 CrossRef.
- M. S. Bacchu, M. R. Ali, S. Das, S. Akter, H. Sakamoto, S. I. Suye, M. M. Rahman, K. Campbell and M. Z. H. Khan, Anal. Chim. Acta, 2022, 339332, DOI:10.1016/j.aca.2021.339332.
- S. Sakamoto, W. Putalun, S. Vimolmangkang, W. Phoolcharoen, Y. Shoyama, H. Tanaka and S. Morimoto, J. Nat. Med., 2018, 72, 32–42 CrossRef CAS PubMed.
- S. Mahari and S. Gandhi, Biosensors, 2022, 12, 365 CrossRef CAS PubMed.
- U. Farooq, M. W. Ullah, Q. Yang, A. Aziz, J. Xu, L. Zhou and S. Wang, Biosens. Bioelectron., 2020, 112163, DOI:10.1016/j.bios.2020.112163.
- S. Hameed, L. Xie and Y. Ying, Trends Food Sci. Technol., 2018, 81, 61–73 CrossRef CAS.
- L. Zhang, W. Guo, C. Lv, X. Liu, M. Yang, M. Guo and Q. Fu, Adv. Sensor Energy Mater., 2023, 2, 100081 CrossRef.
- A. Azmi, A. A. Azman, S. Ibrahim and M. A. M. Yunus, Int. J. Smart Sens. Intell. Syst., 2017, 10, 223–261 CAS.
- B. A. Prabowo, P. D. Cabral, P. Freitas and E. Fernandes, Chemosensors, 2021, 9, 299 CrossRef CAS.
- Y. L. Oon, Y. S. Oon, M. Ayaz, M. Deng, L. Li and K. Song, Front. Microbiol., 2023, 14, 1286923 CrossRef PubMed.
- A. C. G. Foddai and I. R. Grant, Appl. Microbiol. Biotechnol., 2020, 104, 4281–4288 CrossRef CAS PubMed.
- B. Serra, M. Gamella, A. J. Reviejo and J. M. Pingarrón, Anal. Bioanal. Chem., 2008, 391, 1853–1860 CrossRef CAS PubMed.
- M. Gagliardi, M. Agostini, F. Lunardelli, L. Lamanna, A. Miranda, A. Bazzichi, A. G. Luminare, F. Cervelli, F. Gambineri, M. Totaro, M. Lai, G. Maisetta, G. Batoni, M. Pistello and M. Cecchini, Sens. Actuators, B, 2023, 133299, DOI:10.1016/j.snb.2023.133299.
- J. Kuzhandai Shamlee, V. V. L. Swamy, A. S Rajamani, S. Mukherji, J. Satija, V. Janakiraman and V. V. R. Sai, Biosens. Bioelectron. X, 2022, 100271, DOI:10.1016/j.biosx.2022.100271.
- L. Pasquardini, N. Cennamo, F. Arcadio, C. Perri, A. Chiodi, G. D'agostino and L. Zeni, Sci. Rep., 2023, 22832, DOI:10.1038/s41598-023-50344-5.
- I. Palchetti, M. Mascini, Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems, in Amperometric Biosensor for Pathogenic Bacteria Detection, ed. M. Zourob, S. Elwary, A. Turner, Springer, New York, NY, 2008, DOI:10.1007/978-0-387-75113-9_13.
- C. Ercole, M. Del Gallo, L. Mosiello, S. Baccella and A. Lepidi, Sens. Actuators, B, 2003, 91, 163–168 CrossRef CAS.
- J. Liu, I. Jasim, A. Abdullah, Z. Shen, L. Zhao, M. El-Dweik, S. Zhang and M. Almasri, Sci. Rep., 2018, 16109, DOI:10.1038/s41598-018-33972-0.
- I. Bobrinetskiy, M. Radovic, F. Rizzotto, P. Vizzini, S. Jaric, Z. Pavlovic, V. Radonic, M. V. Nikolic and J. Vidic, Nanomaterials, 2021, 11(10), 2700 CrossRef CAS PubMed.
- Z. Kotsiri, J. Vidic and A. Vantarakis, J. Environ. Sci., 2022, 111, 367–379 CrossRef.
- C. Ferrag and K. Kerman, Front. Sensors, 2020, 583822, DOI:10.3389/fsens.2020.583822.
- M. M. Shanbhag, G. Manasa, R. J. Mascarenhas, K. Mondal and N. P. Shetti, Chem. Eng. J. Adv., 2023, 16, 100516 CrossRef CAS.
- H. Sharma, M. Agarwal, M. Goswami, A. Sharma, S. K. Roy, R. Rai and M. S. Murugan, Vet. World, 2013, 6, 968–973 CrossRef CAS.
- M. R. Ali, M. S. Bacchu, M. R. Al-Mamun, M. I. Hossain, A. Khaleque, A. Khatun, D. D. Ridoy, M. A. S. Aly and M. Z. H. Khan, Crit. Rev. Anal. Chem., 2022, 2115286 Search PubMed.
- M. Z. H. Khan, M. R. Hasan, S. I. Hossain, M. S. Ahommed and M. Daizy, Biosens. Bioelectron., 2020, 166, 112431 CrossRef CAS PubMed.
- M. Santhanam, I. Algov and L. Alfonta, Sensors, 2020, 20, 1–15 CrossRef PubMed.
- A. Ahmed, J. V. Rushworth, N. A. Hirst and P. A. Millner, Clin. Microbiol. Rev., 2014, 27, 631–646 CrossRef CAS PubMed.
- C. Dincer, R. Bruch, E. Costa-Rama, M. T. Fernández-Abedul, A. Merkoçi, A. Manz, G. A. Urban and F. Güder, Adv. Mater., 2019, 1806739, DOI:10.1002/adma.201806739.
- M. A. Khaleque, M. I. Hossain, M. R. Ali, M. S. Bacchu, M. Aly Saad Aly and M. Z. H. Khan, RSC Adv., 2023, 13, 22973–22997 RSC.
- M. S. Mannoor, H. Tao, J. D. Clayton, A. Sengupta, D. L. Kaplan, R. R. Naik, N. Verma, F. G. Omenetto and M. C. McAlpine, Nat. Commun., 2012, 763, DOI:10.1038/ncomms1767.
- Z. Xiong, S. Achavananthadith, S. Lian, L. Edward Madden, Z. X. Ong, W. Chua, V. Kalidasan, Z. Li, Z. Liu, P. Singh, H. Yang, S. P. Heussler, S. M. P. Kalaiselvi, M. B. H. Breese, H. Yao, Y. Gao, K. Sanmugam, B. C. K. Tee, P. Y. Chen, W. Loke, C. T. Lim, G. S. H. Chiang, B. Y. Tan, H. Li, D. L. Becker and J. S. Ho, Sci. Adv., 2021, eabj1617, DOI:10.1126/sciadv.abj1617.
- S. L. Percival, S. McCarty, J. A. Hunt and E. J. Woods, Wound Repair Regen., 2014, 22, 174–186 CrossRef PubMed.
- L. Manjakkal, W. Dang, N. Yogeswaran and R. Dahiya, Biosensors, 2019, 14, DOI:10.3390/bios9010014.
- A. Shafiee, E. Ghadiri, J. Kassis, N. Pourhabibi Zarandi and A. Atala, Curr. Stem Cell Rep., 2018, 4, 105–115 CrossRef.
- V. Naresh and N. Lee, Sensors, 2021, 21, 1–35 CrossRef PubMed.
- I. A. Modenez, D. E. Sastre, F. C. Moares and C. G. C. Marques Netto, Molecules, 2018, 2230, DOI:10.3390/molecules23092230.
- F. Bettazzi, G. Marrazza, M. Minunni, I. Palchetti and S. Scarano, Compr. Anal. Chem., 2017, 77, 1–33 Search PubMed.
- C. M. Heckmann and F. Paradisi, ChemCatChem, 2020, 12, 6082–6102 CrossRef CAS PubMed.
- P. Mehrotra, J. Oral Biol. Craniofac. Res., 2016, 6, 153–159 CrossRef PubMed.
- T. Wang, X. Fan, R. Li, J. Xu and J. Liu, Chem. Eng. J., 2018, 129082, DOI:10.1016/j.cej.2021.129082.
- A. Ijaz Gul, M. Sheeraz Ahmad, S. M. Saqlan Naqvi, Ansar Hussain, R. Wali, A. A. Farooqi and I. Ahmed, J. Appl. Biol. Biotechnol., 2017, 72, DOI:10.7324/jabb.2017.50313.
- S. Arora, N. Ahmed and S. Siddiqui, Int. J. Chem. Stud., 2018, 1031–1039 CAS.
- H. Ooka, Y. Chiba and R. Nakamura, Nat. Commun., 2023, 4860, DOI:10.1038/s41467-023-40471-y.
- C. Barisch, J. C. M. Holthuis and K. Cosentino, Biol. Chem., 2023, 404, 467–490 CrossRef CAS PubMed.
- G. Sun, X. Wei, D. Zhang, L. Huang, H. Liu and H. Fang, Biosensors, 2023, 13, 886 CrossRef CAS PubMed.
- D. K. Soni, R. Ahmad and S. K. Dubey, Crit. Rev. Microbiol., 2018, 44, 590–608 CrossRef PubMed.
- J. Chen, Z. Jiang, J. D. Ackerman, M. Yazdani, S. Hou, S. R. Nugen and V. M. Rotello, Analyst, 2015, 140, 4991–4996 RSC.
- X. Yang, J. Kirsch and A. Simonian, J. Microbiol. Methods, 2013, 95, 48–56 CrossRef CAS PubMed.
- L. Alvarado-Ramírez, M. Rostro-Alanis, J. Rodríguez-Rodríguez, J. E. Sosa-Hernández, E. M. Melchor-Martínez, H. M. N. Iqbal and R. Parra-Saldívar, Biosensors, 2021, 11, 410 CrossRef PubMed.
- P. Banerjee, S. Kintzios and B. Prabhakarpandian, Toxins, 2013, 5, 2366–2383 CrossRef CAS PubMed.
- L. Xu, X. Bai, S. Tenguria, Y. Liu, R. Drolia and A. K. Bhunia, Front. Microbiol., 2020, 575615, DOI:10.3389/fmicb.2020.575615.
- S. Shukla, Y. Haldorai, S. K. Hwang, V. K. Bajpai, Y. S. Huh and Y. K. Han, Front. Microbiol., 2017, 8, 2398 CrossRef PubMed.
- A. Hasan, M. Nurunnabi, M. Morshed, A. Paul, A. Polini, T. Kuila, M. Al Hariri, Y. K. Lee and A. A. Jaffa, BioMed Res. Int., 2014, 307519, DOI:10.1155/2014/307519.
- E. E. Ferapontova, Curr. Opin. Electrochem., 2017, 5, 218–225 CrossRef CAS.
- A. Kausaite-Minkstimiene, A. Popov and A. Ramanaviciene, ACS Appl. Mater. Interfaces, 2022, 14, 20720–20728 CrossRef CAS PubMed.
- Y. Zhou, A. Marar, P. Kner and R. P. Ramasamy, Anal. Chem., 2017, 89, 5734–5741 CrossRef CAS PubMed.
- M. D. Marazuela and M. C. Moreno-Bondi, Anal. Bioanal. Chem., 2002, 372, 664–682 CrossRef CAS PubMed.
- Y. Li, G. Xie, J. Qiu, D. Zhou, D. Gou, Y. Tao, Y. Li and H. Chen, Sens. Actuators, B, 2018, 258, 803–812 CrossRef CAS.
- R. R. Al-Hindi, A. D. Teklemariam, M. G. Alharbi, I. Alotibi, S. A. Azhari, I. Qadri, T. Alamri, S. Harakeh, B. M. Applegate and A. K. Bhunia, Biosensors, 2022, 12, 905 CrossRef CAS PubMed.
- J. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS PubMed.
- A. V Lakhin, V. Z. Tarantul and L. V Gening, Aptamers: Problems, Solutions and Prospects, 2013, 5, 34–43 Search PubMed.
- S. Minchin and J. Lodge, Essays Biochem., 2019, 63, 433–456 CrossRef CAS PubMed.
- V. Bosco-Drayon, M. Poidevin, I. G. Boneca, K. Narbonne-Reveau, J. Royet and B. Charroux, Cell Host Microbe, 2012, 12, 153–165 CrossRef CAS PubMed.
- G. L. Popa and M. I. Popa, Germs, 2021, 11, 88–96 CrossRef PubMed.
- A. W. Musser and P. R. Beamer, J. Indiana State Med. Assoc., 1961, 54, 1627–1634 CAS.
- A. C. de M. Santos, F. F. Santos, R. M. Silva and T. A. T. Gomes, Front. Cell Infect. Microbiol., 2020, 10, 1–11 CrossRef PubMed.
- M. Vading, P. Nauclér, M. Kalin and C. G. Giske, PLoS One, 2018, 13, 1–13 CrossRef PubMed.
- M. R. Ali, M. S. Bacchu, D. D. Ridoy, P. L. Mozumder, M. N. Hasan, S. Das, M. F. H. Palash, S. Akter, N. Sakib, A. Khaleque, D. Chakrobortty and M. Z. H. Khan, RSC Adv., 2022, 12, 31497–31505 RSC.
- C. Baker-Austin, J. D. Oliver, M. Alam, A. Ali, M. K. Waldor, F. Qadri and J. Martinez-Urtaza, Nat. Rev. Dis. Primers, 2018, 1–19, DOI:10.1038/s41572-018-0005-8.
- V. Ramaswamy, V. M. Cresence, J. S. Rejitha, M. U. Lekshmi, K. S. Dharsana, S. P. Prasad and H. M. Vijila, J. Microbiol., Immunol. Infect., 2007, 40, 4–13 CAS.
- D. N. Taylor, A. C. Trofa, J. Sadoff, C. Chu, D. Bryla, J. Shiloach, D. Cohen, S. Ashkenazi, Y. Lerman, W. Egan, R. Schneerson and J. B. Robbins, Infect. Immun., 1993, 61, 3678–3687 CrossRef CAS PubMed.
- R. R. Brubaker, Yersinia pestis, Elsevier Ltd, 2014 Search PubMed.
- M. A. Haque, F. Wang, Y. Chen, F. Hossen, M. A. Islam, M. A. Hossain, N. Siddique, C. He and F. Ahmed, Front. Microbiol., 2022, 783103, DOI:10.3389/fmicb.2021.783103.
- A. L. Kau, S. M. Martin, W. Lyon, E. Hayes, M. G. Caparon and S. J. Hultgren, Infect. Immun., 2005, 73, 2461–2468 CrossRef CAS PubMed.
- S. M. Num and N. M. Useh, Jordan J. Biol. Sci., 2014, 7, 81–94 CrossRef.
- M. K. McLendon, M. A. Apicella and L. A. H. Allen, Annu. Rev. Microbiol., 2006, 60, 167–185 CrossRef CAS PubMed.
- A. Strašková and J. Stulík, Mil. Med. Sci. Lett., 2012, 81, 27–39 CrossRef.
- S. M. Nolan, Pediatric Infectious Diseases E-Book: Requisites, 2008, 198–208 Search PubMed.
- J. D. Langereis and M. I. De Jonge, Emerging Infect. Dis., 2015, 21, 1711–1718 Search PubMed.
- T. P. Primm, C. A. Lucero and J. O. Falkinham, Clin. Microbiol. Rev., 2004, 17, 98–106 CrossRef PubMed.
- S. J. Cavalieri and F. C. Knoop, Corynebacterium Infections, Elsevier Inc., 2014 Search PubMed.
- A. von Graevenitz, Annu. Rev. Microbiol., 1977, 31, 447–471 CrossRef CAS.
- S. Elahi, D. R. Thompson, S. Strom, B. O'Connor, L. A. Babiuk and V. Gerdts, J. Infect. Dis., 2008, 198, 384–392 CrossRef.
- N. J. Avire, H. Whiley and K. Ross, Pathogens, 2021, 10, 1–18 CrossRef PubMed.
- K. Tilly, P. A. Rosa and P. E. Stewart, Infect. Dis. Clin. North Am., 2008, 22, 217–234 CrossRef PubMed.
- C. A. Gaydos, Chlamydia trachomatis, Elsevier Inc., 1st edn, 2012, vol. 1 Search PubMed.
- B. M. W. Diederen, J. Infect., 2008, 56, 1–12 CrossRef CAS.
- E. Rukambile, V. Sintchenko, G. Muscatello, R. Kock and R. Alders, Zoonoses Public Health, 2019, 66, 562–578 CrossRef PubMed.
- D. Bravo, A. Hoare, C. Soto, M. A. Valenzuela and A. F. Quest, World J. Gastroenterol., 2018, 24, 3071–3089 CrossRef CAS PubMed.
- J. Buckle, in Clinical Aromatherapy, Elsevier, 2015, pp. 130–167 Search PubMed.
- D. T. Leung, S. K. Das, M. A. Malek, A. S. G. Faruque, F. Qadri, M. J. Chisti and E. T. Ryan, Am. J. Trop. Med. Hyg., 2015, 93, 831–835 Search PubMed.
- Q. Huang, X. Lin, L. Tong and Q. X. Tong, ACS Sustain. Chem. Eng., 2020, 8, 1644–1650 CrossRef.
- M. S. Bacchu, M. R. Ali, S. Das, S. Akter, H. Sakamoto, S. Suye, M. M. Rahman, K. Campbell and M. Z. H. Khan, Anal. Chim. Acta, 2021, 339332 Search PubMed.
- A. Singh, G. Sinsinbar, M. Choudhary, V. Kumar, R. Pasricha, H. N. Verma, S. P. Singh and K. Arora, Sens. Actuators, B, 2013, 185, 675–684 CrossRef CAS.
- R. Das, M. K. Sharma, V. K. Rao, B. K. Bhattacharya, I. Garg, V. Venkatesh and S. Upadhyay, J. Biotechnol., 2014, 188, 9–16 CrossRef CAS PubMed.
- X. Ma, Y. Jiang, F. Jia, Y. Yu, J. Chen and Z. Wang, J. Microbiol. Methods, 2014, 98, 94–98 CrossRef CAS PubMed.
- W. N. Suryapratiwi, V. I. Paat, S. Gaffar and Y. W. Hartati, in AIP Conference Proceedings, American Institute of Physics Inc., 2017, vol. 1848 Search PubMed.
- A. Singh, M. Choudhary, M. P. Singh, H. N. Verma, S. P. Singh and K. Arora, Bioelectrochemistry, 2015, 105, 7–15 CrossRef CAS PubMed.
- S. Ranjbar, S. Shahrokhian and F. Nurmohammadi, Sens. Actuators, B, 2018, 255, 1536–1544 CrossRef CAS.
- M. Barreiros dos Santos, S. Azevedo, J. P. Agusil, B. Prieto-Simón, C. Sporer, E. Torrents, A. Juárez, V. Teixeira and J. Samitier, Bioelectrochemistry, 2015, 101, 146–152 CrossRef CAS PubMed.
- H. Huang, M. Liu, X. Wang, W. Zhang, D. P. Yang, L. Cui and X. Wang, Nanoscale Res. Lett., 2016, 11, 1–7 CrossRef PubMed.
- A. Güner, E. Çevik, M. Şenel and L. Alpsoy, Food Chem., 2017, 229, 358–365 CrossRef PubMed.
- M. Xu, R. Wang and Y. Li, Analyst, 2016, 141, 5441–5449 RSC.
- A. Pangajam, K. Theyagarajan and K. Dinakaran, Sens. Bio-Sens. Res., 2020, 29, 100317 CrossRef.
- N. Razmi, M. Hasanzadeh, M. Willander and O. Nur, Anal. Methods, 2022, 14, 1562–1570 RSC.
- X. Lin, Y. Mei, C. He, Y. Luo, M. Yang, Y. Kuang, X. Ma, H. Zhang and Q. Huang, Front. Chem., 2021, 9, 4–11 Search PubMed.
- R. D. A. A. Rajapaksha, U. Hashim, M. N. Afnan Uda, C. A. N. Fernando and S. N. T. De Silva, Microsyst. Technol., 2017, 23, 5771–5780 CrossRef CAS.
- P. Geng, X. Zhang, Y. Teng, Y. Fu, L. Xu, M. Xu, L. Jin and W. Zhang, Biosens. Bioelectron., 2011, 26, 3325–3330 CrossRef CAS.
- S. Xu, Y. Zhang, K. Dong, J. Wen, C. Zheng and S. Zhao, Int. J. Electrochem. Sci., 2017, 12, 3443–3458 CrossRef CAS PubMed.
- D. Ozkan-Ariksoysal, Y. U. Kayran, F. F. Yilmaz, A. A. Ciucu, I. G. David, V. David, M. Hosgor-Limoncu and M. Ozsoz, Talanta, 2017, 166, 27–35 CrossRef CAS PubMed.
- R. B. Queirós, N. De-Los-Santos-Álvarez, J. P. Noronha and M. G. F. Sales, Sens. Actuators, B, 2013, 181, 766–772 CrossRef.
- R. F. Wang and R. Wang, Bioelectrochemistry, 2022, 148, 108229 CrossRef CAS PubMed.
- J. Xu, C. Zhao, Y. Chau and Y. K. Lee, Biosens. Bioelectron., 2020, 151, 111914 CrossRef CAS PubMed.
- A. Shabani, C. A. Marquette, R. Mandeville and M. F. Lawrence, Talanta, 2013, 116, 1047–1053 CrossRef CAS PubMed.
- C. Tlili, E. Sokullu, M. Safavieh, M. Tolba, M. U. Ahmed and M. Zourob, Anal. Chem., 2013, 4893–4901 CrossRef CAS PubMed.
- D. Wang, J. Chen and S. R. Nugen, Anal. Chem., 2017, 89, 1650–1657 CrossRef CAS PubMed.
- M. R. Ali, M. S. Bacchu, M. A. A. Setu, S. Akter, M. N. Hasan, F. T. Chowdhury, M. M. Rahman, M. S. Ahommed and M. Z. H. Khan, Biosens. Bioelectron., 2021, 188, 113338 CrossRef CAS PubMed.
- P. R. Solanki, M. K. Patel, A. Kaushik, M. K. Pandey, R. K. Kotnala and B. D. Malhotra, Electroanalysis, 2011, 23, 2699–2708 CrossRef CAS.
- M. K. Patel, P. R. Solanki, S. Khandelwal, V. V. Agrawal, S. G. Ansari and B. D. Malhotra, J. Phys.: Conf. Ser., 2012, 012009, DOI:10.1088/1742-6596/358/1/012009.
- D. Vijian, S. V. Chinni, L. S. Yin, B. Lertanantawong and W. Surareungchai, Biosens. Bioelectron., 2016, 77, 805–811 CrossRef CAS.
- M. Rahman, L. Y. Heng, D. Futra and T. L. Ling, Nanoscale Res. Lett., 2017, 474, DOI:10.1186/s11671-017-2236-0.
- P. D. Tam and C. X. Thang, Mater. Sci. Eng. C, 2016, 58, 953–959 CrossRef CAS PubMed.
- O. C. Ozoemena, T. Maphumulo, J. L. Shai and K. I. Ozoemena, ACS Omega, 2020, 5, 5762–5771 CrossRef CAS PubMed.
- P. Tshikalaha and O. A. Arotiba, Int. J. Electrochem. Sci., 2015, 10, 10083–10092 CrossRef CAS.
- P. K. Gupta, Z. H. Khan and P. R. Solanki, J. Electrochem. Soc., 2016, 163, B309–B318 CrossRef CAS.
- P. K. Gupta, A. Gupta, S. R. Dhakate, Z. H. Khan and P. R. Solanki, J. Appl. Polym. Sci., 2016, 133, 1–9 CrossRef.
- Q. Palomar, C. Gondran, M. Holzinger, R. Marks and S. Cosnier, Biosens. Bioelectron., 2017, 97, 177–183 CrossRef CAS PubMed.
- M. R. Ali, M. S. Bacchu, S. Das, S. Akter, M. M. Rahman, M. A. Saad Aly and M. Z. H. Khan, Talanta, 2023, 253, 123909 CrossRef CAS PubMed.
- S. S. Zarei, S. Soleimanian-Zad and A. A. Ensafi, Microchim. Acta, 2018, 185, 1–9 CrossRef CAS PubMed.
- Y. Hu, X. Xu, Q. Liu, L. Wang, Z. Lin and G. Chen, Anal. Chem., 2014, 86, 8785–8790 CrossRef CAS PubMed.
- Z. Bagheryan, J. B. Raoof, M. Golabi, A. P. F. Turner and V. Beni, Biosens. Bioelectron., 2016, 80, 566–573 CrossRef CAS PubMed.
- G. Zhao, X. Zhan and W. Dou, Anal. Biochem., 2011, 408, 53–58 CrossRef CAS PubMed.
- G. Zhao and X. Zhan, Electrochim. Acta, 2010, 55, 2466–2471 CrossRef CAS.
- Z. Izadi, M. Sheikh-Zeinoddin, A. A. Ensafi and S. Soleimanian-Zad, Biosens. Bioelectron., 2016, 80, 582–589 CrossRef CAS PubMed.
- M. S. Yoo, M. Shin, Y. Kim, M. Jang, Y. E. Choi, S. J. Park, J. Choi, J. Lee and C. Park, Chemosphere, 2017, 175, 269–274 CrossRef CAS PubMed.
- H. Wu, S. Liu, J. Jiang, G. Shen and R. Yu, Analyst, 2012, 137, 4829–4833 RSC.
- X. Kang, G. Pang, Q. Chen and X. Liang, Sens. Actuators, B, 2013, 177, 1010–1016 CrossRef CAS.
- E. B. Setterington and E. C. Alocilja, Biosensors, 2012, 2, 15–31 CrossRef CAS PubMed.
- X. Qian, Q. Qu, L. Li, X. Ran, L. Zuo, R. Huang and Q. Wang, Sensors, 2018, 1878, DOI:10.3390/s18061878.
- F. Cui, Z. Zhou, H. Feng and H. Susan Zhou, ACS Appl. Nano Mater., 2020, 3, 357–363 CrossRef CAS.
- R. Singh, G. Sumana, R. Verma, S. Sood, M. K. Pandey, R. K. Gupta and B. D. Malhotra, J. Biotechnol., 2010, 150, 357–365 CrossRef CAS PubMed.
- X. Liu, C. Du, D. Ni, Q. Ran, F. Liu, D. Jiang and X. Pu, Anal. Methods, 2016, 8, 8280–8287 RSC.
- D. Jiang, F. Liu, L. Zhang, L. Liu, C. Liu and X. Pu, RSC Adv., 2014, 4, 57064–57070 RSC.
- N. Gupta, D. Kumar, A. Das, S. Sood and B. D. Malhotra, Biosensors, 2023, 486, DOI:10.3390/bios13040486.
- R. Verma, S. Sood, R. Singh, G. Sumana, M. Bala, V. K. Sharma, J. C. Samantaray, R. M. Pandey and B. D. Malhotra, Diagn. Microbiol. Infect. Dis., 2014, 78, 16–23 CrossRef CAS PubMed.
- M. K. Patel, P. R. Solanki, A. Kumar, S. Khare, S. Gupta and B. D. Malhotra, Biosens. Bioelectron., 2010, 25, 2586–2591 CrossRef CAS PubMed.
- M. Tak, V. Gupta and M. Tomar, Biosens. Bioelectron., 2014, 59, 200–207 CrossRef CAS PubMed.
- S. R. Torati, V. Reddy, S. S. Yoon and C. G. Kim, Biosens. Bioelectron., 2016, 78, 483–488 CrossRef CAS PubMed.
- R. A. Omar, N. Verma and P. K. Arora, Front. Immunol., 2021, 653853, DOI:10.3389/fimmu.2021.653853.
- D. Ramos-Sono, R. Laureano, D. Rueda, R. H. Gilman, A. La Rosa, J. Ruiz, R. León, P. Sheen and M. Zimic, PLoS One, 2020, e0241067, DOI:10.1371/journal.pone.0241067.
- S. Taufiq, M. Waqar, M. N. Sharif and S. R. Abbas, Bioelectrochemistry, 2023, 108353, DOI:10.1016/j.bioelechem.2022.108353.
- C. Liu, D. Jiang, G. Xiang, L. Liu, F. Liu and X. Pu, Analyst, 2014, 139, 5460–5465 RSC.
- C. Thiruppathiraja, S. Kamatchiammal, P. Adaikkappan, D. J. Santhosh and M. Alagar, Anal. Biochem., 2011, 417, 73–79 CrossRef CAS PubMed.
- M. H. M. Zaid, C. E. N. Che-Engku-Chik, N. A. Yusof, J. Abdullah, S. S. Othman, R. Issa, M. F. Md Noh and H. Wasoh, Molecules, 2020, 3373, DOI:10.3390/molecules25153373.
- F. S. Mohamad, M. H. M. Zaid, J. Abdullah, R. M. Zawawi, H. N. Lim, Y. Sulaiman and N. A. Rahman, Sensors, 2017, 2789, DOI:10.3390/s17122789.
- U. Z. M. Azmi, N. A. Yusof, J. Abdullah, S. A. A. Ahmad, F. N. M. Faudzi, N. H. A. Raston, S. Suraiya, P. S. Ong, D. Krishnan and N. K. Sahar, Mikrochim. Acta, 2021, 188, 20 CrossRef PubMed.
- U. Z. M. Azmi, N. A. Yusof, N. Kusnin, J. Abdullah, S. Suraiya, P. S. Ong, N. H. A. Raston, S. F. A. Rahman and M. F. M. Fathil, Sensors, 2018, 3926, DOI:10.3390/s18113926.
- U. Z. M. Azmi, N. A. Yusof, J. Abdullah, F. Mohammad, S. A. A. Ahmad, S. Suraiya, N. H. A. Raston, F. N. M. Faudzi, S. K. Khiste and H. A. Al-Lohedan, Nanomaterials, 2021, 2446, DOI:10.3390/nano11092446.
- M. Roushani, Z. Rahmati, M. Golchin, Z. Lotfi and M. Nemati, Microchim. Acta, 2020, 567, DOI:10.1007/s00604-020-04547-6.
- G. Dai, Z. Li, F. Luo, Y. Lu, Z. Chu, J. Zhang, F. Zhang, Q. Wang and P. He, Microchim. Acta, 2021, 39, DOI:10.1007/s00604-020-04698-6.
- K. Bekir, H. Barhoumi, M. Braiek, A. Chrouda, N. Zine, N. Abid, A. Maaref, A. Bakhrouf, H. Ben Ouada, N. Jaffrezic-Renault and H. Ben Mansour, Environ. Sci. Pollut. Res., 2015, 22, 15796–15803 CrossRef CAS PubMed.
- V. Escamilla-Gómez, S. Campuzano, M. Pedrero and J. M. Pingarrón, Electroanalysis, 2007, 19, 1476–1482 CrossRef.
- T. Majumdar, R. Chakraborty and U. Raychaudhuri, Food Biosci., 2013, 4, 38–45 CrossRef CAS.
- S. Ranjbar and S. Shahrokhian, Bioelectrochemistry, 2018, 123, 70–76 CrossRef CAS PubMed.
- A. Abbaspour, F. Norouz-Sarvestani, A. Noori and N. Soltani, Biosens. Bioelectron., 2015, 68, 149–155 CrossRef CAS PubMed.
- Z. Sun, Y. Peng, M. Wang, Y. Lin, M. Jalalah, S. A. Alsareii, F. A. Harraz, J. Yang and G. Li, Anal. Chem., 2021, 8994, DOI:10.1021/acs.analchem.1c01763.
- R. Cai, S. Zhang, L. Chen, M. Li, Y. Zhang and N. Zhou, ACS Appl. Mater. Interfaces, 2021, 13, 4905–4914 CrossRef CAS PubMed.
- F. Jia, N. Duan, S. Wu, X. Ma, Y. Xia, Z. Wang and X. Wei, Microchim. Acta, 2014, 181, 967–974 CrossRef CAS.
- T. T. Q. Nguyen, E. R. Kim and M. B. Gu, Biosens. Bioelectron., 2022, 198, 113835 CrossRef CAS PubMed.
- P. Guarín and J. Cristancho, J. Colomb. Acad. Phys. Nat. Sci., 2020, 44, 835–844 Search PubMed.
- D. Patel, Y. Zhou and R. P. Ramasamy, J. Electrochem. Soc., 2021, 168, 057523 CrossRef CAS.
- N. Bhardwaj, S. K. Bhardwaj, J. Mehta, G. C. Mohanta and A. Deep, Anal. Biochem., 2016, 505, 18–25 Search PubMed.
- A. Yaghoobi, R. Abiri, A. Alvandi, E. Arkan, G. Mohammadi, T. Farshadnia and A. R. Jalalvand, Sens. Bio-Sens. Res., 2022, 36, 100494 CrossRef.
- A. Mobed, M. Hasanzadeh, S. Hassanpour, A. Saadati, M. Agazadeh and A. Mokhtarzadeh, Microchem. J., 2019, 148, 708–716 CrossRef CAS.
- J. Wang, M. C. Leong, E. Z. W. Leong, W. Sen Kuan and D. T. Leong, Anal. Chem., 2017, 89, 6900–6906 CrossRef CAS PubMed.
- N. Malinowska, W. Białobrzeska, T. Łęga, K. Pałka, K. Dziąbowska, S. Żołędowska, E. Czaczyk, K. Pala and D. Nidzworski, Sensors, 2020, 20, 1–10 CrossRef PubMed.
- N. Li, A. Brahmendra, A. J. Veloso, A. Prashar, X. R. Cheng, V. W. S. Hung, C. Guyard, M. Terebiznik and K. Kerman, Anal. Chem., 2012, 84, 3485–3488 CrossRef CAS PubMed.
- J. J. Ezenarro, N. Párraga-Niño, M. Sabrià, F. J. Del Campo, F. X. Muñoz-Pascual, J. Mas and N. Uria, Biosensors, 2020, 102, DOI:10.3390/bios10090102.
- A. Laribi, S. Allegra, M. Souiri, R. Mzoughi, A. Othmane and F. Girardot, Talanta, 2020, 215, 120904 CrossRef CAS PubMed.
- V. Rai, J. Deng and C. S. Toh, Talanta, 2012, 98, 112–117 CrossRef CAS PubMed.
- S. Lv, J. Sheng, S. Zhao, M. Liu and L. Chen, Biosens. Bioelectron., 2018, 117, 138–144 CrossRef CAS PubMed.
- A. Rahi, N. Sattarahmady and H. Heli, Sci. Rep., 2015, 5, 18060 CrossRef CAS PubMed.
- G. Bayramoglu, V. C. Ozalp, M. Oztekin and M. Y. Arica, Talanta, 2019, 200, 263–271 CrossRef CAS PubMed.
- J. Xu, J. Ma, Y. Li, L. Kang, B. Yuan, S. Li, J. Chao, L. Wang, J. Wang, S. Su and Y. Yuan, Sens. Actuators, B, 2022, 364, 131864 Search PubMed.
- A. Rahi, N. Sattarahmady and H. Heli, Anal. Biochem., 2016, 510, 11–17 CrossRef CAS.
- L. Chen, S. Zhao, M. Liu, P. Wu and C. Chen, Sens. Actuators, B, 2019, 287, 510–516 CrossRef CAS.
- H. Wu, Y. Zuo, C. Cui, W. Yang, H. Ma and X. Wang, Sensors, 2013, 13, 8551–8563 CrossRef CAS PubMed.
- H. Jaradat, A. Al-Hamry, M. Ibbini, N. Fourati and O. Kanoun, Biosensors, 2023, 527, DOI:10.3390/bios13050527.
- M. M. Lv, S. F. Fan, Q. L. Wang, Q. Y. Lv, X. Song and H. F. Cui, Microchim. Acta, 2020, 187, 12–16 Search PubMed.
- A. Asadzadeh-Firouzabadi, H. R. Zare and N. Nasirizadeh, J. Electrochem. Soc., 2016, 163, B43–B48 CrossRef CAS.
- K. Saxena, A. Kumar, N. Chauhan, M. Khanuja, B. D. Malhotra and U. Jain, ACS Omega, 2022, 7, 32292–32301 CrossRef CAS PubMed.
- K. Saxena, N. Chauhan, B. D. Malhotra and U. Jain, Process Biochem., 2023, 130, 87–95 CrossRef CAS.
- L. Wang, K. Cui, P. Wang, M. Pei and W. Guo, Anal. Bioanal. Chem., 2021, 413, 4353–4362 CrossRef CAS PubMed.
- P. Peng, F. Xu, Y. Xu and S. Sun, Int. J. Electrochem. Sci., 2017, 12, 9478–9487 CrossRef CAS.
- S. Hajihosseini, N. Nasirizadeh, M. S. Hejazi and P. Yaghmaei, Mater. Sci. Eng. C, 2016, 61, 506–515 CrossRef CAS PubMed.
- S. Gupta, U. Jain, B. T. Murti, A. D. Putri, A. Tiwari and N. Chauhan, Sci. Rep., 2020, 10, 1–14 CrossRef PubMed.
- S. Gupta, A. Tiwari, U. Jain and N. Chauhan, Mater. Sci. Eng. C, 2019, 103, 109733 CrossRef PubMed.
- N. Chauhan, S. Gupta, D. K. Avasthi, R. Adelung, Y. K. Mishra and U. Jain, ACS Appl. Mater. Interfaces, 2018, 10, 30631–30639 CrossRef CAS PubMed.
- U. Jain, S. Gupta, S. Soni, M. P. Khurana and N. Chauhan, Helicobacter, 2020, 25, 1–11 CrossRef PubMed.
- F. Li, Q. Ye, M. Chen, B. Zhou, J. Zhang, R. Pang, L. Xue, J. Wang, H. Zeng, S. Wu, Y. Zhang, Y. Ding and Q. Wu, Biosens. Bioelectron., 2021, 179, 113073 CrossRef CAS PubMed.
- I. H. Cho, D. H. Kim and S. Park, Biomater. Res., 2020, 24, 1–12 CrossRef PubMed.
- G. Doyon, J. Arch, G. Doyon, D. Twede, B. Drasner, A. M. Baylis, D. R. Kopsilk, E. Fisher, F. J. Sweeney, M. W. Leonard, B. Finnigan, L. Liu, J. Kost, P. Takhistov, P. Takhistov, C. Irwin, A. S. Mandel, G. A. Foster, L. Lynch, R. M. Brasington, R. Quinn and P. Henningsen, in The Wiley Encyclopedia of Packaging Technology, Wiley, 2009, pp. 71–180 Search PubMed.
|
| This journal is © The Royal Society of Chemistry 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *d,
Hala S. Abuelmakareme,
Muhammad Shamim Al Mamunf and
Md. Zaved Hossain Khan*ab
*d,
Hala S. Abuelmakareme,
Muhammad Shamim Al Mamunf and
Md. Zaved Hossain Khan*ab



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 Salmonella typhimurium/E. coli O157:H7 medium. Many of these impedimetric immunosensors displayed high sensitivity and selectivity. Other antibody modified immunosensors were developed and reported for the detection of Escherichia spp., and they are listed in Table 4. Furthermore, Xu et.al. reported on NH2-ssDNA modified ssDNA/GO/CS/GCE electrode for the electrochemical detection of E. coli O157:H7 using EIS and DPV. In this study, the developed sensor showed an excellent performance on the detection of E. coli O157:H7.118 A T4 phagosensor was decorated on micro-electrochemical sensor with a 3-mercaptoreopionic acid modified gold electrode.122 Utilizing DPV, the modified sensor showed a limit of detection of 14 ± 5 CFU mL−1 and a wide dynamic range of 1.9 × 101 to 1.9 × 108 CFU mL−1. This phagosensor was successfully able to selectively distinguish between viable and dead bacteria cells. The schematic representation of the AuE/Cys/PDCIT/T4 phage electrode fabrication aided with applied chemistry is shown in Fig. 5.
500 Salmonella typhimurium/E. coli O157:H7 medium. Many of these impedimetric immunosensors displayed high sensitivity and selectivity. Other antibody modified immunosensors were developed and reported for the detection of Escherichia spp., and they are listed in Table 4. Furthermore, Xu et.al. reported on NH2-ssDNA modified ssDNA/GO/CS/GCE electrode for the electrochemical detection of E. coli O157:H7 using EIS and DPV. In this study, the developed sensor showed an excellent performance on the detection of E. coli O157:H7.118 A T4 phagosensor was decorated on micro-electrochemical sensor with a 3-mercaptoreopionic acid modified gold electrode.122 Utilizing DPV, the modified sensor showed a limit of detection of 14 ± 5 CFU mL−1 and a wide dynamic range of 1.9 × 101 to 1.9 × 108 CFU mL−1. This phagosensor was successfully able to selectively distinguish between viable and dead bacteria cells. The schematic representation of the AuE/Cys/PDCIT/T4 phage electrode fabrication aided with applied chemistry is shown in Fig. 5.



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)



