Open Access Article
Open Access ArticleLiposome leakage and increased cellular permeability induced by guanidine-based oligomers: effects of liposome composition on liposome leakage and human lung epithelial barrier permeability†
Yeonjeong Ha a,
Yerim Kooa,
Seon-Kyung Parka,
Ga-Eun Kimb,
Han Bin Oh
a,
Yerim Kooa,
Seon-Kyung Parka,
Ga-Eun Kimb,
Han Bin Oh c,
Ha Ryong Kim*b and
Jung-Hwan Kwon
c,
Ha Ryong Kim*b and
Jung-Hwan Kwon *a
*a
aDivision of Environmental Science and Ecological Engineering, Korea University, 145 Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea. E-mail: junghwankwon@korea.ac.kr
bCollege of Pharmacy, Daegu Catholic University, 13-13 Hayang-ro, Hayang-eup, Gyeongsan 38430, Republic of Korea. E-mail: kimhr@cu.ac.kr
cDepartment of Chemistry, Sogang University, 35 Baekbeom-ro, Mapo-gu, Seoul 04107, Republic of Korea
First published on 28th September 2021
Abstract
Over the decades, guanidine-based oligomer groups have been one of the most widely used antimicrobial agents. Reportedly, these cationic oligomers cause serious damage to microorganisms but have low toxicity to humans. However, public concerns regarding the guanidine group have rapidly grown after the fatal misuse of these oligomers as humidifier disinfectants, which resulted in thousands of fatalities in South Korea. Herein, we investigated liposome leakage and cellular permeability changes caused by polyhexamethylene guanidine (PHMG) and polyhexamethylene biguanide (PHMB), both representative guanidine-based oligomers. The leakage of zwitterionic liposomes, induced by cationic oligomers, was more extensive than that of negative liposomes, indicating that oligomer adsorption onto lipid head groups via electrostatic interaction cannot fully explain the induced lipid membrane damage. Furthermore, lipid packing parameters, including intrinsic curvature, cholesterol content, and lipid phases, affected liposome leakage, particularly for PHMG. Cellular permeability tests were performed using an A549 cell monolayer model and a respiratory 3D tissue model, revealing that PHMG and PHMB damaged cell membranes and reduced cell barrier function. Furthermore, liposome leakage induced by PHMG and PHMB was higher in human lung surfactant-mimicking liposomes than that observed in Escherichia coli-mimicking liposomes. These results indicated that human cells are susceptible to guanidine-based oligomers. Considering that the interaction of oligomers and cell membranes is a major mechanism of toxicity initiation, this study provides crucial insights into the action of these disinfectants on mammalian cells.
Introduction
Chemical disinfectants play a crucial role in sterilization and disinfection, and a broad range of disinfectants ranging from small molecules (e.g., hypochlorite, triclosan, and quaternary ammonium salts) to macromolecules (e.g., antimicrobial peptides and cationic polymers) have been developed and widely employed. Among various chemical disinfectants, biocide polymers are considered promising owing to their long-lasting resistance against microorganisms, excellent stability, low toxicity toward host cells, and low production cost.1Guanidine-based oligomers such as polyhexamethylene guanidine (PHMG) and polyhexamethylene biguanide (PHMB) are well-known representative biocidal polymers. As positively charged guanidine moieties in guanidine-based oligomers demonstrate a high binding affinity toward negatively charged cell membranes, they can be strongly adsorbed onto negatively charged bacterial membranes, eventually resulting in membrane disruption.2 Thus, these oligomers have been widely employed in various sterilization and disinfection protocols.3,4 In early studies, PHMB highly interacted with negatively charged lipid bilayers, demonstrating negligible impact on zwitterionic lipid membranes.5–7 Considering that mammalian or fish cells are mainly composed of zwitterionic phospholipids, it is believed that guanidine-based oligomers destroy only microorganisms and thus possess low toxicity toward host cells. However, recent studies have revealed that polyguanidines can penetrate mammalian lipid membranes8,9 or cause membrane damage,2,4,10 raising concerns regarding possible harmful effects on humans. In 2011, the Korean Centers for Disease Control and Prevention (KCDC) reported that exposure to PHMG in humidifier disinfectants might induce unidentified lung diseases.11 To date, more than 1000 deaths from lung injuries have been caused by the fatal misuse of humidifier disinfectants.12 Although extensive studies on the toxicity of humidifier disinfectants have been conducted considering different aspects,13–17 only a few have examined the interaction and disruption mechanism of active disinfectant ingredients with human cell membranes.2,18,19
Previously,19 we developed a quantitative method to determine the distribution constant (Klipw) of PHMG between solid-supported lipid membranes and water and evaluated various lipid membrane compositions. Unexpectedly, PHMGs were distributed into zwitterionic lipid membranes and negatively charged lipid membranes. This finding implies that guanidine-based oligomers may have harmful effects even toward higher-level organisms, including humans.19 However, in these studies using solid-supported lipid membranes, a few critical issues could not be appropriately addressed, including membrane leakage and disruption by the cationic oligomers and the subsequent transport of those chemicals across the cell monolayer. Furthermore, gel phase lipids could not be subjected to this methodology, as surface cracks or defects were ubiquitously reported when solids were covered by gel phase lipid membranes.20–22
In the present study, we attempted to overcome the limitations of solid-supported lipid membranes. Accordingly, a liposome, which is considered a better human cell-mimicking model, was adopted as a platform to investigate the leakage caused by guanidine-based oligomers (PHMG and PHMB). In particular, a variety of lipid membrane compositions were evaluated considering the leakage and disruption induced by guanidine-containing oligomers. Liposome leakage assays were performed by assessing the increase in fluorescence owing to the release of fluorescent dye, from inside the liposome, into the solvent.23,24 This assay has been successfully employed to investigate lipid bilayer leakage induced by nanoparticles24–27 and macromolecules28–30 including positively charged oligomers31 and protein transduction domains of virus.32 Furthermore, the effects of head group charges, lipid packing parameters, and lipid phases, including gel and liquid crystalline phases, on liposome leakage were investigated. Liposome leakage was also evaluated using lung surfactants and Escherichia coli-mimicking liposomes. Finally, A549 human lung adenocarcinoma cells and respiratory 3D tissue (SoluAirway human respiratory cells) were subjected to the cell monolayer transport assay as a human lung model system. To the best of our knowledge, this is the first study to investigate liposome leakage and cellular permeability induced by PHMG and PHMB using human lung surfactant mimic liposomes and human lung cells, respectively.
Experimental
Materials
PHMG powder (SKYBIO 1100, +95.5%) was kindly supplied in 2011 by SK Chemical Co., Ltd. (Seongnam, Republic of Korea). SKYBIO 1100 was one of the active ingredients that caused the humidifier disinfectant tragedy in South Korea and is no longer commercially available. PHMB powder was purchased from the Shanghai TECH Chemical Industry Testing Co., Ltd. (Shanghai, China). All lipids were obtained from Avanti Polar Lipids (Alabaster, AL, USA). Molecular structures of PHMG and PHMB are shown in Fig. S1.† Characteristics of all lipids used in this study, including acyl chain lengths, main transition temperatures, and lipid head charges, are summarized in Table 1. 5(6)-Carboxyfluorescein, Triton X-100, 1 M HEPES buffer, cholesterol, and FITC-dextran were purchased from Sigma-Aldrich (St. Louis, MO, USA). RPMI 1640 was purchased from Thermo Fisher Scientific (Gibco Laboratories, Grand Island, NY, USA). Fetal bovine serum (FBS) was purchased from Atlas Biologicals, Inc. (Fort Collins, CO, USA).| Liposome component abbreviation (full name) | Carbon chain: double bond | Main transition temperature (Tm, °C)a | Physical state at room temperature | Lipid head charge |
|---|---|---|---|---|
| a Avanti Polar Lipids, Inc. (https://avantilipids.com/tech-support/physical-properties/phase-transition-temps). | ||||
| DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) | (C18:1, 18:1) | −17 | Liquid crystalline | Zwitterion |
| POPC (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine) | (C16:0, 18:1) | −2 | Liquid crystalline | Zwitterion |
| DOPG (1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)) | (C18:1, 18:1) | −18 | Liquid crystalline | Negative |
| DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine) | (C16:0, 16:0) | 41 | Gel | Zwitterion |
| DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) | (C18:1, 18:1) | −16 | Liquid crystalline | Zwitterion |
| DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) | (C18:1, 18:1) | ∼−0 | Liquid crystalline | Positive |
Characterization of PHMG and PHMB
The weight-average molecular mass (Mw) was determined using gel permeation chromatography (GPC) using an Agilent 1260 Infinity high-performance liquid chromatography (HPLC) system (Agilent Technologies, Santa Clara, CA, USA), equipped with a refractive index (RI) detector. A tandem of two Ultrahydrogel 120 columns (7.8 × 300 mm; 6 μm; Waters, Milford, MA, USA) was employed. HPLC grade water was eluted at a flow rate of 0.5 mL min−1 at 30 °C. Polyethylene glycol samples were used as mass calibrants. Aqueous solutions of PHMG and PHMB were prepared at 1000 mg L−1 for characterization.Liposome preparation
To prepare 5(6)-carboxyfluorescein-encapsulating liposomes, 100 mM 5(6)-carboxyfluorescein was prepared in deionized water by adjusting the pH of the solution between 7.0 and 7.5 by adding 1 M NaOH. One milliliter of lipid or lipid mixture dissolved in chloroform solution (25 mg mL−1) was transferred to clean glass vials, and a thin lipid film was formed after chloroform was evaporated by nitrogen purging. Then, the lipid film was hydrated with 5 mL of 100 mM 5(6)-carboxyfluorescein solution by vigorous vortexing followed by overnight storage. Next, the resultant solutions were extruded more than 12 times using an Avanti Mini-Extruder (Alabaster, AL, USA) with 100 nm polycarbonate membranes. Free 5(6)-carboxyfluorescein, not encapsulated inside liposomes, was removed using Zeba spin desalting columns (7k MWCO; Thermo Fisher Scientific, Waltham, MA, USA). As centrifugal forces can disrupt the structure of 5(6)-carboxyfluorescein encapsulating liposomes, spin desalting columns were used as gravitational columns, passaging 100 μL liposome solutions through the columns after three rounds of column buffer exchange. Then, 10 mM HEPES buffer containing 100 mM NaCl (pH 7.4) was used for buffer exchange. Liposomes (0.5 mL) were collected and used immediately for liposome leakage assays. For unsaturated lipids, lipid hydration and extrusion were assessed at room temperature. For saturated lipids such as a DPPC lipid and a DPPC lipid mixture used to represent lung surfactant liposomes, lipid hydration and extrusion were performed at 55 °C, a temperature more than 10 °C above the main transition temperature of DPPC (41 °C).The liposome composition of the lung surfactant (DPPC/DOPC/DOPG/DOPE = 70/10/10/10) was selected based on the phospholipid composition of adult human lung surfactant reported by Shelley et al.33 and we modified the composition based on the principal component used in this study. Lipid mixtures DOPE/DOPG (80/20) were used as E. coli-mimicking liposomes.
Liposome leakage assays
To assess liposome leakage caused by guanidine-based oligomers (PHMG and PHMB), 50 μL of 5(6)-carboxyfluorescein loaded liposomes was added to 50 μL of 10 mM HEPES buffer with 100 mM NaCl (pH 7.4), followed by the addition of 30 μL of PHMG or PHMB solution prepared in the same buffer solution. The solution fluorescence was detected using a Hidex Sense microplate reader (Hidex, Finland) at room temperature every 2 min, until 40 min after the addition of PHMG or PHMB. The excitation (λex) and emission (λem) wavelengths were set to 485 ± 10 nm and 510 ± 10 nm, respectively. At 40 min, 30 μL Triton X-100 was added for complete liposome disruption. For each liposome leakage assay, the fluorescence of liposomes only (50 μL 5(6)-carboxyfluorescein loaded liposomes + 50 μL buffer solution) and liposomes with Triton X-100 (50 μL 5(6)-carboxyfluorescein loaded liposomes + 50 μL buffer solution + 30 μL Triton X-100 solution) were simultaneously monitored to detect fluorescence signals of liposomes and fully ruptured liposomes, respectively. Liposome leakage was calculated using eqn (1).
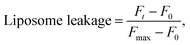 | (1) |
Liposome size measurements
DOPC and DOPG liposome sizes were measured after incubation with four different concentrations of PHMG and PHMB (23, 115, 231, and 461 mg L−1). In order to measure the liposome size, DOPC and DOPG liposomes without a fluorescent dye were used to avoid any interference of the dye on size measurements. Liposome solutions were prepared by hydration with 10 mM HEPES buffer overnight, followed by size extrusion, as described above. Then, 0.5 mL of liposome solution, 10 mM HEPES buffer (0.5 mL), and 0.3 mL PHMG or PHMB solutions (each mL) were mixed and allowed to stand for 30 min. Liposome sizes with and without guanidine-based oligomers were measured using dynamic light scattering (DLS, Malvern Zetasizer, Westborough, MA, USA).Cell cultures
A549 human lung epithelial cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in RPMI 1640, supplemented with 5% FBS and penicillin–streptomycin solution (100 U mL−1). The respiratory 3D tissue model was purchased from Biosolution (Seoul, Republic of Korea). Tissues were maintained in the assay medium (Biosolution). Cells and tissues were cultured in an atmosphere of 5% CO2/95% air under saturated humidity at 37 °C.Cellular permeability tests
FITC-dextran flux was evaluated to determine permeability from the apical side to the basal side induced by PHMG or PHMB in the A549 cell monolayer model or respiratory 3D tissue model. FITC-dextran was dissolved in 1 mg mL−1 of P buffer (10 mM HEPES [pH 7.4], 1 mM sodium pyruvate, 10 mM glucose, 3 mM CaCl2, and 145 mM NaCl) and added to the apical surface. After exposing the apical surface to 23 mg L−1 of PHMG or PHMB for 1, 3, 6, 12, and 24 h, the supernatant was removed from both apical and basal compartments. In the A549 cell monolayer model, 500 μL of FITC-dextran was added to the apical surface, and 2 mL of P buffer was added to the basolateral surface and incubated for 6 h. In the respiratory 3D tissue model, 200 μL of FITC-dextran was added to the apical surface, and 300 μL of P buffer was added to the basolateral surface and incubated for 24 h. Then, P buffer (200 μL) was collected from the basolateral chamber, and the amount of FITC-dextran was measured using a Fluostar Omega (BMG LABTECH, Offenburg, Germany) microplate fluorometer at excitation/emission wavelengths of 485 nm/520 nm.Statistical analysis
Cellular permeability test was performed at least three times. Data were analyzed using Microsoft® Office Excel (Microsoft, Redmond, WA, USA) and expressed as the mean ± standard deviation. Statistical analysis was performed using the Statistical Package for Social Sciences version 21.0 (IBM Co., Armonk, NY, USA). Comparisons between groups were assessed using Duncan's post hoc test following a one-way analysis of variance. p < 0.01 or 0.05 was considered statistically significant.Results and discussion
Development of liposome leakage assays for PHMG and PHMB
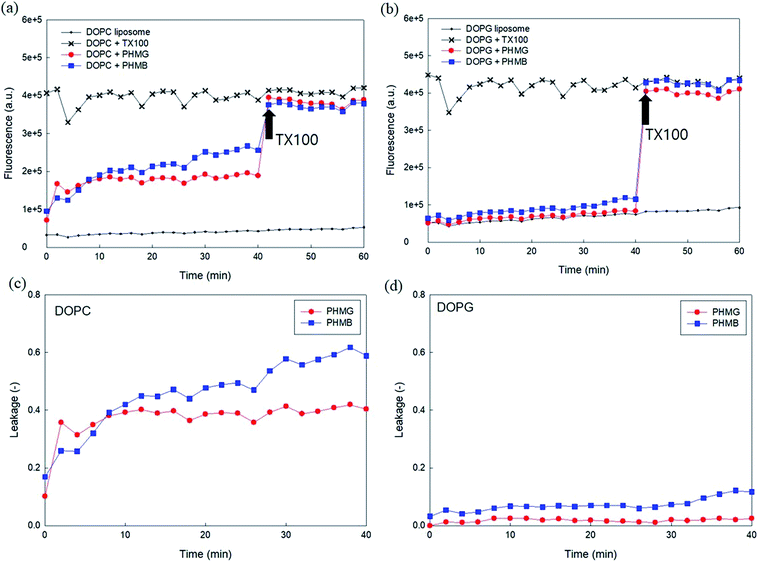
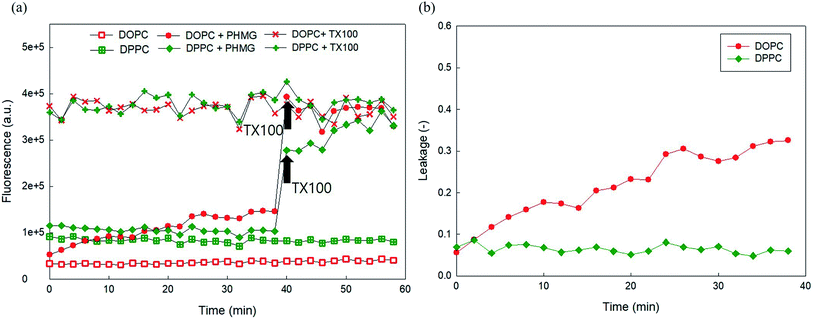
Owing to the self-quenching property, the high concentration of fluorescent dye inside the liposome presents minimal fluorescence. However, once the dyes are released from liposomes, they are highly diluted, and the fluorescence signal tends to increase. To establish the liposome leakage assay, we first constructed a calibration curve of 5(6)-carboxyfluorescein in the low concentration range between 0 and 0.00625 mM and assessed the self-quenching property of this fluorescent dye at high concentrations in 10 mM HEPES buffer. As shown in Fig. S2(a),† the fluorescence signal linearly increased with the addition of 5(6)-carboxyfluorescein in the concentration range of 0–0.00625 mM. However, at higher dye concentrations (i.e., >0.125 mM), the fluorescence signal dramatically decreased due to the self-quenching property of the dye. When the dye concentration reached 100 mM, at which dye was prepared for encapsulation within liposomes, the fluorescence signal appeared negligible (Fig. S2(b)†).Fig. 1(a) and (b) show the fluorescence signal of 100 mM 5(6)-carboxyfluorescein for the two encapsulating liposomes of DOPC and DOPG, with the selective addition of 23 mg L−1 PHMG or PHMB and TX-100 (0.5 M). As shown in Fig. 1(a) and (b), the fluorescence signal was negligible in the absence of PHMG or PHMB due to self-quenching of the dye. In contrast, the fluorescence signal increased and remained steady with the addition of TX-100, indicating that 5(6)-carboxyfluorescein completely leaked out of liposomes following TX-100 addition.
On adding PHMG or PHMB solution (23 mg L−1) to the liposomes ((a) DOPC and (b) DOPG), it was observed that the fluorescence signal significantly increased owing to attenuated self-quenching, which in turn can be attributed to liposome leakage caused by PHMG or PHMB (Fig. 1(c) and (d)). This was confirmed in another experiment, wherein Triton X-100 was added at 40 min to the liposome solution incubated with PHMG and PHMB. Following Triton X-100 addition, the fluorescence intensity significantly increased and promptly reached steady-state values, indicating that the partial rupture of liposomes by PHMG or PHMB was also the leading cause underlying the increased fluorescence signal. However, it should be mentioned that liposome leakage caused by partial perturbation with cationic oligomers does not necessarily indicate permanent pore formation at the liposome shell.27
Characterization of guanidine-based oligomers
The molecular distributions of PHMG and PHMB at 1000 mg L−1, as used in the present study, were characterized in terms of molecular mass using GPC. Fig. S3† shows the GPC spectra obtained for 1000 mg L−1 PHMG and PHMB. The Mw values of PHMG and PHMB were determined based on the first peak of the GPC spectrum, determined as 5674 and 5503 for PHMG and PHMB, respectively, showing similar Mw values. In several previous studies, the antimicrobial efficiency of cationic polymers was found to be strongly dependent on the average molecular weight of polymers.1,34–36 In general, PHMB prepared as an antimicrobial agent showed a molecular weight distribution ranging between 400 and 8000 (i.e., representing a polymer repeating unit (n) ranging from 2 to 40 (ref. 37)). Thus, Mw values of PHMG and PHMB used in this study lie in the Mw range of PHMB, which have reported antimicrobial activities in the literature.The average PHMG Mw value obtained in this GPC-based measurement was approximately one order of magnitude higher than that by MALDI-TOF MS (matrix-assisted laser desorption/ionization time-of-flight mass spectrometer) experiments in previous studies, although the same PHMG sample was used for analysis with an interval of eight years.38,39 Hwang et al.38 have reported that the Mw of PHMG ranged between m/z 524.2–810.7, and Park et al.39 indicated that the degree of PHMG oligomerization (n) ranged from 2 to 4, with a molecular weight distribution of m/z 441.0–678.0. Likewise, the average Mw value for PHMG acquired using MALDI-TOF MS was also significantly smaller than that measured by the National Industrial Chemicals Notification and Assessment Scheme (NICNAS), i.e., Mw = 137![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, for the risk assessment of PHMG used in this study (SKYBIO 1100).40
000, for the risk assessment of PHMG used in this study (SKYBIO 1100).40
A significant discrepancy in the average Mw values obtained by GPC and MALDI-TOF MS has been previously reported.41 Liu et al.41 have reported that the Mw value of MALDI-TOF MS for poly(dimethylsiloxane) (PDMS) oligomers was substantially underestimated when compared with that of GPC analysis. The underestimation by MALDI-TOF MS analysis was attributed to the over-representation of low-molecular-weight oligomers owing to the high volatility of low-mass PDMS polymers under MALDI ionization conditions. A few other factors may affect the discrepancy between MALDI-TOF MS- and GPC-measured Mw values. GPC measurements provide information regarding the relative molecular weight distribution, whereas MALDI-TOF MS provides absolute mass information. GPC mass distribution measurements are often calibration-sensitive (i.e., calibrant-sensitive); GPC analysis assumes that the detection sensitivity and mobility of analytes under examination are identical to those of the calibrant (in this case, polyethylene glycol). Moreover, RI detection, specifically differential RI, often adopted in GPC analysis, tends to overestimate high mass oligomers (or polymers); RI increases nonlinearly with mass density for dense liquids and solids.42 Accordingly, the MALDI-TOF MS detection signal is responsive to the number of ions that arrive on the detector surface, whereas the RI detector provides a higher output signal in response to heavier molecules owing to the increased change in the RI even for the same number of ions under detection. However, the discrepancy between Mw values measured by GPC and MALDI-TOF MS for PHMG and PHMB needs to be comprehensively elucidated in future investigations.
Effects of head group charges on liposome leakage
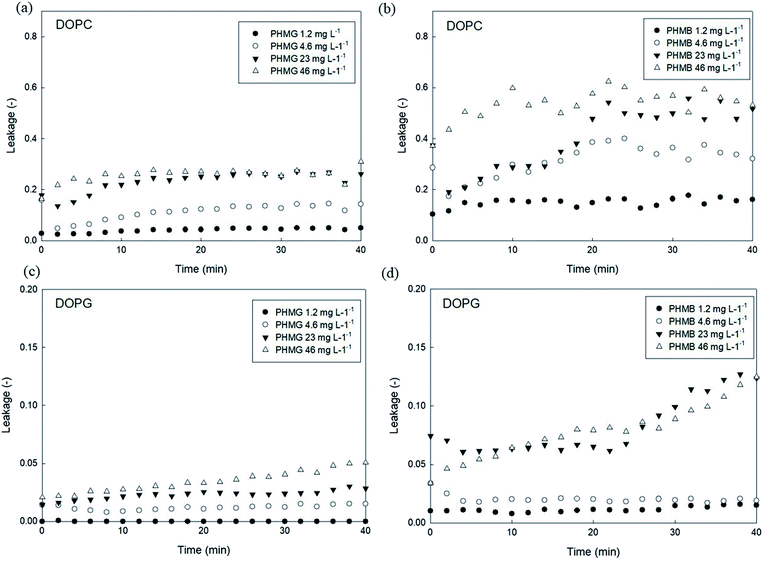
To investigate the effects of head group charges of lipid bilayers on liposome leakage, zwitterionic DOPC lipids and negatively charged DOPG underwent the leakage assay with 23 mg L−1 of PHMG and PHMB, with results shown in Fig. 1. For both PHMG and PHMB, liposome leakage was significantly higher with DOPC liposomes (Fig. 1(c)) than with DOPG liposomes (Fig. 1(d)). The liposome leakage with DOPC liposomes ranged from 0.10 to 0.42 and from 0.17 to 0.62 for PHMG and PHMB, respectively. With DOPG lipids, liposome leakages were estimated as 0–0.025 and 0.032–0.12 for PHMG and PHMB, respectively. In our previous study, we calculated the affinity constant (K) between PHMG and solid-supported lipid membranes coated with lipid bilayers.19 The affinity constant with the negatively charged DOPG lipids was significantly higher than that with the zwitterionic DOPC lipids, and adsorption kinetics with DOPG lipids were faster than other lipids, such as zwitterionic DOPC and cationic DOTAP lipids. These results suggest that PHMG strongly and rapidly interacts with negatively charged lipid heads owing to electrostatic interactions. However, interestingly, based on results shown in Fig. 1, lipid perturbation caused by cationic guanidine-based oligomers occurred more frequently with zwitterionic lipids than with negative lipids, indicating that electrostatic interactions cannot solely explain lipid leakage.Furthermore, we prepared different concentrations of PHMG and PHMB ranging from 1.2 mg L−1 to 46 mg L−1, and liposome leakage tests were conducted with DOPC and DOPG lipids at each concentration. As shown in Fig. 2, from 1.2 mg L−1 to 23 mg L−1, more liposome leakage occurred with increasing cationic oligomer concentrations with both DOPC and DOPG liposomes. These results are consistent with those of previous studies, which revealed that higher oligomer concentrations caused more liposome leakages.30,43 It is noted that liposome leakage occurred very rapidly as the leakage levels were significantly higher at 0 min shown in Fig. 2. Since the initial stage of liposome damage is affected by the concentration ratios of liposomes to chemicals,44 further study can be needed to investigate the effects of liposome/oligomer concentration ratios on the initial dye leakage from liposomes.
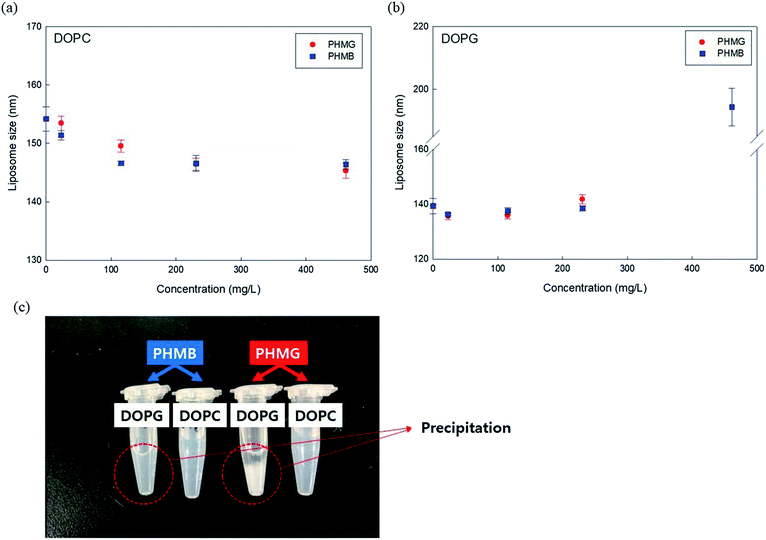
To better understand the mechanism of liposome leakage, we measured DOPC and DOPG liposome size changes after incubation with PHMG and PHMB (concentrations ranging from 23 mg L−1 to 461 mg L−1) for 30 min. With DOPC liposomes, although the liposomes were incubated with markedly high concentrations of PHMG or PHMB (461 mg L−1), the liposome size decreased marginally (less than 5 nm) (Fig. 3(a)), indicating that PHMG and PHMB did not entirely disrupt DOPC liposomes. When DOPG liposomes were incubated with the guanidine-based oligomers, the liposome size was not significantly altered until 230 mg L−1. However, at 461 mg L−1 of PHMG or PHMB, precipitation was observed, and the debris size was significantly larger than the liposome size. The debris generated by DOPG and PHMB interaction reached approximately 200 nm, and the size of that generated by DOPG and PHMG interaction could not be measured by DLS, suggesting that the debris is exceeded micrometers. This indicates that the negatively charged liposomes were irreversibly and entirely destabilized because of strong electrostatic interactions between the cationic oligomers and negatively charged lipid heads. The results in Fig. 1 and 3(b) suggest that cationic oligomers used in this study predominantly adhered to the negative head of DOPG in the range from 23 mg L−1 to 230 mg L−1; however, at 461 mg L−1, DOPG liposomes were fully ruptured by both oligomers.
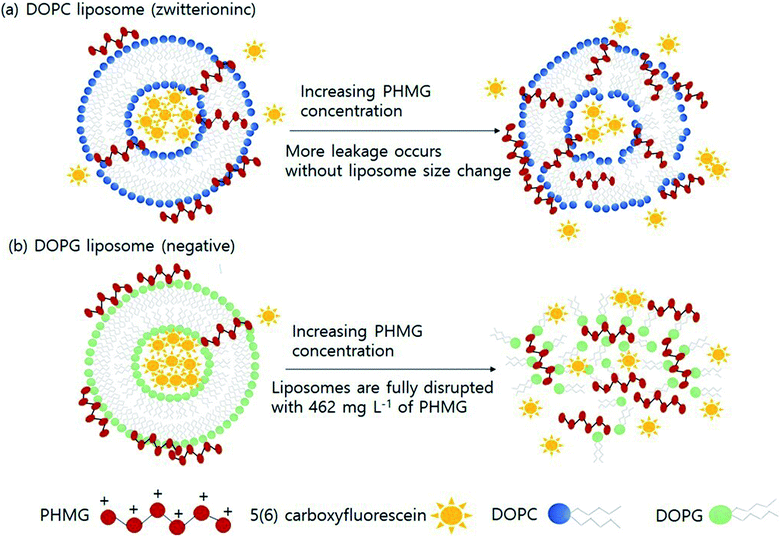
Based on the findings of this study, we demonstrated the possible mechanisms of liposome leakage caused by the guanidine family and the impact of liposome composition (Fig. 4). Considering the electrostatic interactions between PHMG and liposomes, considerably more PHMG adsorbs on negatively charged liposomes than on zwitterionic liposomes. However, more leakage was found to occur with zwitterionic lipids, possibly due the more substantial PHMG insertion inside lipid bilayers. With zwitterionic liposomes, more liposome leakage occurred at higher PHMG concentrations without altering the liposome size (Fig. 4(a)). In contrast, at high PHMG concentrations, negative liposomes were completely disrupted (Fig. 4(b)).
Previously, we have reported that PHMG is also adsorbed onto the positively charged lipid membranes (e.g., DOTAP). Accordingly, we conducted a lipid leakage assay using DOTAP liposomes. However, it was impossible to encapsulate 5(6)-carboxyfluorescein inside DOTAP liposomes, possibly because of the electrostatic interaction between positive DOTAP and negative 5(6)-carboxyfluorescein at pH ∼ 7. Alternatively, we could synthesize fluorescent dye-entrapped DOPC/DOTAP (80/20) liposomes, but liposome leakage caused by PHMG with DOPC/DOTAP liposomes did not show a significant difference when compared with leakage with DOPC liposomes (Fig. S4†).
Effects of liposome packing parameters and phases on liposome leakage
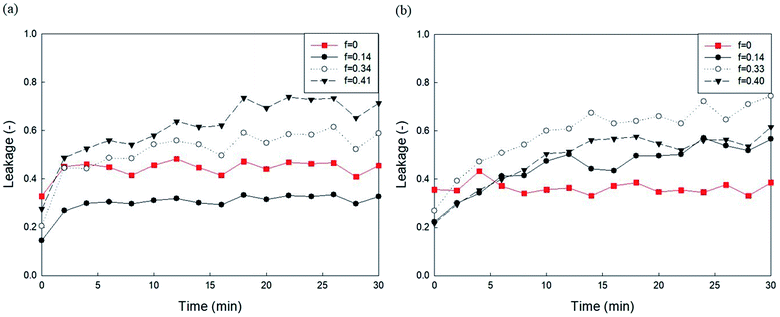
It has been established that phosphoethanolamine (PE) and cholesterol illustrate major differences in lipid membrane composition between microorganisms and mammalian cells,10 as lipid membranes of microorganisms are rich in PE but lack cholesterol, whereas mammalian cell membranes contain approximately 25 mol% cholesterol and a relatively small PE content.45 PE and cholesterol are also the main factors affecting lipid membrane packing. PE in biological membranes is inverted cone-shaped with negative intrinsic curvature (C0 < 0) because the head group of PE is smaller than the size of the acyl chains. However, other lipid components such as PC and PG are cylindrical in shape with zero intrinsic curvature (C0 ≈ 0).28 Cholesterol allows a much tighter packing membrane by increasing the orderly state of membrane surfaces.10Fig. 5 shows the effects of DOPE (18:1 PE) content in two zwitterionic liposomes, DOPC and POPC, on the leakage caused by 23 mg L−1 PHMG. As shown in Fig. 4, in both DOPC and POPC liposomes, DOPE content decreased fluorescein leakage. As DOPE has a negative intrinsic curvature with small head groups and splayed acyl chains,46 less PHMG can enter the DOPE lipid tails when compared with DOPC or POPC lipids. In addition, Paliienko et al.10 have suggested that PHMG can penetrate PE-containing lipid membranes, forming weakly cation-selective transmembrane pores. Furthermore, we assessed the DOPE effects on liposome leakage of negative liposomes (DOPG), but PE effects were found to be insignificant (Fig. S5†).
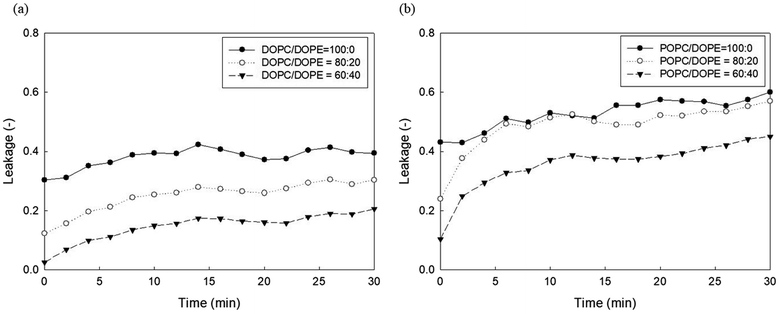 | ||
| Fig. 5 Effects of DOPE contents in (a) DOPC and (b) POPC liposomes on the 5(6)-carboxyfluorescein leakage caused by PHMG. PHMG, polyhexamethylene guanidine. | ||
The effects of cholesterol in zwitterionic liposomes, DOPC and POPC, were investigated and are shown in Fig. 6. Interestingly, the liposome leakage caused by PHMG generally increased with increasing cholesterol content. This result is in agreement with Paliienko et al.10 who reported that cholesterol facilitates the membrane partitioning of PHMG into lipid membranes owing to the cholesterol-induced lipid phase segregation. In the case of melittin, a well-known peptide that forms pores inside lipid membranes, Wessman et al.47 suggested that the lipid bilayer interaction mechanisms of melittin are modulated by cholesterol, and once melittin is associated with lipid membranes, the potential for membrane leakage is increased in the presence of cholesterol. In our previous study, we found that cholesterol decreased the distribution coefficients of PHMG between solid-supported lipid membranes and water,19 possibly due to the decrease in surface area covered by lipid molecules in the presence of cholesterol. Thus, we can also conclude that adsorption itself cannot be a critical factor in liposome leakage.
Finally, to investigate the phase of the lipid membrane, 5(6)-carboxyfluorescein leakage of DOPC liposomes (liquid crystalline phase at room temperature) caused by 23 mg L−1 PHMG was compared with the leakage of DPPC (gel phase at room temperature). DPPC is the main component of lung surfactant and is closely associated with lung injuries caused by PHMG in humidifier disinfectants. As shown in Fig. 7(a), we observed that the encapsulated concentration of 5(6)-carboxyfluorescein inside DPPC differed from that inside DOPC, possibly because of the different phases of DPPC and DOPC. When 23 mg L−1 PHMG was applied to the DPPC liposome, the fluorescence signal increased marginally (Fig. 7(a)), which resulted in less than 10% liposome leakage (Fig. 7(b)). Conversely, DOPC liposome leakage reached approximately 30% after 30 min, which was significantly higher than DPPC liposome leakage. To the best of our knowledge, no previous study has elaborated on the effects of liposome phases on liposome leakage by cationic polymers. Reportedly, ZnO causes DOPC liposome leakage due to the local gelation of liposomes via nanoparticle adsorption, while no DPPC liposome leakage by the nanoparticle was detected. Similarly, PHMG can strongly bind to the hydrophilic headgroups of both DOPC and DPPC liposomes.18,19 This adsorption onto the DOPC creates local gelation of DOPC liposomes, resulting in significant liposome leakage, whereas adsorption onto DPPC does not alter the DPPC phase, with minimal or no leakage.
Cellular permeability
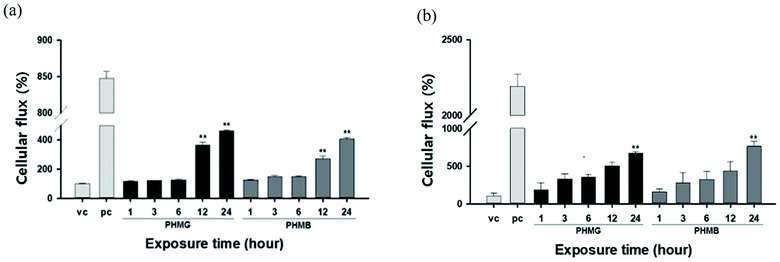
The lung epithelial barrier permeability altered by PHMG and PHMB was evaluated by measuring the FITC-dextran flux in the A549 cell monolayer model and the respiratory 3D tissue model. Both models were exposed to 23 mg L−1 of PHMG or PHMB for 1, 3, 6, 12, and 24 h. Notably, A549 cells exposed to PHMG or PHMB for 12 and 24 h exhibited reduced barrier function, as demonstrated by the significantly increased permeability with FITC-dextran when compared to vehicle-treated cells (Fig. 8(a)). As cell monolayers are insufficient to reflect airways, a respiratory 3D tissue model, presenting a high degree of similarity with human tissue, was used. In the respiratory 3D tissue model, the permeability of FITC-dextran was significantly increased following exposure for 24 h (Fig. 8(b)). These results suggest that both PHMG and PHMB increased FITC-dextran permeability through the cell membranes and reduced the lung epithelial barrier function.The lipid membranes of A549 cells are predominantly composed of cholesterol and phosphatidylcholine,48 with the unsaturated degree of membrane fatty acids reaching approximately 70%. Thus, the lipid membrane of A549 cells is mainly composed of unsaturated phospholipids,49 such as DOPC and POPC used in this study. For the respiratory 3D tissue model, DPPC, a saturated gel phase lipid, could be the main component as the cell line demonstrates a higher similarity with human respiratory cells, and other components such as PC, PG, and PE can be considered minor components of 3D tissue cells. For A549 cells, the increased ratio of the cellular membrane transport of FITC-dextran induced by PHMG to the positive control reached almost 0.28 after 24 h incubation, which is similar to the level of cellular leakage observed with DOPC/cholesterol and POPC/cholesterol mixture liposomes, as shown in Fig. 6. For the respiratory 3D tissue model, the ratio of cellular membrane transport of FITC-dextran to positive control reached approximately 0.30, which is significantly higher than the liposome leakage using DPPC liposomes (Fig. 7). This suggests that paracellular transport can be the primary cellular permeability transport or other lipid membrane components, such as unsaturated lipids, and negatively charged PG can facilitate the membrane transport of guanidine-based disinfectants.
Toxicological implications
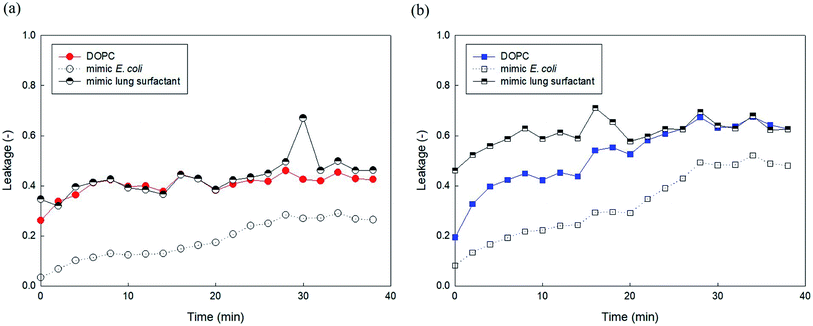
We attempted to compare the liposome leakage observed in mammalian and microorganism cells. Accordingly, liposomes consisting of major components of human lung surfactant and E. coli were prepared and used to assess liposome leakage tests of PHMG and PHMB (Fig. 9). With E. coli-mimicking liposomes, liposome leakage caused by PHMG and PHMB was lower than that by DOPC. This finding is consistent with the results shown in Fig. 5, indicating that the PE content decreased liposome leakage. Conversely, human lung surfactant mimic liposomes revealed similar (PHMG) and even higher (PHMB) liposome leakage than DOPC liposome leakage. Although DPPC, the gel phase saturated lipid, is the predominant component, other minor components such as unsaturated DOPC lipids with negatively charged groups can promote oligomer adsorption and insertion inside the acyl chains, inducing liposome leakages. This is also in agreement with the cellular transport results observed using respiratory 3D tissues, as shown in Fig. 8, which indicates that a significant amount of fluorescent dye can be transported through human lung tissue cells after incubation with PHMG and PHMB. Thus, in terms of lipid membrane damage, human lung cells might be more vulnerable than microorganism cells, and lipid leakage by guanidine-based disinfectants is undoubtedly affected by the membrane composition.Conclusions
In the present study, we demonstrated liposome leakage and increased cellular permeability induced by PHMG and PHMB, both representative guanidine-based disinfectants. Liposome leakage caused by these cationic oligomers is higher with zwitterionic lipids than with negatively charged lipids, and the leakage increased with increasing PE content and decreasing cholesterol content in unsaturated lipids. Typically, microorganisms have higher contents of negatively charged lipids and PE, presenting less cholesterol than the host (e.g., mammalian and fish) cells; guanidine-based disinfectants are believed to destroy microorganism cells with negligible effects on host cells. However, the present study indicates that host cells may be more vulnerable than microorganism cells, considering liposome leakage. In addition, although the DPPC (i.e., the main component of human lung surfactant) liposome leakage was negligible, liposome leakage using lung surfactant mimic liposomes was higher than that using microorganism-mimicking liposomes. In the A549 cell monolayer and respiratory 3D tissue model, the observed cellular permeability indicated that both PHMG and PHMB disrupt the cell monolayer and reduce epithelial barrier function, allowing increased FITC-dextran transport through the monolayer. Thus, this study can provide insights into the possible harmful effects of guanidine-based oligomers on living organisms, including humans, which is not been explicitly investigated.Considering that characteristics of guanidine-based oligomers such as molecular weight and charges are significant for investigating cellular interactions of those chemicals, future research on standardization methods for characterization should be conducted. In addition, liposome leakage and cellular permeability assay used in this study can be effectively applied to various materials (e.g., various disinfectants, microplastics, particulate matters, etc.) for elucidating the plausible cytotoxicity of those chemicals.
Author contributions
Yeonjeong Ha: conceptualization, methodology, data curation, writing-original draft, funding acquisition; Yerim Koo: methodology, data curation (liposome leakage assay); Seon-Kyung Park: data curation (GPC analysis); Ga-Eun Kim: data curation (cell permeability test); Han Bin Oh: formal analysis, writing-review & editing; Ha Ryong Kim: methodology, supervision, writing-review & editing; Jung-Hwan Kwon: supervision, writing-review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2019R1I1A1A01059970) and by the Korea Environment Industry & Technology Institute (KEITI) through “Technology Development Project for Safety Management of Household Chemical Products” funded by the Korea Ministry of Environment (MOE) (No. 2020002970001 and 1485017105).References
- A. Chen, H. Peng, I. Blakey and A. K. Whittaker, Polym. Rev., 2017, 57, 276–310 CrossRef CAS.
- J. Song, K. J. Jung, S.-j. Yoon, K. Lee and B. Kim, Toxicology, 2019, 414, 35–44 CrossRef CAS PubMed.
- K. O. Paliienko, T. O. Veklich, O. Y. Shatursky, O. A. Shkrabak, A. O. Pastukhov, M. O. Galkin, N. V. Krisanova, A. J. Chunikhin, A. V. Rebriev, A. V. Lysytsya, T. A. Borisova and S. O. Kosterin, Toxicol. In Vitro, 2019, 60, 389–399 CrossRef CAS PubMed.
- Y. J. Park, M. H. Jeong, I. J. Bang, H. R. Kim and K. H. Chung, Inhalation Toxicol., 2019, 31, 161–166 CrossRef CAS PubMed.
- T. Ikeda, A. Ledwith, C. H. Bamford and R. A. Hann, Biochim. Biophys. Acta, 1984, 769, 57–66 CrossRef CAS.
- T. Ikeda, S. Tazuke and M. Watanabe, Biochim. Biophys. Acta, 1983, 735, 380–386 CrossRef CAS.
- P. Broxton, P. Woodcock, F. Heatley and P. Gilbert, J. Appl. Bacteriol., 1984, 57, 115–124 CrossRef CAS PubMed.
- H.-R. Kim, G.-W. Hwang, A. Naganuma and K.-H. Chung, J. Toxicol. Sci., 2016, 41, 711–717 CrossRef CAS PubMed.
- H.-N. Jung, T. Zerin, B. Podder, H.-Y. Song and Y.-S. Kim, Toxicol. In Vitro, 2014, 28, 684–692 CrossRef CAS PubMed.
- K. O. Paliienko, T. O. Veklich, O. Y. Shatursky, O. A. Shkrabak, A. O. Pastukhov, M. O. Galkin, N. V. Krisanova, A. J. Chunikhin, A. V. Rebriev and A. V. Lysytsya, Toxicol. In Vitro, 2019, 60, 389–399 CrossRef CAS PubMed.
- J. H. Lee, Y. H. Kim and J. H. Kwon, Environ. Sci. Technol., 2012, 46, 2498–2500 CrossRef CAS PubMed.
- Asian Citizen's Center for Environment and Health (ACCEH), http://eco-health.org/, accessed on February 20, 2021.
- J. Lee, S.-J. Choi, J.-S. Jeong, S. Y. Kim, S.-H. Lee, M. J. Yang, S.-J. Lee, Y.-J. Shin, K. Lee and E. J. Jeong, J. Hazard. Mater., 2021, 404, 124007 CrossRef CAS PubMed.
- J. D. Lee, H. Y. Kim, K. Kang, H. G. Jeong, M.-K. Song, I. H. Tae, S. H. Lee, H. R. Kim, K. Lee and S. Chae, Arch. Toxicol., 2020, 94, 887–909 CrossRef CAS PubMed.
- S. Kim, S. Park, H. Jo, S. Song, S. Ham and C. Yoon, Environ. Res., 2020, 182, 109078 CrossRef CAS PubMed.
- H. Kim and K. Ji, Ecotoxicol. Environ. Saf., 2019, 184, 109663 CrossRef CAS PubMed.
- H. E. Shim, J. Y. Lee, C. H. Lee, S. Mushtaq, H. Y. Song, L. Song, S. J. Choi, K. Lee and J. Jeon, Chemosphere, 2018, 207, 649–654 CrossRef CAS PubMed.
- C. Lim, S. Park, J. Park, J. Ko, D. W. Lee and D. S. Hwang, J. Hazard. Mater., 2018, 353, 271–279 CrossRef CAS PubMed.
- Y. Ha and J.-H. Kwon, RSC Adv., 2020, 10, 22343–22351 RSC.
- Z. V. Feng, T. A. Spurlin and A. A. Gewirth, Biophys. J., 2005, 88, 2154–2164 CrossRef CAS PubMed.
- F. Tokumasu, A. J. Jin, G. W. Feigenson and J. A. Dvorak, Ultramicroscopy, 2003, 97, 217–227 CrossRef CAS PubMed.
- Y. Ha, L. E. Katz and H. M. Liljestrand, Environ. Sci. Technol., 2015, 49, 14546–14553 CrossRef CAS PubMed.
- H. Patel, C. Tscheka and H. Heerklotz, Soft Matter, 2009, 5, 2849–2851 RSC.
- B. Y. Moghadam, W.-C. Hou, C. Corredor, P. Westerhoff and J. D. Posner, Langmuir, 2012, 28, 16318–16326 CrossRef CAS PubMed.
- A. Hirano, K. Uda, Y. Maeda, T. Akasaka and K. Shiraki, Langmuir, 2010, 26, 17256–17259 CrossRef CAS PubMed.
- C. M. Goodman, C. D. McCusker, T. Yilmaz and V. M. Rotello, Bioconjugate Chem., 2004, 15, 897–900 CrossRef CAS PubMed.
- Y. Liu and J. Liu, Langmuir, 2020, 36, 810–818 CrossRef CAS PubMed.
- A. Som and G. N. Tew, J. Phys. Chem. B, 2008, 112, 3495–3502 CrossRef CAS PubMed.
- Y. Wang, Y. Tang, Z. Zhou, E. Ji, G. P. Lopez, E. Y. Chi, K. S. Schanze and D. G. Whitten, Langmuir, 2010, 26, 12509–12514 CrossRef CAS PubMed.
- R. F. Epand, B. P. Mowery, S. E. Lee, S. S. Stahl, R. I. Lehrer, S. H. Gellman and R. M. Epand, J. Mol. Biol., 2008, 379, 38–50 CrossRef CAS PubMed.
- T. Hitz, R. Iten, J. Gardiner, K. Namoto, P. Walde and D. Seebach, Biochemistry, 2006, 45, 5817–5829 CrossRef CAS PubMed.
- A. Ziegler, X. Li Blatter, A. Seelig and J. Seelig, Biochemistry, 2003, 42, 9185–9194 CrossRef CAS PubMed.
- S. Shelley, J. Balis, J. Paciga, C. Espinoza and A. Richman, Lung, 1982, 160, 195–206 CrossRef CAS PubMed.
- T. Ikeda, H. Hirayama, H. Yamaguchi, S. Tazuke and M. Watanabe, Antimicrob. Agents Chemother., 1986, 30, 132–136 CrossRef CAS PubMed.
- R. Kenawy el, S. D. Worley and R. Broughton, Biomacromolecules, 2007, 8, 1359–1384 CrossRef PubMed.
- T. Tashiro, Macromol. Mater. Eng., 2001, 286, 63–87 CrossRef CAS.
- M. J. Allen, A. P. Morby and G. F. White, Biochem. Biophys. Res. Commun., 2004, 318, 397–404 CrossRef CAS PubMed.
- H. J. Hwang, J. Nam, S. I. Yang, J.-H. Kwon and H. B. Oh, Bull. Korean Chem. Soc., 2013, 34, 1708–1714 CrossRef CAS.
- D.-U. Park, J. Park, K. W. Yang, J.-H. Park, J.-H. Kwon and H. B. Oh, Molecules, 2020, 25, 3301 CrossRef CAS PubMed.
- National Industrial Chemicals Notification and Assessment Scheme (NICAS), Polyhexamethyleneguanidine Phosphate Search PubMed.
- X. M. Liu, E. P. Maziarz, D. J. Heiler and G. L. Grobe, J. Am. Soc. Mass Spectrom., 2003, 14, 195–202 CrossRef CAS PubMed.
- Y. Liu and P. H. Daum, J. Aerosol Sci., 2008, 39, 974–986 CrossRef CAS.
- L. Zhang, T. Peng, S.-X. Cheng and R.-X. Zhuo, J. Phys. Chem. B, 2004, 108, 7763–7770 CrossRef CAS.
- P. Walde, J. Sunamoto and C. J. O'Connor, Biochim. Biophys. Acta, Biomembr., 1987, 905, 30–38 CrossRef CAS.
- G. van Meer and A. I. de Kroon, J. Cell Sci., 2011, 124, 5–8 CrossRef CAS PubMed.
- N. W. Schmidt and G. C. Wong, Curr. Opin. Solid State Mater. Sci., 2013, 17, 151–163 CrossRef CAS PubMed.
- P. Wessman, A. A. Strömstedt, M. Malmsten and K. Edwards, Biophys. J., 2008, 95, 4324–4336 CrossRef CAS PubMed.
- Y. Zhang, W. Zeng, F. Jia, J. Ye, Y. Zhao, Q. Luo, Z. Zhu and F. Wang, Surf. Interface Anal., 2020, 52, 256–263 CrossRef CAS.
- X. Liang and Y. Huang, Int. J. Biochem. Cell Biol., 2002, 34, 1248–1255 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1 shows representative molecular structures of PHMG and PHMB; fluorescence intensity of low concentration and high concentration of 5(6)-carboxyfluorescein at pH 7.4, as shown in Fig. S2; GPC spectra of PHMG and PHMB (Fig. S3); 5(6)-carboxyfluorescein leakage of DOPC and DOPC/DOTAP (80/20) liposomes (Fig. S4); 5(6)-carboxyfluorescein leakage of DOPG and DOPG/DOPE (50/50) liposomes (Fig. S5). See DOI: 10.1039/d1ra05478c |
| This journal is © The Royal Society of Chemistry 2021 |